Abstract
Several Bacillus strains secrete phytase, an enzyme catalyzing dephosphorylation of myo-inositol hexakisphosphate (phytate). We identified the phyC (phytase) gene from environmental Bacillus amyloliquefaciens FZB45 as a member of the phosphate starvation-inducible PhoPR regulon. In vivo and in vitro assays revealed that PhoP∼P is essential for phyC transcription. The transcriptional start site was identified downstream of a σA-like promoter region located 27 bp upstream of the probable translation ATG start codon. Inspection of the phyC promoter sequence revealed an unusual structure. The −35 and −10 regions are separated by a window of 21 bp. A pair of tandemly repeated PhoP TT(T/A/C)ACA binding boxes was located within and upstream of the −35 consensus promoter region. A single PhoP box was found within the −10 consensus promoter region. DNase I footprinting experiments performed with isolated PhoP confirmed that PhoP∼P binds at two sites overlapping with the phyC −35 and −10 consensus promoter region. While binding of dimeric PhoP∼P at −35 is essential for activation of the phyC promoter, binding of PhoP∼P at −10 suppresses promoter activity. A sixfold enhancement of phyC gene expression was registered after T:G substitution of nucleotide −13 (mutant MUT13), which eliminates PhoP binding at the single PhoP box without impairing the −10 consensus sequence. Moreover, MUT13 also expressed phyC during phosphate-replete growth, suggesting that the repressing effect due to binding of PhoP∼P at −10 was abolished. A model is presented in which transcription initiation of phyC is positively and negatively affected by the actual concentration of the PhoP∼P response regulator.
Phytases are enzymes that sequentially remove phosphate groups from myo-inositol 1,2,3,4,5,6-hexakisphosphate (phytate), the main storage form of phosphate in plants. Based on sequence homology, phytases (EC 3.1.3.8 for 3-phytase and EC 3.1.8.26 for 6-phytase) can be classified into histidine acid phosphatases, plant purple acid phosphatases, and Bacillus β-propeller phytases (36). Besides their ability to make phytate phosphorus available, elimination of chelate-forming phytate, which is known to bind nutritionally important minerals (Zn2+, Fe2+, and Ca2+), is another beneficial effect of extracellular phytase activities of several soil bacteria, such as Pseudomonas (24), Klebsiella (18), and Bacillus spp. (25). Phytase activities of bacteria inhabiting the plant rhizosphere may contribute to their plant growth-promoting effect (23, 44). Although phytases from various microbial sources are now widely used in biotechnology, mainly in the animal feed industry to improve the bioavailability of phosphate locked in the phytins (32, 38), reports about the molecular mechanisms directing phytase expression in bacteria are scarce. One of the few studied examples is Escherichia coli appA, which encodes a histidine acid phosphatase with high phytase activity. The appA gene is a member of the cyx appA operon, which is regulated by anaerobiosis, phosphate starvation, and growth phase (5, 13).
Phytase genes of several Bacillus species have recently been cloned and characterized as single genes apparently not involved in operon structures (23-26, 48). We observed that, in contrast to the phytase genes of Bacillus wild-type strains, the phytase gene of Bacillus subtilis 168 is cryptic, most likely due to the absence of a functional promoter structure (O. Makarewicz and R. Borriss, unpublished observations).
In order to reveal the regulation network controlling phytase expression on a genetic level, we fused the environmental Bacillus amyloliquefaciens FZB45 phytase gene promoter and the lacZ reporter gene and transformed the construct as a single copy into the genetic background of Bacillus subtilis 168 and its derivatives. We demonstrate now that the B. amyloliquefaciens FZB45 phytase is a member of the phosphate starvation-induced regulon controlled by the PhoPR signal transduction system, which is directing gene expression by a combination of positive and negative interactions of the response regulator with the phyC promoter sequence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani (LB) medium, low-phosphate medium (LPM), consisting of 0.1% casein peptone, 0.045% soya peptone, 0.4% glucose, 0.05% glutamate, 0.5% NaCl, 1.7 mM MgCl2, 1.4 mM MgSO4, 0.47 mM KCl, 0.3 mM CaCl2, and 50 mM Tris-HCl at pH 7.5, and high-phosphate medium (HPM), consisting of LPM plus 10 mM phosphate. When appropriate, antibiotics were added in the following concentrations: for E. coli, 100 mg/liter of ampicillin (Ap) and 5 mg/liter of kanamycin (Km); for B. subtilis, 5 mg/liter of chloramphenicol (Cm).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | supE44ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | Lab strain |
| C41(DE3) | F ompT hsdS(r m) gal dcm (DE3) | 35 |
| C41PhoP | F ompT hsdS(r m) gal dcm (DE3) pPHOP | This work |
| C41PhoR231 | F ompT hsd(r m) gal dcm (DE3) pPHOR* | This work |
| Bacillus | ||
| FZB45 | Wild type | FZB Berlin |
| Bacillus subtilis | ||
| MF1 | trpC2 pheA1 NeorrpoCHis6 | M. Fujita |
| 168 | trpC2 | Laboratory stock |
| 1A254 | trpC2 pheA1 phoP | BGSC |
| OM211 | trpC2 amy::pOM2 Cmr | This work |
| OM611 | trpC2 amy::pOM6 Cmr | This work |
| OM621 | trpC2 pheA1 phoP amy::pOM6 Cmr | This work |
| OM711 | trpC2 amy::pOM7 Cmr | This work |
| OM145 | trpC2 amy::pCUT1 Cmr | This work |
| OM245 | trpC2 amy::pCUT2 Cmr | This work |
| OM345 | trpC2 amy::pCUT3 Cmr | This work |
| OM445 | trpC2 amy::pCUT4 Cmr | This work |
| OM545 | trpC2 amy::pCUT5 Cmr | This work |
| MUT7 | trpC2 amy::pMUT10 Cmr | This work |
| MUT11 | trpC2 amy::pMUT6 Cmr | This work |
| MUT13 | trpC2 amy::pMUT12 Cmr | This work |
| MUT17 | trpC2 amy::pMUT11 Cmr | This work |
| MUT27 | trpC2 amy::pMUT4 Cmr | This work |
| MUT37 | trpC2 amy::pMUT1 Cmr | This work |
| MUT47 | trpC2 amy::pMUT21 Cmr | This work |
| MUT49 | trpC2 amy::pMUT2 Cmr | This work |
| Plasmids | ||
| pDG268 | Integrative vector amy::lacZ Cmr | 4 |
| pOM2 | pDG268 containing a 559-bp insert of phyC of B. subtilis | This work |
| pOM6 | pDG268 containing a 500-bp insert of phyC of B. amyloliquefaciens | This work |
| pOM7 | pDG268 containing a 315-bp insert of phyC of B. amyloliquefaciens | This work |
| pCUT1 | pDG268 containing a 217-bp insert of phyC of B. amyloliquefaciens | This work |
| pCUT2 | pDG268 containing a 252-bp insert of phyC of B. amyloliquefaciens | This work |
| pCUT3 | pDG268 containing a 279-bp insert of phyC of B. amyloliquefaciens | This work |
| pCUT4 | pDG268 containing a 313-bp insert of phyC of B. amyloliquefaciens | This work |
| pCUT5 | pDG268 containing a 355-bp insert of phyC of B. amyloliquefaciens | This work |
| pMUT1 | pOM6, transition (−37) A→G | This work |
| pMUT2 | pOM6, transversion (−49) T→A | This work |
| pMUT4 | pOM6, transversion (−27) T→A | This work |
| pMUT6 | pOM6, transition (−11) A→G | This work |
| pMUT10 | pOM6, transversion (−7) T→G | This work |
| pMUT11 | pOM6, transition (−17) T→C | This work |
| pMUT12 | pOM6, transition (−13) T→G | This work |
| pMUT21 | pOM6, transversion (−47) T→G | This work |
| pET15b | Expression vector, Apr | Novagen |
| pET28b(+) | Expression vector, Kmr | Novagen |
| pPHOP | pET15b containing a 728-bp insert of phoP of B. subtilis | This work |
| pPHOR231 | pET28b(+) containing a 1,051-bp insert of phoR of B. subtilis | This work |
DNA manipulations and general methods.
Isolation of plasmid and chromosomal DNA, restriction endonuclease digestion, agarose gel electrophoresis, PCR, and transformation of E. coli and B. subtilis were performed as described previously (23).
Construction of plasmids and bacterial strains.
Specific DNA fragments were amplified from the phyC promoter region of B. amyloliquefaciens FZB45 or B. subtilis 168 using the primer pairs listed in Table 2. The promoter-lacZ fusions derived from B. amyloliquefaciens FZB45 were cloned into the EcoRI/BamHI-digested integration vector pDG268 (4); the B. subtilis promoter-lacZ fusions were cloned into the EcoRI/HindIII-digested vector pDG268. The plasmids were linearized by XhoI digestion and transformed into competent B. subtilis cells (2). The transformants were screened for Cm resistance and amylase-negative phenotype. The cloned DNA regions were confirmed by sequencing.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Position relative to transcription start site |
|---|---|---|
| Cut1 | TATGTATTTTAGAATTCAAGTGAAGG | −21 to +5 |
| Cut2 | TTCACGAATTCTTAACACTGAACTTCC | −50 to −23 |
| Cut3 | TCTCCGTGAATTCTCACATGC | −81 to −58 |
| Cut4 | TATTCATTTGAATTCTTTGCTCACG | −115 to −90 |
| Cut5 | TCCGATTAATAGAATTCAAACAC | −158 to −135 |
| F2for | AATATTTGCTCACGTCAATTTTTTTTCTCC | −104 to −75 |
| F2rev | GTGTTTTTGAATGATTCATTTTCCTTCC | +13 to + 44 |
| F1for | GCGAGTTAATGAAAGAAACC | −234 to −215 |
| F3rev | GTGATAAGGATCAGACAGCTTATGC | +107 to +131 |
| MutF7 | CTTCCTGTATGTATTTTACAAGTAAAGTGAACG | −31 to +5 |
| MutF17 | CTTCCTGTATGCATTTTACAATTAAAGTGAACG | |
| MutR7 | GAACGTTCACTTTACTTGTAAAATACATACAGG | −25 to +8 |
| MutR17 | GAACGTTCACTTTAATTGTAAAATGCATACAGG | |
| MutF11 | TTCCTGTATGTATTTTGCAATTAAAGTGAACG | −30 to +5 |
| MutR11 | CGTTCACTTTAATTGCAAAATACATACAGG | −30 to +5 |
| MutF13 | TGTATGTATTGTACAATTAAAGTGAACG | −26 to +5 |
| MutR13 | TTCACTTTAATTGTACAATACATACAGG | −28 to +3 |
| MutF27 | CACTGAACATCCTGTATGTATTTTAC | −35 to −10 |
| MutR27 | TACATACAGGATGTTCAGTGTTAAG | −40 to −16 |
| MutF37 | CGGACAATCTTCACAAAAACTTGACACTGAACTTCC | −58 to −23 |
| MutF49 | CGGACAATCTACACAAAAACTTAACACTGAACTTCC | |
| MutR37 | GAAGTTCAGTGTCAAGTTTTTGTGAAGATTGTCCGC | −61 to −22 |
| MutR49 | GAAGTTCAGTGTTAAGTTTTTGTGTAGATTGTCCGC | |
| MutF47 | ACAATCTTCGCAAAAACTTAACACTGAAC | −47 to −27 |
| MutR47 | AGTGTTAAGTTTTTGCGAAGATTGTCCG | −59 to −32 |
| Om01 | ATGAATTCCTCCAACTCTCGTTTCTCTACCATGC | −289 to −257 |
| Om02 | AATGGCAAGCTTATCTGCTGCATCATCGC | +172 to +200 |
| Om08 | CAATTAAAGTGAAGCTTCATTAAAAGGAGG | −2 to +19 |
| Om09 | ATTCTTGGGATCCAGCCAAATCG | +199 to +221 |
| Om14 | TGTTTTTGAAGGATCCATTTTCCTTCCTCC | +14 to +43 |
| Om11 | TCTTCTTCTTCTGCGATAAAGACTGCC | +684 to +709 |
| Om16 | GGATCAGACAGCTTATGCTTGCCC | +101 to +124 |
| Pho4 | GGCACCATATGAACAAAAAAATTTTAGTTG | −8 to +21 |
| Pho5 | CGCACTCGAGCTTTATTCATTCATT | +710 to 734 |
| Om15 | ATATGCGCGAATTCCTGTAGAACGAACACTAG | |
| RNA linker | GAUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA |
The plasmids pMUT7, pMUT17, pMUT27, pMUT37, pMUT48, and pMUT50, which carry mutations in the putative PhoP binding sites or promoter regions, were generated using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The pOM6 plasmid was used for the amplification reactions. The mutations were introduced with the primer pairs listed in Table 2 (the changed base pairs are shown in bold). The plasmids bearing the mutations were integrated into the amyE locus of B. subtilis 168 as described above.
Overexpression and purification of PhoP, PhoR, and RNA polymerase.
The phoP gene was amplified from B. subtilis 168 chromosomal DNA using the primers Pho4 and Pho5. The PCR product was cloned into pGEMT (Promega) to construct pGEM-phoP. The phoP gene was isolated from pGEM-phoP by NdeI and XhoI digestion and cloned into NdeI/XhoI-digested pET15b (Novagen), yielding pPHOP. E. coli C41(DE3) (34) served as a host for overexpressing the PhoP and PhoR231 proteins. Overexpression and purification of PhoP was as described previously (28). The His6 tag was removed using the Thrombin CleanCleave kit (Sigma) according to manufacturer's instructions.
The His6-PhoR231 protein was cloned, overexpressed, and purified as described previously (39). The expression plasmid was named pPHOR231. LB with Km was used for expression of pPHOR231.
The σA-containing RNA polymerase holoenzyme (RNAP) was purified as described previously (17). Bacillus subtilis MF1 was grown at 37°C in LB until an optical density at 600 nm (OD600) of 0.8 to 1 was reached. The cells were lysed by sonification, and the holoenzyme was purified by Ni-agarose. The protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting. SDS-polyacrylamide gel electrophoresis revealed that RNA polymerase core subunits were copurified with σA. No other σ factors were detected (data not shown).
Enzyme assays.
Overnight cultures grown without shaking in LB-Cm at 37°C were diluted in a volume of 20 ml fresh LPM or HPM to obtain an OD600 of 0.1 and grown at 37°C with shaking at 200 rpm. Samples (0.5 ml) were collected for the determination of optical density at 600 nm, alkaline phosphatase (APase) activity (supernatant), and β-galactosidase activity (cell pellets).
For the APase assay, 80-μl samples were solubilized with 300 μl 1 M Tris-HCl (pH 8.0) containing lysozyme (200 μg/ml), benzonase (0.1 U/ml), chloramphenicol (100 μg/ml), and 0.0005% SDS for 10 min at 30°C. Subsequently, 300 μl prewarmed p-nitrophenyl phosphate (1 mg/ml in 1 M Tris HCl, pH 8.0) was added to each lysed sample, and the mixture was incubated at 30°C for 5 to 15 min. The assay was stopped with 400 μl 2 M NaOH when the color had changed to yellow. Cell debris were removed by centrifugation for 5 min at 13,000 rpm, and the absorbance was measured at 410 nm. Specific APase activity was calculatedas described previously (35): U = (E410 × 235 × Vtotal)/(t [min] × Vsample × OD600).
The β-galactosidase assay (29) was modified as follows: 100 μl of the cell suspensions were resuspended in 800 μl Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, and 0.001 M MgSO4, 50 U/liter benzonase, 100 μg/ml chloramphenicol, 4 μg/μl lysozyme, and 0.0005% SDS) and incubated for 10 min at 30°C. The reaction was started with 200 μl prewarmed 2-o-nitrophenyl-β-d-galactopyranoside (4 mg/ml Z-buffer), and the mixtures were incubated at 30°C for 5 to 15 min. The assay was stopped by the addition of 400 μl 1 M Na2CO3 when the color had changed to yellow. The samples were spun for 5 min, and the absorbance was measured at 420 nm and 550 nm. Specific β-galactosidase activity was calculated according to the method of Miller (33): MU = 1,000 × (E420 − 1.755) × E550/(t [min] × Vsample [ml] × OD600).
RNA analysis.
Total RNA of B. amyloliquefaciens FZB45 was prepared using the NucleoSpin kit (Macherey-Nagel). The transcriptional start site was determined by 5′ rapid amplification of cDNA ends (RACE), following the method of Bensing et al. (7). Five micrograms of total RNA was treated with tobacco acid pyrophosphatase (Epicenter), followed by phenol-chloroform-isoamylalcohol extraction. The RNA linker (Table 2) was ligated with RNA-ligase (Epicenter). After a second extraction, the pellet was resuspended in 20 μl RNase-free water. Reverse transcription was carried out according to the Fermentas protocol using 5-μl aliquots of treated RNA, the Om09 primer (+199 to +221), and Moloney murine leukemia virus reverse transcriptase (RT) (Fermentas GmbH). The subsequent PCR was performed with 5-μl aliquots of the RT mixture, the forward primer Om15, and the nested reverse primer Om16 (+101 to +124). The PCR product was cloned into pGEMT (Promega), transformed into E. coli DH5α, and analyzed by sequencing.
The primer extension analysis was performed using the Moloney murine leukemia virus reverse transcriptase (Fermentas GmbH) and the [γ-32P]Om16 primer according to the protocol given by the manufacturer. Total RNA of FZB45 (LPM culture) was used for the RT reaction. The sequencing reaction was performed using the Thermo-Sequenase-Cycle sequencing kit (General Electrics).
For Northern blot analysis, a phyC-specific DNA probe was synthesized with primers Om08 and Om11. Labeling was performed using digoxigenin and the Ready-To-Go DNA-labeling kit (Roche Diagnostics GmbH). Total RNA was separated on denaturated agarose gels and hybridized with the probe.
DNase I footprinting.
DNase I footprinting experiments were essentially performed as previously described (14). A 150-bp DNA fragment corresponding to the phyC promoter region was obtained using primers F2for and F2rev and Pwo polymerase and purified with the QIAquick PCR purification kit. The PCR product was labeled on the coding strand with 5′ [γ-32P]F2for and the noncoding strand by 5′ [γ-32P]F2rev in separated amplification reactions and purified with the QIAquick PCR purification kit (QIAGEN). Efficiency of labeling was in the range of 300,000 to 600,000 cpm. For the DNA binding reactions, a solution of 5 mM ATP, 0.05 μg/μl bovine serum albumin, and 0.1 μg/μl poly(dI-dC) was incubated with 0, 0.05, 0.1, 0.25, 0.5, 1, or 1.5 μM PhoP in the presence or absence of 0.4 μM PhoR231 for 20 min at room temperature in binding buffer. After addition of one μl of the diluted DNA probe (adjusted to 50,000 cpm), the mixture was incubated for a further 20 min at room temperature. DNase I (0.1 U in 10 mM MgCl2, 5 mM CaCl2) was added to the reaction mixture, and digestion was carried out for 1 min. The reactions were stopped with DNase I stop solution (0.4 M Na acetate, 50 μg/ml calf thymus DNA [Gibco], and 2.5 mM EDTA). The samples were analyzed on a 6% polyacrylamide gel containing 7 M urea. A Maxam and Gilbert sequencing reaction mixture (cleavage reactions at purine residues A and G) (45) was loaded on the same gel.
Gel shift assay.
A labeled 511-bp DNA fragment corresponding to the phyC promoter region was amplified using primers Om01 and 5′[γ-32P]Om9 using the conditions described for footprinting. The fragment was preincubated for 10 min at room temperature with PhoP (0.2, 0.4, 0.8, or 1.6 μM), 0.1 μM PhoR231, RNAP (10, 20, or 40 nM), and 5 mM ATP in binding buffer (20 mM Tris-HCl buffer [pH 8], 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol). The binding reaction (10 μl) was initiated by addition of 15 nmol of the DNA probe (20,000 cpm) and performed for 20 min at room temperature. The reaction mixtures were separated on 6% polyacrylamide gels, prerun for 30 min at 100 V, under nondenaturing conditions in 1.5× TBE buffer (133 mM Tris base, 133 mM boric acid, 2.8 mM EDTA) at 60 V for 180 to 240 min.
In vitro transcription.
The linear 511-bp templates used for in vitro transcription assays were amplified by PCR by using primers Om01 and Om09. PCR products were purified with the QIAquick PCR purification kit (QIAGEN). The in vitro transcription buffer contained 100 mM Tris-HCl, pH 8, 50 mM NaCl, 50 mM MgCl2, 250 mM KCl, 5 mM CaCl2, 100 μM EDTA, 5 mM dithiothreitol, and 10% glycerine. RNAP was incubated with 80 ng of template in 16 μl of transcription buffer at 37°C for 5 min. Previously phosphorylated PhoP (1.3 μM), which had been generated in binding buffer in the presence of 0.3 μM PhoR and 5 mM ATP, was added to the transcription reaction to final concentrations of 0.03, 0.06, 0.12, 0.18, 0.24, and 0.300 μM. The reaction was started by adding 4 μl of a nucleoside triphosphate mix (300 μM ATP/CTP/GTP, 0.45 μM UTP, 2 μCi [γ-32P]UTP, and 40 U RNasin [Fermentas]). After incubation at 37°C for 20 min, 5 μl of stop solution (95% formamide, 30 mM EDTA, 5% glycerol, 0.05% bromophenol blue) was added. Transcripts were analyzed on 6% polyacrylamide-urea gels. A low-range RNA marker was prepared, following the protocol of Fermentas for radioactive labeling.
Sequence determination.
The Thermo Sequenase Cy5 dye terminator kit (Amersham Biosciences) was used. The samples were run on ALFexpress II (Amersham Biosciences) using ReproGel High Resolution (Amersham Biosciences) and analyzed by using OMIGA 2 (Oxford Molecular) and NCBI BLAST(http://www.ncbi.nlm.nih.gov/BLAST/).
RESULTS
Transcription start and promoter sequence of the phyC gene of B. amyloliquefaciens FZB45.
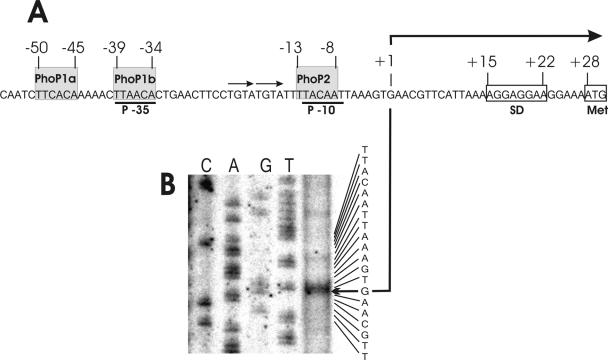
The coding region, including the flanking regions of the FZB45 phyC gene, had previously been cloned and sequenced (23). Transcription initiation was investigated by 5′ RACE as described in experimental procedures. Only RNA of the low-phosphate culture yielded a unique 124-bp PCR product consisting of the first nucleotides of the transcript at the 5′ end and ending up with the sequence complementary to the OM16 primer (Table 2) at the 3′ end. The fragment was cloned, sequenced, and confirmed to be of phyC origin. A transcription initiation site 27 bp upstream from the putative translation initiation codon was detected. The results obtained by 5′ RACE were corroborated by primer extension, yielding G as the first nucleotide of the phyC transcript (Fig. 1). The σA-like promoter sequence displayed an unusual structure bearing TTAACA (5/6 of −35 consensus) and TACAAT (5/6 of −10 consensus) but separated by an exceptionally large window of 21 bp which harbored in its 3′ part two direct repetitions of the sequence TGTA. At the −35 promoter sequence, two direct repeats separated by 5 bp perfectly matched the TT(C/A/T)A(C/A)A consensus PhoP binding box sequence of B. subtilis (16). Another putative PhoP binding box sequence was present at the −10 consensus promoter sequence, but a repeat of this sequence in an appropriate distance was missing (Fig. 1A). Almost no striking differences were detected when the sequences of the PhoP response regulators from B. subtilis and B. amyloliquefaciens were compared (85% identity). Especially, the functional domains involved in DNA binding and phosphorylation were found perfectly preserved (see Fig. SM1 in the supplemental material), suggesting that the B. amyloliquefaciens phyC promoter might also interact with the heterologous B. subtilis PhoP response regulator. The promoter structure of the FZB45 phyC gene is completely conserved within the upstream regions of other B. subtilis (25) and B. amyloliquefaciens (26) phyC genes, except that of the silent B. subtilis 168 phyC gene. Here the two tandemly arranged PhoP binding boxes were absent, while the single PhoP binding box located around the −10 promoter sequence remained conserved. Interestingly, within the Bacillus licheniformis phyC (48) promoter region the single PhoP box at −10 does not exist, while the two upstream-located PhoP binding boxes at −35 are well preserved (Fig. 1B).
FIG. 1.
Promoter structure of the FZB45 phyC gene. (A) Architecture of the phyC promoter of FZB45. Gray shading identifies the putative PhoP binding boxes. Transcriptional start site is indicated by a bent arrow. Consensus sequences (−35 and −10) are underlined. (B) Mapping of the 5′ end of the phyC transcript by primer extension. The CAGT sequence ladder corresponding to the nucleotide sequence of the noncoding strand is indicated at the right. The initiation of transcription (G) is marked by a bent arrow.
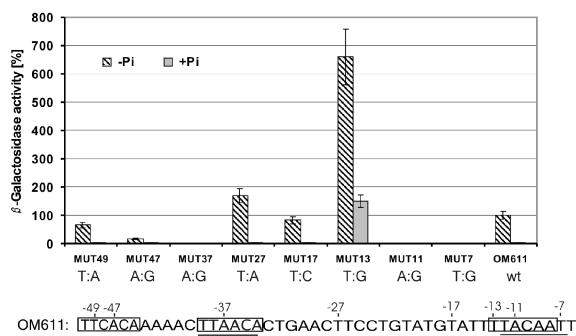
An alternative candidate −10 promoter region was detected between −17 and −12 (TATTTT). To test the functionality of both candidate −10 regions, two different base-pair substitutions were performed and checked with the appropriate lacZ fusion constructs (see Fig. 7). The transversion at −7 (T→G), representing the last nucleotide of the TACAAT sequence, completely abolished the promoter activity, while the transition at −17 (T→C), representing the first nucleotide of the alternative −10 region, did not significantly affect promoter activity (see Fig. 6). These results supported the idea that the sequence TACAAT is the −10 region, which matches exactly with the experimentally verified transcription initiation site.
FIG. 7.
Base substitution analysis within the promoter region upstream of phyC. The following substitutions were made: −49 (T→A), −47 (A→G), −37 (A→G), −27 (T→A), −17 (T→C), −13 (T→G), −11 (A→G), and −7 (T→G). The β-galactosidase activities of the clones were measured after 6 h of growth under high- and low-phosphate conditions. The reporter activity of the wild-type (wt) promoter corresponds to 100%; the activities of the other promoters were calculated as average percentages of expression relative to that of the wt promoter. The average mean deviation (±) was calculated from three independent experiments. Bottom: sequence of PphyC. Substitutions are numbered, and the −35 and −10 regions are indicated. The putative PhoP-binding sites are contrasted by a gray background.
FIG. 6.
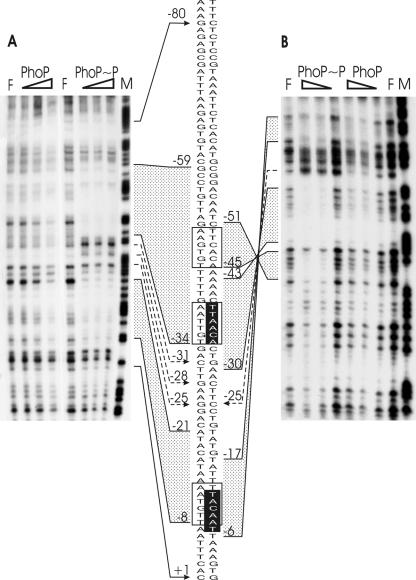
DNase I footprinting analysis of the FZB45 phyC promoter bound by the PhoP or PhoP∼P protein. A promoter fragment amplified by the F2for and F2rev primers was used to prepare the probe. Various amounts of PhoP incubated with or without PhoR231 (0.4 μM) in the presence of 5 mM ATP were mixed with the 150-bp phyC promoter probe, and DNase I footprinting experiments were performed with both the end-labeled noncoding (A) and coding (B) strands. F, control without protein; M, A+G-sequencing reaction. The concentrations of PhoP used in each reaction were 0.5 μM, 1.0 μM, and 1.5 μM. The gray areas represent the PhoP- and PhoP∼P-protected regions at the coding (right) and the noncoding (left) strands, and the hypersensitive sites are marked with dashed arrows. The transcription start +1 site is indicated by an arrow. The corresponding sequence section is shown in the center, and the positions of interest are numbered. The PhoP recognition sites are framed, and the −10 and −35 regions are shown in white letters on a black background.
Expression of the phyC gene is dependent on phosphate starvation and PhoP.
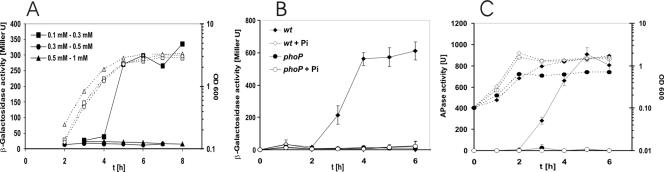
To study the regulation of phyC gene expression in B. subtilis, lacZ fusions to the phyC promoters of B. subtilis 168 and B. amyloliquefaciens FZB45 were ectopically integrated at the amyE locus. The strains were grown at three different phosphate concentrations, and the phyC promoter-driven β-galactosidase and the APase activities were measured throughout growth. Under low-phosphate conditions, strain OM611, harboring the FZB45 phyC promoter region ranging from −287 to +208, expressed β-galactosidase (Fig. 2A and B). Under medium- and high-phosphate conditions, OM611 did not express β-galactosidase. Levels of phosphate higher than 0.3 mM caused a complete arrest in formation ofβ-galactosidase (Fig. 2A), suggesting that expression of the phyC gene is strictly dependent on phosphate starvation. Similar results were obtained for activity of APase, whose induction is PhoPR dependent (22) (Fig. 2C). Northern analysis confirmed that the expression of the phyC gene in Bacillus amyloliquefaciens FZB45 is also regulated by the phosphate level. The 1.3-kb monocistronic phyC gene transcript was expressed only at phosphate starvation (results not shown). No reporter activity of OM211 harboring the B. subtilis 168 phyC promoter sequence was detected under any of the conditions tested.
FIG. 2.
Induction of APase and β-galactosidase in phyC-lacZ fusion strains. (A) Expression of β-galactosidase by strain OM611, which contains an FZB45 phyC-lacZ fusion, under three different phosphate concentrations (filled symbols). Growth of the cultures is indicated by open symbols linked by dashed lines. The symbols are given as an average range of phosphate concentrations according to measurements performed during cultivation. (B) phyC promoter activity in strain OM611 (wild type) (diamonds) or strain OM621 (phoP) (circles). Both strains were grown in LPM (solid symbols) and HPM containing 10 mM phosphate (open symbols). (C) Growth and APase activity of OM611 (diamonds) or OM622 (phoP) (circles) in HPM (open symbols) and LPM (solid symbols). APase activity was measured as a control for the PhoP-negative phenotype. The experiments were repeated at least three times, with similar results.
For strain OM621, containing a phoP mutation rendering the strain unable to express the transcription regulator PhoP, no β-galactosidase activity was detected under low-phosphate conditions (Fig. 2B). Similarly, APase activity was totally repressed in the phoP mutant background, suggesting that the expression of phytase, like that of APase, is under control of the PhoP/PhoR two-component system (Fig. 2C).
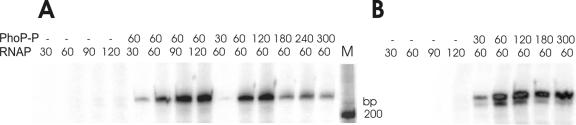
We performed in vitro transcription using a purified B. subtilis σA-saturated RNA polymerase holoenzyme (see experimental procedures) and a 10 nM concentration of the phyC promoter fragment from pOM6 as a template to confirm the promoter-lacZ fusion data. The results, presented in Fig. 3A, demonstrated that transcription depends on the presence of PhoP in its phosphorylated state, as previously shown for the phoA gene of B. subtilis (42). In vitro transcription with RNAP alone (30 nM to 120 nM) yielded no visible product, but RNAP concentration-dependent transcripts with the expected size of 223 nucleotides formed in the presence of 60 nM Pho∼P. At a 60 nM concentration of RNAP and increasing concentrations of PhoP∼P (30 to 120 nM), a gradual increase of transcription efficiency was registered, but amounts of PhoP∼P exceeding 120 nM caused a sudden decrease in transcription efficiency (Fig. 3A, right). This suggested that binding of PhoP∼P at secondary sites might impede binding and/or transcription by RNAP (see later sections).
FIG. 3.
In vitro transcription analysis of phyC. Transcription was carried out for 20 min at 37°C with purified RNAP in various amounts and a 10 nM concentration of a 511-bp phyC template of the wild-type (A) or MUT13 (B) promoter. PhoP (1.25 μM) was phosphorylated by 0.3 μM PhoR in binding buffer in the presence of 5 mM ATP for 20 min as described previously and was then added to the transcription reaction. The final concentrations (nM) of RNAP and PhoP-P are given at the top. M, molecular standard.
Promoter mapping.
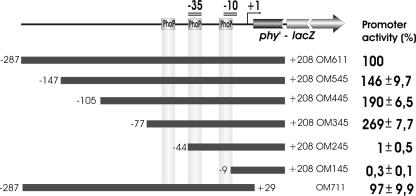
To define the regions important for activation of the FZB45 phyC promoter, 5′ deletions were introduced within the original 486-bp DNA fragment that contained the promoter region and the 5′ end of the coding region of the phyC gene (Fig. 4). These truncated phyC promoters were individually fused with a promoterless lacZ gene in pDG268. The plasmid was linearized and transformed into B. subtilis 168. Transformants were analyzed to ensure that the fusion was integrated as a single copy at the amyE locus. The activity of each promoter was determined under phosphate starvation. Deletions up to −77 did not negatively affect gene expression, while almost no activity was detected in strains where deletions were ranging up to −45 and further down. Although strain OM245 retained the complete −35 and −10 consensus boxes and two of the three PhoP recognition sites at the −35 and −10 sequence, it did not express β-galactosidase. This indicated that the presence of the PhoP box upstream of the −35 consensus sequence was necessary to confer full promoter activity. The highest level of expression was detected in strain OM345, harboring the −77 promoter deletion. This fragment contained the −35 and −10 consensus sites and all potential PhoP binding sites. Increasing the length of the 5′ upstream region gradually led to decreased reporter gene activity. Sites for a transcription repressor(s) may therefore be present within the promoter upstream region. Neither β-galactosidase nor APase activities were observed in strains growing in HPM.
FIG. 4.
Deletion analysis of the FZB45 phyC promoter. Top: fusion product consisting of the phyC promoter linked at +208 with the lacZ gene. Position of the PhoP boxes, of the −35 and −10 promoter sequences, and of the start point of transcription (+1) are indicated. The filled boxes represent the various lengths of the phyC promoter fragments used in this assay. The 5′ and 3′ ends of each fragment were labeled relative to the transcription start site, +1. The strains carrying the various truncated phyC promoters were grown in LPM, and the promoter activity was determined every hour. The highest activity of the reporter was obtained after 6 h and was used for calculating the relative promoter activity. The reporter activity of the full-length promoter corresponds to 100%; the activities of the other promoters are calculated as the average percentages of expression relative to that of the full-length promoter. The average mean deviation (±) was calculated from three independent experiments.
A 3′ deletion that removed almost the complete coding sequence did not affect promoter activity. A −287 to +29 promoter fragment displayed the same activity as the strain harboring the entire fragment, suggesting that there are no additional regulatory sites within the coding region (Fig. 4).
PhoP∼P binds to the phyC promoter.
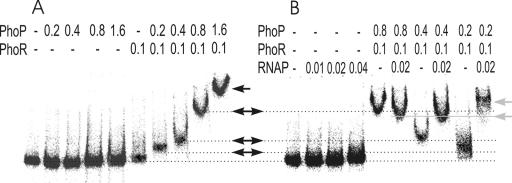
Genetic and in vitro analyses described above indicated that PhoP is necessary for transcription of the B. amyloliquefaciens FZB45 phyC gene. Gel shift assays were subsequently used to analyze the binding of purified PhoP to an end-labeled 511-bp DNA fragment covering residues −290 to +221, relative to the phyC transcriptional start site. In these experiments, purified B. subtilis 168 PhoP, PhoR, and RNA polymerase were used. The functional activities of these proteins were determined with an in vitro phosphorylation assay, which confirmed that His6-PhoR231 is autophosphorylated in the presence of [γ32]ATP and that it can phosphorylate PhoP (see Fig. SM2 in the supplemental material).
Protein binding, indicated by a shift of the 511-bp promoter fragment in the presence of unphosphorylated PhoP, was not apparent even at PhoP concentrations of up to 1.6 μM, although the slight U-shaped migration in the presence of 0.8 and 1.6 μM PhoP possibly indicates some signs of binding (Fig. 5A). Interactions between unphosphorylated PhoP and DNA were demonstrated with other PhoP-dependent promoters, although these interactions were weaker than those with PhoP∼P (30). In our experiments, the complex between nonphosphorylated PhoP and the phyC promoter may be too labile to retard fragment migration, whereas phosphorylation of PhoP may stabilize this binding. Phosphorylated PhoP∼P bound to the promoter DNA in a concentration-dependent manner. The fragment was shifted at 0.2 μM and 0.4 μM concentrations of PhoP∼P, and even more dramatic changes in mobility were observed at 0.8 μM and 1.6 μM PhoP∼P (Fig. 5A). The increased reduction of mobility may be caused by stepwise binding of PhoP∼P at PhoP boxes with different affinities and/or nonspecific binding of polymeric PhoP∼P molecules at the promoter fragment. In the absence of PhoP∼P, binding of purified RNAP (0.04 μM) at the promoter DNA was not detected (Fig. 5B). While no mobility shift occurred in the presence of unphosphorylated PhoP and RNAP (data not shown), 0.2 μM PhoP∼P shifted the promoter fragment in the presence of RNAP to a greater extent than without RNAP, suggesting that PhoP∼P and RNAP might interact cooperatively. The gel mobility shift in the presence of 0.2 μM PhoP∼P and RNAP was reproducibly more pronounced than with 0.4 or 0.8 μM PhoP∼P and RNAP (Fig. 5B). This is in agreement with the results obtained by in vitro transcription (Fig. 3A) and might suggest that higher concentrations of PhoP∼P negatively affect RNAP binding.
FIG. 5.
Gel retardation analysis of the FZB45 phyC promoter by PhoP, PhoP∼P, and RNAP. The promoter DNA fragment was γ-32P labeled at the 5′ end of the reverse strand. Each lane contained a 15 nM concentration of the labeled probe and 5 mM ATP. The proteins were purified as described in the text. Phosphorylation of PhoP was performed in the presence of PhoR231 and ATP. After the binding reaction with the DNA fragment, the samples were loaded on a native polyacrylamide gel in order to separate the free DNA and the DNA-protein complex. The concentrations (μM) of the proteins added to the lanes are indicated on top. The arrows indicate the different complexes.
Interaction of PhoP and PhoP∼P with the phyC promoter.
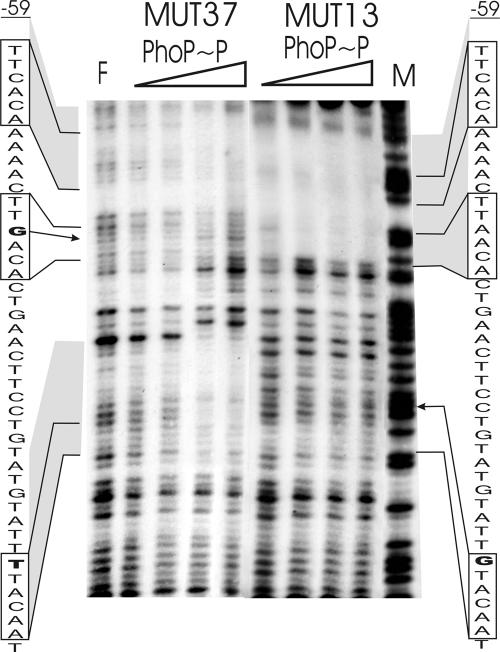
DNase I footprintings were performed to define the binding sites of PhoP at the phyC promoter. The experiments were carried out with DNA fragments amplified from the phyC promoter region—150 bp corresponding to the region −107 to +45—and with the purified PhoP and His6-PhoR231 proteins. Areas of protection were only weak when unphosphorylated PhoP (≥1 μM) was added. In contrast, PhoP∼P protected two distinct promoter areas, corroborating the results obtained by mobility shifts and in vitro transcription. One region ranged from −21 to −8 at the noncoding strand and −17 to −6 at the coding strand. The second PhoP∼P-protected region was located between nucleotides −51 and −30 at the coding strand and nucleotides −59 and −34 at the noncoding strand. Two hypersensitive sites were identified at −31 and −28 on the noncoding strand and at −25 on the coding strand (Fig. 6A and B). Existence of a further binding region at around −80 cannot be ruled out, but this possibility was not substantially supported by the DNase I footprinting experiment shown in Fig. 6, since the control lane, F, was also weaker in the same area. Sequence analysis did not reveal any PhoP binding boxes in the sequence upstream of 51. In addition, the results of promoter mapping provided no evidence for the existence of additional PhoP binding sites within regions further upstream (Fig. 4). Therefore, we concluded that the main binding region of PhoP was located around the two PhoP boxes tandemly arranged at −50 to −45 and −39 to −34. Another binding area of PhoP∼P was experimentally verified at −8 to −21, although only one PhoP binding box nearly matching the −10 consensus was detected in that area.
These results suggested that a pair of dimeric PhoP molecules might cover both promoter consensus sequences. Binding of PhoP at a single PhoP box covering the −10 consensus seems to be a unique feature of the phyC promoter structure and has to our knowledge not previously been reported for any other member of the PhoP/R regulon. The presence of PhoP∼P-hypersensitive sites may indicate PhoP∼P-dependent DNA bending.
PhoP binding around −35 is crucial for phyC transcription activation.
To analyze the functional importance of the two PhoP binding boxes tandemly arranged at around −47 and −35, three single-base-pair substitutions were introduced by site-directed mutagenesis (Fig. 7). As expected, nucleotide changes introduced into the −35 (MUT37) and −10 (MUT7) σA promoter consensus regions abolished the normal phyC gene expression under phosphate deprivation. Replacement of the −37 A by a G converted the conserved PhoP-binding region into a perfect promoter consensus TTGACA motif. However, no β-galactosidase activity was observed under high- or low-phosphate conditions. When nucleotide substitutions were introduced into the first PhoP binding box (MUT49 and MUT47), including the change of the A at −47 to G, only moderate reduction of transcription was observed. A→G substitutions act as one of the most deleterious substitutions in other PhoP-dependent promoters (16). According to these data, functional integrity of the second PhoP-binding site, which overlaps the consensus at −35, seems to be crucial for transcription activation, while the presence of the consensus −35 motif is less important. Conclusively, PhoP binding at −35 appears to be of more importance than RNAP binding for transcription activation.
This conclusion was examined by a DNase I footprinting assay performed with the DNA promoter fragment harboring the −37 mutation. Binding of PhoP∼P at around −35 was completely abolished. Binding of PhoP∼P at the upstream PhoP box at −47 was also negatively affected, which might indicate cooperativity in binding of PhoP dimers at this site (Fig. 8). It could be speculated that binding of the dimeric PhoP molecule occurs first at the second PhoP box located at −35, and accordingly, binding at around −47 is secondary and may be supported by protein-protein interactions after the molecule has bound at −35. However, this conclusion needs further verification by additional experiments with site-directed mutagenesis.
FIG. 8.
DNase I footprinting analysis of mutagenized FZB45 phyC promoter fragments bound with the PhoP∼P protein. The promoter fragment amplified by F1for and F3rev primers was used to prepare a 364-bp probe. Various amounts of PhoP incubated with PhoR231 (0.4 μM) in the presence of 5 mM ATP were mixed with the phyC promoter probe, and DNase I footprinting experiments were performed with the end-labeled noncoding strand. F = without protein; M = A+G-sequencing reaction lane. The concentrations of PhoP used in each reaction were 0.1 μM, 0.25 μM, 0.5 μM, and 1.0 μM. The gray areas represent PhoP∼P-protected regions, the boxes indicate the PhoP recognition sites, and the letters in bold mark the substituted bases.
PhoP binding around the −10 region negatively affects phyC promoter activity.
In order to test if the sequence TTCC located at around −27 can function as a complementary PhoP binding site, substitution of T to A at −27 was accomplished. The resulting mutant, MUT27, displayed a higher level of promoter activity (180%) than the wild-type OM611, excluding functional importance of the −27 region as a PhoP binding site (Fig. 7).
The DNase I footprinting experiments described in previous sections revealed that binding of PhoP also occurs at the −10 promoter region despite its singular PhoP box structure. In contrast to the −35 region, the single PhoP site spanning −13 to −8 does not completely match the −10 consensus, which is spanning the area −12 to −7. To dissect the functionality of this PhoP box without affecting the promoter consensus sequence, we substituted the −13 T with a G. The resulting clone, MUT13, produced more than sixfold the amount of β-galactosidase activity in LPM compared to that produced by the wild-type OM611. MUT13 also displayed phyC gene expression during phosphate-replete growth (Fig. 7), where the PhoP∼P concentration is very low (40). Without additional experimental data, we can only speculate that binding of a few PhoP∼P molecules is sufficient for gene expression if binding at the single PhoP box at −10 is prevented. In addition, it is possible that binding of unphosphorylated PhoP at the −35 region might support activation of the mutant phyC promoter under high-phosphate conditions.
DNase I footprinting analysis of this substitution revealed that PhoP∼P was not bound at the mutated PhoP box around the −10 promoter region, while the PhoP boxes at around −35 and −47 were perfectly protected (Fig. 8). This suggested a dual function of the PhoP transcriptional regulator. While occupation of the PhoP boxes at the −35 region is essential for gene activation, PhoP∼P binding at −10 does not support promoter activation but instead obstructs promoter RNAP interaction. Upon elimination of this additional binding site, transcription activity is strongly enhanced, but also, gene expression in the absence of PhoP∼P is turned on. This idea was supported by the results of the in vitro transcription assay performed with MUT13. While transcription of wild-type phyC DNA was gradually suppressed in the presence of rising concentrations of PhoP∼P (Fig. 3A), the same effect was not observed for MUT13. In addition, the reduction in transcription efficiency observed for the wild type at 300 nM PhoP∼P was not noticed for MUT13 (Fig. 3B).
Destruction of both the −10 promoter consensus and the overlapping PhoP box prevented any transcription activity, as demonstrated with mutant MUT11, in which −11T was replaced by G. The same observation was made for the mutant MUT7, designed to selectively destroy the −10 consensus without impairing the PhoP binding box (see the previous section). An intact −10 promoter sequence is therefore crucial for phyC gene expression.
DISCUSSION
We show that expression of FZB45 phyC is controlled by the PhoPR two-component system. Generally, the PhoPR signal transduction system is induced under phosphate starvation and controls several reactions that increase the cellular supply of soil-living microorganisms with the limiting nutrient phosphate (20). including liberation of phosphate groups from myo-inositol hexakisphosphate by phytase. Known examples of PhoP∼P-directed transcription activation in B. subtilis are the alkaline phosphatase genes (21) phoA (22) and phoB (12), the phosphodiesterase genes phoD (15) and glpQ (3), the gene for a high-affinity phosphate transport system, pstS (41), genes of the teichuronic acid synthesis operon (teichuronic acid is a cell wall polymer lacking phosphate), tuaABCDEFGH (29, 47), and the expression of its own operon, phoPR (37). PhoP∼P has been shown to repress the expression of the tagAB and tagDEF genes, responsible for the production of teichoic acid (a cell wall polymer containing phosphate) (31), presumably to keep phosphate consumption on a minimal level.
Using promoter lacZ fusions, we demonstrated that the phyC gene of B. amyloliquefaciens FZB45 is under control of the phosphate starvation-induced PhoPR two-component system, while the phyC promoter of B. subtilis 168 is silent even under conditions of phosphate starvation. This result is supported by the observation that despite the presence of an intact coding region, the B. subtilis 168 phyC gene product is not detectable in the secretome of B. subtilis 168 grown under phosphate starvation conditions (3). In vitro transcription analysis established that both the EσA RNAP holoenzyme and PhoP∼P are necessary and sufficient to establish transcription from the FZB45 phyC promoter.
Data obtained with several members of the PhoPR regulon support a model in which positive regulation is exerted by binding of PhoP∼P to the upstream high-affinity sites. In addition, internal sites such as those detected in the phoA and pstS genes enhance transcription initiation (42, 30). An interesting example for dual control exerted by PhoP on expression of PhoB (formerly alkaline phosphatase III) was recently reported (1). In B. subtilis, the phoB gene expression during vegetative growth under phosphate deprivation is activated by PhoP acting on an EσA-dependent promoter and repressed by PhoP acting on an EσE-dependent promoter, which is active at stage two of sporulation. As demonstrated here, the phyC gene from environmental Bacillus amyloliquefaciens is regulated by a unique control mechanism in which PhoP∼P positively and negatively affects one EσA-responsive promoter.
Previous studies suggested that PhoP-dependent promoters possess a PhoP core binding region to which both PhoP and PhoP∼P can bind (28). This ability is different from that of target sites of response regulators, such as NarL and ComA, which bind only in the phosphorylated form (43, 49). We were able to detect strong interactions between the phyC promoter and PhoP in its phosphorylated form, while binding between the unphosphorylated PhoP and the phyC promoter DNA fragment appeared to be only weak. It was shown for the resA promoter that unphosphorylated PhoP binds at concentrations higher than 3.4 μM (10).
PhoP binding boxes occurring in most B. subtilis promoters activated by PhoP consist of at least four TTAACA-like sequences repeated at specific intervals of <11 bp (28, 30). The upstream region of the B. amyloliquefaciens FZB45 phyC gene deviates from this general architecture in that there is only one appropriate binding site for the dimeric PhoP protein, which consists of two boxes centered at −47 and −35 and separated by 5 bp. This situation resembles that of the PA4EσA promoter of the PhoPR operon, in which only a single PhoP dimer consensus repeat exists on the noncoding strand (37). A unique feature of the Bacillus phyC promoter is the presence of a functional single PhoP binding box located at −13 to −8, nearly matching the −10 consensus.
There is a striking similarity in promoter anatomy of the B. subtilis spoIIG and B. amyloliquefaciens phyC genes. Despite highly conserved −35 and −10 consensus sequences, both genes are transcribed only if a dimeric phosphorylated transcription activator, Spo0A∼P (11) or PhoP∼P, respectively, binds at two tandemly arranged sites of either seven or six base pairs which are separated only by a few base pairs. At the spoIIG promoter, Spo0A∼P stimulates transcription (6, 8). In vitro, RNAP binds readily, albeit weakly, to this promoter, but on linear templates it requires Spo0A∼P to initiate transcription efficiently (8, 9). Similarity between both promoters is also reflected by the fact that the first of the two activator binding sites is located upstream of the −35 promoter sequence at the nontranscribed strand, while the second one is directly overlapping the −35 consensus sequence.
Optimal spacing in EσA-dependent promoters is 17 to 19 bp. The inability of the RNAP to transcribe spoIIG in the absence of Spo0A∼P may be due to the large window of 22 bp separating the −35 and −10 promoter regions, effectively preventing proper binding of the enzyme to the DNA. In vitro transcription assays performed with heteroduplex templates implied that Spo0A∼P stimulated transcription at least in part by stabilizing the RNA-polymerase-spoIIG complex until contacts between the RNA polymerase and the −10element induced strand separation. Therefore, Seredick and Spiegelman (46) argued that the role of the transcription activator Spo0A∼P is to promote alignment of σA with the downstream promoter elements by two possible mechanisms: (i) stimulation of the release of upstream contacts and (ii) locking of RNAP near the DNA after release from the −35 element contacts. For a recent model, Kumar et al. (27) proposed that activation of the spoIIG promoter is accomplished by direct interaction of the surfaces of the dimeric regulator Spo0A and EσA. According to their model it was unlikely that Spo0A and EσA simultaneously occupy the same binding site at −35. Instead, binding of RNAP at a site with optimal spacing of 17 to 18 bp to the −10 region was favored by protein-protein contacts between dimeric Spo0A located at −35 and the RNAP bound at −10.
The sequence of another Spo0A-activated promoter, spoIIE, is similar to that of spoIIG in that it contains a −35-like box separated by 21 bp from the −10 region sequence (19). The Spo0A binding box overlaps with the −35 sequence as well (50). Due to the similarities mentioned above, we assume that a similar mechanism occurs after binding of PhoP at the −35 sequence of the phyC gene promoter, which possibly overcomes the steric constraints caused by improper spacing between the −35 and −10 regions.
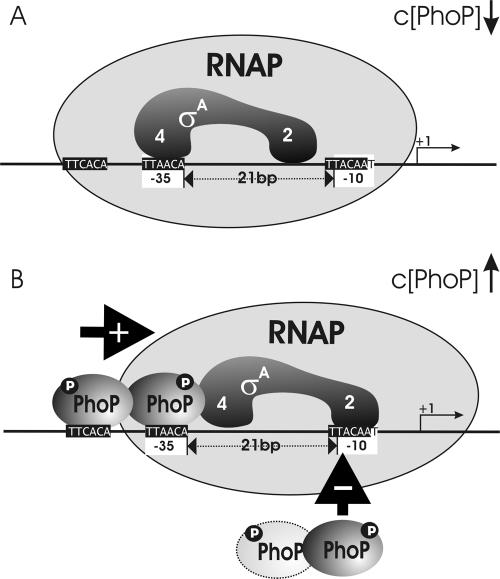
This view is mainly supported by the results of the DNase I footprinting obtained for mutants MUT13 and MUT37 and for the PhoP∼P-dependent in vitro transcription of MUT13. We suggest the following model: after first contact of the RNAP at the −35 consensus, RNAP binds directly at the −10 consensus promoter region. Most likely this event does not occur in the absence of the response regulator PhoP∼P during high-phosphate conditions, due to improper spacing between the two consensus regions. During phosphate limitation, the level of PhoP∼P rises, which results in occupation of the two PhoP boxes around the −35 promoter region. Protein-protein interactions between the bound PhoP∼P dimer and RNAP subsequently stabilize the complex, which is linked with the promoter DNA at −10, and will finally lead to transcription activation. At high-phosphate conditions, without PhoP∼P attached at −35, RNAP binding at the promoter upstream region is not supported and the phyC gene is not expressed. The DNase I footprinting data shown in Fig. 5 and 7 reveal higher affinity of PhoP∼P to the two tandemly arranged PhoP∼P boxes than to the single PhoP box at −10, suggesting first binding of the response regulator at the region adjacent to and upstream from −35. Higher levels of PhoP∼P will then lead to competition between RNAP and PhoP∼P at the −10 binding site and result in decreasing gene transcription. This way, phyC expression is relatively tightly regulated under conditions of phosphate deprivation (Fig. 9). This model is supported by our in vitro transcription experiments, in which a high concentration of PhoP reduced transcription efficiency in promoters harboring a functional PhoP binding box adjacent to −10 but was without effect in MUT13, with a mutated single PhoP box.
FIG. 9.
Model of the interactions of PhoP and RNAP on the phyC promoter of B. amyloliquefaciens FZB45. In the absence of PhoP∼P, EσA RNAP binds preferentially to one of the two possible binding sites at −35 and −10 due to their improper spacing. In the presence of phosphorylated PhoP, the potential binding sites become occupied by the response regulator: the tandem repeat PhoP boxes adjacent to −35 with higher efficiency than the single one at −10. Upon binding of the dimeric response regulator PhoP∼P to a region adjacent to −35, binding of RNAP at −10 is facilitated by protein-protein interactions with the upstream bound PhoP∼P. Formation of open complex and transcription initiation will start. A rising concentration of PhoP∼P, causing competition between PhoP∼P and RNAP at −10, eventually decreases transcription efficiency. The promoter consensus sites are labeled with white, the PhoP binding sites with black boxes. The putative binding sites at subdomains EσA2 and EσA4 are indicated.
Future experiments may include a shortening of the window between the −35 and −10 sites, as well as identification of the amino acid residues involved in surface interactions between PhoP∼P bound at the phyC promoter and specific EσA regions of the RNAP holoenzyme. These will lead to a better understanding of the activation process of this unusual promoter structure. The promoter structure described here is well conserved in the phytase genes of B. subtilis VTT E-68013 (accession no. AF029053), B. amyloliquefaciens (accession no. U85968) and B. licheniformis (accession no. AF469936), suggesting that transcriptional activation of the FZB45 phytase gene is representative of phyC gene regulation in bacilli. In spite of these similarities, it is likely that mutations introduced within the phyC promoter as described here for MUT13 will be important for the design of industrial Bacillus strains engineered for high productivity in phytase gene expression.
Supplementary Material
Acknowledgments
Partial financial support given for the genomic network sponsored by BMBF, the German ministry for education and research, is gratefully acknowledged. Sarah Dubrac's work was supported by research funds from the European Commission (Grant LSHG-CT-2004-503468 BACELL Health), the Centre National de la Recherche Scientifique, and the Institut Pasteur (Grand Programme Horizontal no. 9).
We thank Masaya Fujita for providing the Bacillus subtilis strain MF1 used for isolation of RNAP. We also thank Christiane Müller for technical support and sequencing and Markus Wilhelms for cloning pPHOR231 and purification of the proteins. Steffen Porwollik (Sidney Kimmel Cancer Center, San Diego, Calif.) is especially thanked for valuable hints for improving the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdel-Fattah, W. R., Y. Chen, A. Eldakak, and F. M. Hulett. 2005. Bacillus subtilis phosphorylated PhoP: direct activation of the EσA- and repression of the EσE-responsive phoB-PS+V promoters during Pho response. J. Bacteriol. 187:5166-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniewski, C., B. Savelli, and P. Stragier. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlung, T., and L. Brönsted. 1994. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J. Bacteriol. 176:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldus, J. M., B. D. Green, P. Youngman, and C. P. Moran, Jr. 1994. Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J. Bacteriol. 176:296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensing, B. A., B. J. Meyer, and G. M. Dunny. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 93:7794-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird, T. H., J. K. Grimsley, J. A. Hoch, and G. B. Spiegelman. 1993. Phosphorylation of Spo0A activates its stimulation of in vitro transcription from the B. subtilis spoIIG operon. Mol. Microbiol. 9:741-749. [DOI] [PubMed] [Google Scholar]
- 9.Bird, T. H., J. K. Grimsley, J. A. Hoch, and G. B. Spiegelman. 1996. The Bacillus subtilis response regulator Spo0A stimulates transcription of the spoIIG operon through modification of RNA polymerase promoter complexes. J. Mol. Biol. 256:436-448. [DOI] [PubMed] [Google Scholar]
- 10.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 11.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 12.Chesnut, R. S., C. Bookstein, and F. M. Hulett. 1991. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol. Microbiol. 5:2181-2190. [DOI] [PubMed] [Google Scholar]
- 13.Dassa, J., H. Fsihi, C. Marck, M. Dion, M. Kieffe-Bontemps, and P. L. Boquet. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol. Gen. Genet. 229:341-352. [DOI] [PubMed] [Google Scholar]
- 14.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 15.Eder, S., L. Shi, K. Jensen, K. Yamane, and F. M. Hulett. 1996. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology 142:2041-2047. [DOI] [PubMed] [Google Scholar]
- 16.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, M., and Y. Sadaie. 1998. Rapid isolation of RNA polymerase from sporulating cells of Bacillus subtilis. Gene. 221:185-190. [DOI] [PubMed] [Google Scholar]
- 18.Greiner, R., E. Haller, U. Konietzky, and K.-D. Jany. 1997. Purification and characterization of a phytase from Klebsiella terrigena. Arch. Biochem. Biophys. 341:229-248. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, P., J. Westpheling, and P. Youngman. 1988. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J. Bacteriol. 170:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 21.Hulett, F. M., E. E. Kim, C. Bookstein, N. V. Kapp, C. W. Edwards, and H. W. Wyckoff. 1991. Bacillus subtilis alkaline phosphatases III and IV. Cloning, sequencing, and comparisons of deduced amino acid sequence with Escherichia coli alkaline phosphatase three-dimensional structure. J. Biol. Chem. 266:1077-1084. [PubMed] [Google Scholar]
- 22.Hulett, F. M., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. F. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the Pho regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idriss, E. E., O. Makarewicz, A. Farouk, K. Rosner, R. Greiner, H. Bochow, T. Richter, and R. Borriss. 2002. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth promoting effect. Microbiology 148:2097-2109. [DOI] [PubMed] [Google Scholar]
- 24.Irving, G. J. C., and D. J. Cosgrove. 1971. Inositol phosphate phosphatases of microbiological origin. Some properties of a partially purified bacterial (Pseudomonas sp.) phytase. Aust. J. Biol. Sci. 24:547-557. [DOI] [PubMed] [Google Scholar]
- 25.Kerovuo, J., M. Lauraeus, P. Nurminen, N. Kalkkinen, and J. Apajalahti. 1998. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 64:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, Y.-O., J.-K. Lee, H.-K. Kim, J.-H. Yu, and T.-K. Oh. 1998. Cloning of the thermostable phytase (Phy) from Bacillus sp. DS11 and its overexpression in Escherichia coli. FEMS Microbiol. Lett. 162:185-191. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, A., C. Buckner Starke, M. DeZalia, and C. P. Moran, Jr. 2004. Surfaces of Spo0A and RNA polymerase sigma factor A that interact at the spoIIG promoter in Bacillus subtilis. J. Bacteriol. 186:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W., and F. M. Hulett. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 30.Liu, W., Y. Qi, and F. M. Hulett. 1998. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol. Microbiol. 28:119-130. [DOI] [PubMed] [Google Scholar]
- 31.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucca, P., R. Hurrell, and I. Potrykus. 2002. Fighting iron deficiency anemia with iron-rich rice. J. Am. Coll. Nutr. 21:184-190. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus.. Wiley, Chichester, United Kingdom.
- 36.Oh, B.-C., W.-C. Choi, S. Park, Y.-O. Kim, and T.-K. Oh. 2004. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 63:362-372. [DOI] [PubMed] [Google Scholar]
- 37.Paul, S., S. Birkey, W. Liu, and F. M. Hulett. 2004. Autoinduction of Bacillus subtilis phoPR. Operon transcription results from enhanced transcription from EσA- and EσE-responsive promoters by phosphorylated PhoP. J. Bacteriol. 186:4262-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pen, J., T. C. Verwoerd, P. A. Paridon, R. F. Beudeker, P. J. Elzen, K. Geerse, J. D. Klis, H. A. Versteegh, A. J. Ooyen, and A. Hoekema. 1993. Phytase-containing transgenic seeds as a novel feed additive for improved phosphorus utilization. BioTechnology 11:811-814. [Google Scholar]
- 39.Pragai, Z., N. E. E. Allenby, N. O'Connor, S. Dubrac, G. Rapoport, T. Msadek, and C. R. Harwood. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri-Taneja, A., S. Paul, Y. Chen, and F. M. Hulett. 2006. CcpA causes repression of the phoPR promoter through a novel transcription start site, PA6. J. Bacteriol. 188:1266-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi, Y., and F. M. Hulett. 1998. PhoP∼P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 43.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sajidan, A., A. Farouk, R. Greiner, P. Jungblut, E.-C. Müller, and R. Borriss. 2004. Molecular and physiological characterisation of a 3-phytase from soil bacterium Klebsiella sp. ASR1. Appl. Microbiol. Biotechnol. 65:110-118. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Seredick, S. D., and G. B. Spiegelman. 2004. The Bacillus subtilis response regulator Spo0A stimulates sA dependent transcription prior to the major energetic barrier. J. Biol. Chem. 279:17397-17403. [DOI] [PubMed] [Google Scholar]
- 47.Soldo, B., V. Lazarevic, M. Pagni, and D. Karamata. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol. Microbiol. 31:795-805. [DOI] [PubMed] [Google Scholar]
- 48.Tye, A. J., F. K. Siu, T. Y. C. Leung, and B. L. Lim. 2002. Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl. Microbiol. Biotechnol. 59:190-197. [DOI] [PubMed] [Google Scholar]
- 49.Walker, M. S., and J. A. DeMoss. 1994. NarL-phosphate must bind to multiple upstream sites to activate transcription from the narG promoter of Escherichia coli. Mol. Microbiol. 14:633-641. [DOI] [PubMed] [Google Scholar]
- 50.York, K., T. J. Kenney, S. Satola, C. P. Moran, Jr., H. Poth, and P. Youngman. 1992. Spo0A controls the σA-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J. Bacteriol. 174:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.