Abstract
Enterobactin, the tris-(N-(2,3-dihydroxybenzoyl)serine) trilactone siderophore of Escherichia coli, is synthesized by a three-protein (EntE, B, F) six-module nonribosomal peptide synthetase (NRPS). In this work, the 142-kDa four-domain protein EntF was bisected into two double-domain fragments: a 108-kDa condensation and adenylation construct, EntF C-A, and a 37-kDa peptidyl carrier protein (PCP) and thioesterase protein, EntF PCP-TE. The adenylation domain activity of EntF C-A formed seryl-AMP but lost the ability to transfer the seryl moiety to the cognate EntF PCP-TE in trans. Seryl transfer to heterologous PCP protein fragments, the SrfB1 PCP from surfactin synthetase and Ybt PCP1 from yersiniabactin synthetase, was observed at rates of 0.5 min−1 and 0.01 min−1, respectively. The possibility that these slow acylation rates reflected dissociation of acyl/aminoacyl-AMP followed by adventitious thiolation by the heterologous PCPs in solution was addressed by measuring catalytic turnover of pyrophosphate (PPi) released from the adenylation domain. The holo SrfB1 PCP protein as well as Ybt PCP1 did not stimulate an increase in PPi release from EntF C-A or EntE. In this light, aminoacylations in trans between A and PCP domain fragments of NRPS assembly lines must be subjected to kinetic scrutiny to determine whether transfer is truly between protein domains or results from slow aminoacyl-AMP release and subsequent nonenzymatic thiol capture.
Nonribosomal peptide synthetases (NRPS) activate a broad range of amino acids, including numerous nonproteinogenic amino acids (1, 2) and incorporate them into growing peptidyl chains as an elongating series of peptidyl-S-enzyme intermediates. This is an RNA-independent templated chain-growth process with an assembly line organization of different catalytic and carrier protein domains whose placement and function determine the length and structure of the released peptide natural product. Both the amino acid monomers and the elongating peptide chains are tethered as thioester intermediates (acyl-S-enzymes) to the terminal thiol of phosphopantetheinyl (Ppant) moieties of peptidyl carrier protein (PCP) domains, also known as thiolation (T) domains for their functions as thiol-tethering way stations. The Ppant thiol in the PCP domains is added posttranslationally by priming enzymes (3, 4) to a conserved serine side chain in each PCP domain, converting it from inactive apo form to holo form capable of aminoacylation.
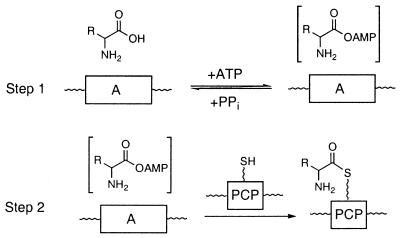
The 8- to 10-kDa peptidyl chain-carrying PCP domains are interspersed among and paired with two catalytic domains that make up the core of each NRPS elongation module: 50- to 60-kDa adenylation (A) domains for each amino acid to be activated and 50-to 60-kDa condensation (C) domains for each peptide bond-forming chain-translocating step. Thus a typical NRPS module consists of the core triad of (C-A-PCP)n domains as the functional unit competent for: selection and activation of a given amino acid as the aminoacyl-AMP (Fig. 1, Step 1), covalent loading of the aminoacyl moiety on the paired PCP domain (Fig. 1, Step 2), and condensing it with the upstream peptidyl-S-PCPn-1 to yield a chain-extended translocated peptidyl-S-PCPn intermediate.
Figure 1.
NRPS A domain activity. In Step 1, the amino acid is activated as the aminoacyl adenylate, whereas in Step 2 the amino acid is covalently transferred to the PCP domain via a thioester linkage.
It has been assumed that the major fidelity or editing functions for amino acid selection and incorporation into the elongating peptide chain reside in the A domains of NRPS, analogous to the major editing functions of the cognate A domains of aminoacyl-tRNA synthetases (5, 6). The classic assay for A domains in both NRPS and aminoacyl-tRNA synthetases is the amino acid-dependent exchange of radiolabel from 32PPi into ATP (7), which measures reversible formation of aminoacyl-AMP, the first half reaction, and allows a clear measure of amino acid selectivity in this step. The assay is uninformative with regard to the second half reaction of A domains, the capture of the aminoacyl portion of the aminoacyl-AMP by the thiol of the Ppant prosthetic group of the paired PCP domain. It is unknown whether paired A-PCP domains communicate with each other largely because of covalently enforced proximity (the in cis fusion within a module), which ensures a high local concentration of Ppant-SH to capture the aminoacyl-AMP lodged in the adjacent A domain, or whether there is editing specificity for the particular aminoacyl moiety to be captured and/or protein–protein recognition features between a given A domain and its immediate downstream PCP domain. The general answer has implications both for the efficiency and design fidelity of any NRPS assembly line as well as for domain swap strategies in combinatorial biosynthesis approaches to create diversity in NRPS antibiotic peptides (8, 9).
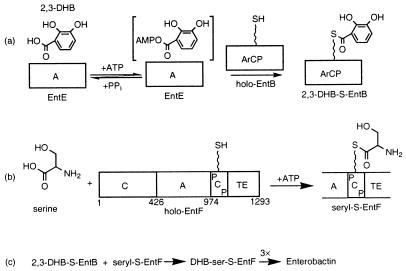
To begin to dissect paired A-PCP domain recognition issues, we have turned to the Escherichia coli enterobactin synthetase system, an NRPS comprised of the three proteins EntE, EntB, and EntF (10, 11) (Fig. 2). Two PCP domains, one in EntB and one in EntF, are primed with Ppant-SH by an Ent operon-dedicated posttranslational transferase enzyme EntD (3). Enterobactin, the E. coli siderophore assembled and secreted during iron starvation (12), is the cyclic trilactone of N-(2,3-dihydroxybenzoyl)serine, and its robust in vitro biosynthesis has been achieved by reconstitution of the purified EntE, B, F system (11). With EntE contributing one A domain, EntB one PCP domain, and the 142-kDa EntF contributing four domains, C-A-PCP- and chain-releasing thioesterase (TE), the six domains, A-PCP-C-A-PCP-TE, spread over three proteins, comprise a simple well-characterized two-module NRPS. The first A domain forms 2,3-dihydroxybenzoyl acid (DHB)-AMP in step one and transfers it to yield DHB-S-EntB in step two. The second A domain, in Ent F, yields seryl-AMP and then seryl-S-EntF. The C domain of Ent F condenses DHB-S-PCP (EntB) onto ser-S-PCP (EntF) to produce the DHB-Ser-S-EntF acyl–S-enzyme intermediate (11, 13). In the Ent synthetase, the first A-PCP pair, A(EntE)-PCP(EntB), exists naturally in trans, whereas the second pair A-PCP(EntF) is naturally in cis.
Figure 2.
Schematic of the proteins involved in enterobactin assembly. (a) DHB is loaded onto the Ppant thiol of the EntB aryl carrier protein domain (ArCP) via the in trans action of the A enzyme EntE. (b) Serine is loaded onto the PCP of holo-EntF via the cis-acting A domain of holo-EntF. (c) To assemble enterobactin, DHB-S-EntB ArCP condenses with seryl-S-EntF PCP to form the EntF-bound DHB-ser monomer. After three cycles of C and loading, enterobactin is released.
In this work, we have assessed the selectivity of both the EntE A domain and the EntF A domain for PCP domain partners by bisecting the four-domain Ent F into C-A and PCP-TE fragments and assaying each A domain in trans with homologous and heterologous PCP domains.
Materials and Methods
Construction of EntF C-A Two-Domain Proteins.
Recombinant DNA techniques were carried out according to established protocols (14). EntF DNA encoding the C-A peptide fragment (corresponding to EntF residues 1–974) was amplified from the pMS22 template (15) with one round of PCR by using the forward primer 5′-GAATTCCATATGAGCCAGCACCTGCCTTTGGTCGCCGCACAG-3′ and the reverse primer 5′-TGACCCAAGCTTTCACGCTTTCGGCGCACGCCC-3′, where the restriction sites used for cloning are underlined. The 2.8-kb PCR product and the vector pET22b (Novagen) were digested with NdeI-HindIII and ligated. The resulting plasmid, pET22b-EntF (1–974), was transformed into E. coli DH5a to propagate the plasmid. All PCR-amplified fragments in this study were confirmed by DNA sequencing (Dana Farber Molecular Biology Core Facility, Boston, MA).
A subsequent construction of the EntF C-A protein was designed with a C-terminal hexahistidine tag. For this, the pET22b-EntF (1–974) plasmid was used as the template for PCR with the forward primer 5′-AGTTCGCCGCGGATCCGGCGCTGTTGTGC-3′ and reverse primer 5′-AGCCGCAAGCTCGAGCGCTTTCGGCGCACG-3′. The 1.7-kb PCR product and the vector pET22b-EntF (1–974) were digested with BamHI and XhoI, ligated, and transformed into E. coli DH5a cells to create the plasmid pET22b-EntF (1–974-His6).
Another version of the EntF C-A protein was cloned with 31 additional C-terminal amino acids and an N-terminal maltose-binding protein (MBP) tag that could bind with an amylose resin (New England Biolabs) during purification. First, site-directed mutagenesis by using the splicing by overlap extension method (16) was performed to remove an internal NdeI site from the EntF gene while retaining the histidine codon. Plasmid pMS22 served as a template for the first round of PCR amplification by using the primers EntFCA-for1 (5′-CCAGGTGAACGGATGTACCGTACCGGAGACG-3′) and EntFCA-rev (5′-CCAC- CGGTACCATGTGTGGTGGCAATGTTTC-3′) to produce a 322-bp oligonucleotide and the primers EntFCA-for2 (5′-GAAACATTGCCACCACACATGGTACCGGTGG-3′) and EntFCA-HindIIIrev (5′-GCCAGTTTCATTGCAAGCTTTCAATGACCGCC-3′) to produce a 257-bp nucleotide. The two PCR-amplified DNA fragment products from round one were gel purified (Qiaquick Kit, Qiagen) and used as the templates during a second round of amplification to yield the final 548-bp DNA product containing the desired mutation. The final PCR product was gel purified and digested along with the vector pMS22 by AvaI-HindIII. Ligations were transformed into E. coli DH5a to yield the product plasmid pMS22-EntF (1–1005). This plasmid was subsequently digested along with pET22b-EntF (1–974) by BamHI-HindIII, and the transformed ligations yielded pET22b-EntF (1–1005). Finally, the plasmid pET22b-EntF (1–1005) was digested by NdeI-HindIII along with pIADL14 (17) containing the MBP gene on a pET28b (Novagen) vector. Successful transformants with this ligation yielded the plasmid pEntF (1–1005)-MBP containing the MBP gene followed by a thrombin cleavage site and the EntF C-A gene encoding residues 1–1005 of EntF.
Overproduction and Purification of Enterobactin Synthetase Proteins.
EntD (3), EntB (10), EntE (10), EntF (11), and EntF PCP-TE (13) were overproduced and purified as previously described. For the overproduction and purification of the EntF C-A proteins, each overexpression plasmid was transformed into E. coli BL21(DE3) and the resulting strain grown at 30°C to an optical density of 0.7 (LB, 100 μg/ml ampicillin for pMS22 or pET22b vectors or 50 μg/ml kanamycin for pIADL14 vector) and induced with 1 mM isopropyl-d-thiogalactoside. Four hours after induction, cells were harvested by centrifugation and lysed by two passages through a French pressure cell. The lysis buffer for the EntF (1–974) 107-kDa protein was 25 mM Tris⋅HCl (pH 8.0)/10 mM MgCl2/5 mM DTT. Ammonium sulfate precipitation, ion exchange, and gel filtration chromatography purification steps for the EntF (1–974) protein were carried out as described for full-length EntF (11).
The His6-tagged protein EntF C-A (1–974-His6) was purified by nickel chelate chromatography according to the manufacturer's instructions (Novagen).
The lysis buffer for the EntF C-A (1–1005)-MBP 153-kDa protein was 20 mM Tris⋅HCl (pH 7.5)/1 mM EDTA/200 mM NaCl/1 mM DTT. The MBP fusion protein was subsequently purified by affinity chromatography by using amylose resin according to the manufacturer's instructions (New England Biolabs). Fractions containing the EntF C-A (1–1005)-MBP fusion were pooled and cleaved with thrombin endonuclease (Novagen) for 1 hr according to the supplier's instructions. Cleavage and purity of the EntF C-A (1–1005)-MBP fusion was assessed by SDS/PAGE, and positive chromatography fractions were pooled, dialyzed against 25 mM Tris⋅HCl (pH 8.0)/10 mM MgCl2/5 mM DTT buffer with 10% glycerol and stored at −80°C.
Protein concentrations were determined by using the calculated extinction coefficients for the absorbance of each protein at 280 nm (18): 138,060 M−1 cm−1 for EntF C-A (1–974 +/− His6) and 202,900 M−1 cm−1 for EntF C-A (1–1005)-MBP.
Overproduction and Purification of Surfactin and Yersiniabactin Synthetase PCP Domains.
The SrfB1 PCP protein was overproduced and purified as described previously (19). The yersiniabactin protein containing PCP1, named S1977A and composed of amino acids 1383–2035 of the high molecular-weight protein 2, was overproduced and purified as described (20).
ATP–[32P]PPi Exchange Assay.
ATP-pyrophosphate (PPi) exchange was assayed as described previously (21). Reactions (100 μL) contained 75 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM tris-(2-carboxyethyl)phosphine (Molecular Probes), 1 mM sodium [32P] PPi (5.0 Ci/mol) (Dupont NEN), 5 mM ATP, and enough of the EntF protein to maintain linear initial velocity conditions (for EntF wild type 10 nM for 5 min, for EntF 1–974-His6 50 nM for 2 min, for EntF 1–974 100 nM for 2 min, and for EntF 1–1005 100 nM for 20 min).
Assay for Covalent Incorporation of Amino or Aryl Acids into PCP Domains.
Serine incorporation into holo-carrier proteins was quantified with a trichloroacetic acid (TCA) precipitation radioassay. Reaction mixtures (100 μl final volume) included 75 mM Tris⋅HCl (pH 8.0)/1 mM tris-(2-carboxyethyl)phosphine/10 mM MgCl2/5 mM ATP/0.5 mM CoASH (Sigma)/550 nM EntD/800 μM l[3H]-serine (320 Ci/mol) (Dupont NEN)/5 μM EntF C-A, and varying concentrations of PCP protein. Apo proteins were preincubated for 45 min at 37°C with EntD to allow phosphopantetheinylation before addition of the radiolabeled serine and ATP. Reactions were initiated by addition of ATP, incubated at 37°C for 30 min, and quenched with 0.8 ml 10% TCA with BSA (375 μg) added as a carrier. Precipitated proteins were centrifuged and washed three times with 10% TCA. The protein pellet was dissolved in 150 μl 1 M Tris base and the amount of incorporated [3H]Ppant quantified by liquid scintillation counting.
For salicylate transfer catalyzed by EntE, reactions were carried out with 6.5 μM EntE, 180 μM [14C] salicylate (56 Ci/mol) (Dupont NEN), and varying concentrations of SrfB1 PCP.
Reactions for analysis by MS were prepared by using final concentrations of 50 μM SrfB1 PCP, 5 μM EntF 1–974-His6, and 10 mM unlabeled serine. Samples were separated by HPLC followed by electrospray MS, both performed by the Howard Hughes Medical Institute Biopolymers Facility at Harvard Medical School.
PPi Release Assay.
PPi levels were measured by using the continuous spectrophotometric assay furnished by the EnzChek Pyrophosphate Assay Kit (Molecular Probes). In this assay, PPi is hydrolyzed to inorganic phosphate by inorganic pyrophosphatase and phosphate production coupled to phosphorolysis of the guanosine analogue, 2-amino-6-mercapto-7-methylpurine ribonucleoside (MesG), catalyzed by the enzyme purine nucleoside phosphorylase (PNP). The chromophoric product, 2-amino-6-mercapto-7-methylpurine, is monitored by absorbance at 360 nm. Additional quantities of MesG were synthesized by the method originally described by Webb (22), whereas yeast inorganic pyrophosphatase and calf spleen PNP were purchased from Sigma.
Reactions (200 μL) listed in Table 3 contained 75 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM tris-(2-carboxyethyl)phosphine, 5 mM ATP, 400 μM MesG, 0.2 units purine nucleoside phosphorylase, and 0.2 units inorganic pyrophosphatase. For the rate of release of acyl/aminoacyl-AMP in the absence of PCPs, 1 μM EntE was used with 2 mM salicylate and 2 μM EntF C-A with 9 mM serine. For experiments in the presence of PCPs, holo PCPs were prepared and added to the reactions to 50 μM. Readings of absorbance at 360 nm were measured over a 6-min interval in a Perkin–Elmer Lambda 6 UV-vis spectrophotometer and slopes correlated with a standard curve created with PPi.
Table 3.
Pyrophosphate release rates
| Adenylation construct | Added carrier protein | PPi release rate,
min-1
|
Acylation rate, min−1
|
||
|---|---|---|---|---|---|
| Salicylate dependent | Serine dependent | Salicylation | Serylation | ||
| EntE | 9 ±1 | ||||
| EntE | EntB ArCP | 95 ± 5 | 130 | ||
| EntE | SrfB1 PCP | 9 ± 1 | 0.5 | ||
| EntF C-A | 2.3 ± 0.4 | ||||
| EntF C-A | SrfB1 PCP | 2.0 ± 0.3 | 0.5 | ||
| EntF C-A | Ybt PCP1 | 2.2 ± 0.3 | 0.01 | ||
Results
Construction of EntF Fragments.
Our first attempt to create an amino acid-activating A domain fragment protein from EntF was an excised module, amino acids 426–974, which failed to yield soluble active protein (23). To address this failure, in the next generation of EntF fragments, the native N terminus of EntF was retained and breakage points selected between the A and PCP domains of the four-domain EntF. The breakage points were chosen on the basis of known structural data for an A domain (24, 25) and boundaries previously noted for active individual domain fragments (26–28). Two EntF fragment proteins containing the C and A domains (EntF C-A) were constructed, differing in their C termini. The first fragment protein, EntF 1–974, terminated at the proposed A-PCP junction, whereas the second fragment, EntF 1–1005, continued into the PCP domain without including its conserved serine. In addition, a version of EntF 1–974 was created with a C-terminal hexahistidine affinity tag to facilitate purification. Even though the His6 tag could potentially interfere with interactions between the A and PCP domains, its location was chosen to maintain a native N terminus for a C domain-mediated interaction with the 2,3-dihydroxybenzoyl-carrying protein, EntB. The start of the fragment comprising the back half of EntF, the PCP and TE domains (EntF PCP-TE) was chosen approximately 40 amino acids upstream of the PCP's conserved serine (S1006). Beginning at amino acid 960 and ending at the native C terminus of EntF, the 37-kDa EntF PCP-TE fragment was also purified via a C-terminal His6 tag.
The A and PCP Domains Retain Activity.
To verify the structural and functional integrity of the fragment proteins, individual domain functions were assessed. We have not focused on the C or TE domains in this work. A domain activity was measured by ATP–[32P]PPi exchange. All of the EntF C-A fragments displayed a reduced catalytic efficiency for serine-dependent A compared with native EntF (Table 1). This reduction, derived from Kms for serine 20- to 30-fold higher than intact full-length EntF, suggests some disruption of A domain architecture for serine binding in the fragment C-A proteins. The higher kcat observed for EntF 1–974-His6 may result from its purification in fewer steps than the other A domain-containing proteins. EntF 1–974-His6, retained the most robust activity compared with intact EntF and was thus used for further experiments. In addition, the apo PCP domain of EntF PCP-TE could be converted to holo form (data not shown) after phosphopantetheinylation by the phosphopantetheinyl transferase EntD and cosubstrate [3H]CoASH (13).
Table 1.
Kinetic parameters for serine-dependent ATP/[32P]PPi exchange
| Construct | kcat, min−1 | Km, mM | kcat/Km min−1 mM−1 |
|---|---|---|---|
| EntF full length | 670 ± 20 | 0.6 ± 0.03 | 1,120 |
| EntF C-A 1-974-His6 | 990 ± 80 | 14 ± 2 | 70 |
| EntF C-A 1-974 | 340 ± 10 | 22 ± 3 | 15 |
| EntF C-A 1-1005 | 16 ± 3 | ND | ND |
ND, not determined.
Aminoacylation of Noncognate PCP Domains by EntF C-A and EntE.
With the A activity of EntF C-A retained and phosphopantetheinylation of EntF PCP-TE demonstrated, the ability to transfer seryl-AMP from EntF C-A to EntF PCP-TE was examined. The assay entailed detecting radioactive serine incorporation by TCA precipitation or autoradiography. Unfortunately, no covalent loading of PCP-TE with radioactive serine was observed under any conditions assayed (data not shown). Although failure to transfer may have stemmed from a faulty A domain C terminus, transfer catalyzed by untagged EntF 1–974 and EntF 1–1005 was also undetectable.
At this point, heterologous PCP domain acceptors were investigated. Previous work had shown that the 16.8-kDa SrfB1 PCP fragment from the surfactin synthetase of Bacillus subtilis could be aminoacylated with valine by its cognate A domain in trans (27), therefore this PCP was tested for serylation by EntF C-A. Holo SrfB1 PCP was indeed loaded with serine as detected by TCA precipitation with radiolabeled serine (not shown), autoradiography (not shown), and MS of the 16.8-kDa fragment, with a gain of 88 mass units detected as an index of serinylation. Variation of rate vs. SrfB1 PCP concentration revealed saturation kinetics with an aminoacylation rate of 0.5 min−1 (Table 2). Another heterologous PCP protein fragment, Ybt PCP1, a 75.1-kDa protein fragment [residues 1383–2035 of the high molecular-weight protein 2 subunit (20)] containing PCP1 from the synthetase of the Yersinia pestis siderophore yersiniabactin, was also serylated by EntF C-A, but at an even slower rate of 0.01 min−1 (Table 2).
Table 2.
In trans aminoacylation rates
| Adenylation construct | Carrier protein construct | Acylation rate |
|---|---|---|
| EntF C-A | EntF PCP-TE | — |
| EntF C-A | SrfB1 PCP | 0.5 min−1 |
| EntF C-A | Ybt PCP1 | 0.01 min−1 |
| EntE | EntB ArCP | 130 min−1 |
| EntE | SrfB1 PCP | 0.5 min−1 |
These slow rates prompted a comparison to the action of the A protein EntE, whose carrier protein, EntB, is naturally acylated in trans (Fig. 2). Covalent loading of the aryl carrier protein domain of EntB (EntB ArCP) by EntE has been measured for the alternate substrate salicylate and found to be 130 min−1 (10). Loading of SrfB1 PCP with salicylate by EntE was assessed and found to occur at a rate of 0.5 min−1 (Table 2), over 200 times slower than the natural in trans rate to EntB ArCP.
PPi Release Rates.
To determine whether the slow observed aminoacylation rates were a result of specific interdomain recognition and loading or instead merely scavenging of released aminoacyl-AMP by the thiol-bearing holo-PCPs, PPi release rates were measured for the A domains. PPi release represents a complementary assay for the interdomain transfer of the acyl or aminoacyl moieties from AMP by measuring simultaneous release of the ATP cleavage products, AMP and PPi. We have chosen to measure PPi production by a coupled continuous spectrophotometric assay using the guanosine analogue MesG (22). PPi release rates were previously measured for EntE by using a phosphomolybdate-based spectrophotometric assay and found to be 0.4 min−1 for DHB and 10 min−1 for salicylate (21). By the MesG-based assay, PPi release from EntE was determined to be 0.7 min−1 for DHB and 9 min−1 for salicylate (Table 3), in good agreement. Addition of SrfB1 PCP to EntE did not increase the release rate for salicyl-AMP, whereas addition of holo EntB ArCP spurred PPi release 10-fold to a rate of 95 min−1. This assay clearly reveals the expected tight binding of aminoacyl-AMP in the absence of partner holo PCP and also can sensitively measure covalent transfer to the holo PCP.
The other enterobactin-dedicated A domain, from full-length EntF, displayed a serine-dependent release rate of 0.2 min−1, whereas the fragment EntF C-A released seryl-AMP 10-fold faster, at a rate of 2.3 min−1. Addition of holo SrfB1 PCP or holo Ybt PCP1 did not increase PPi release rates from EntF C-A.
Thiol-Dependent Stimulation of PPi Release.
To address whether alkyl thiols could act as surrogates for the Ppant arm of a holo PCP, the accessibility of acyl-/aminoacyl-AMP intermediates sequestered in their respective EntE or EntF A domains was assessed by measuring thiol-dependent stimulation of PPi release in the presence of aryl/amino acid. Thiols CoA (CoASH), pantetheine, cysteine, and DTT were used. EntE with DHB exhibited a saturable increase in PPi release upon addition of CoASH, pantetheine, and DTT (data not shown), with pantetheine the most efficient nucleophile. The EntF A domain, both in the four-domain full-length EntF and the two-domain C-A fragment, was much less accessible to seryl-AMP capture by the low molecular-weight thiols. PPi release from EntF, under the conditions measured, was detectable only with pantetheine and with a catalytic efficiency two orders of magnitude lower than that of EntE. None of the thiols tested stimulated an increase in PPi release from EntF C-A.
Discussion
In the assembly of nonribosomal peptides and depsipeptides by multimodular synthetases, there are many unresolved questions about the selectivity and editing functions of the core domains. Although the amino acid-activating A domains, like their functional counterparts in the aminoacyl-tRNA synthetases, have been proven to exercise discrimination in the initial recognition of substrate amino acid and conversion to the aminoacyl-AMP mixed anhydride (5), it is less clear to what extent there is selectivity in the second half reaction, the transfer of the activated aminoacyl group to the next downstream domain, the paired holo PCP domain (Fig. 1).
Because most amino acid-activating A domains are fused in cis to their paired PCP domains (2, 29), one way to probe selectivity would be to assess the ability of the excised A domains to transfer the aminoacyl moiety to holo forms of various PCP domains in trans, a strategy requiring both excised domains to fold and function autonomously. In initial efforts, the valine-activating A domain of module 4 of the surfactin synthetase (SrfB1) has been reported to valinate its cognate SrfB1 holo PCP in trans but not detectably the next, holo SrfB2 PCP, as assessed by radioactive covalent transfer (27). However, even the cognate in trans transfer rate was slow, and there was no evidence of saturation by the holo SrfB1 at up to 90 μM concentration. The TycA phenylalanine-specific A domain could also function in trans as assessed by radioactive SDS gel analysis of the holo PCP fragment, but labeling was weak, and the data were only qualitative (28). A particularly interesting recent report from the bleomycin synthetase system underscores the potential utility of probing the recognition of A domains for PCP domains. Du and Shen (30) found a free-standing PCP-type subunit of 90 aa, BlmI, that could be primed with Ppant and then aminoacylated with a noncognate valine-activating A domain from the same organism, Streptomyces verticillus. This is an example of a type II PCP domain, in analogy to the type II polyketide synthases, where separate subunits function iteratively to assemble aromatic polyketides (31), and would suggest a physiologic in trans recognition of such separate PCP subunits by one or more A domains.
In this work, we have turned to the E. coli enterobactin synthetase system for multiple reasons. First, it is an NRPS system that can be robustly reconstituted in vitro from four pure protein components, EntB, D, E, F, to produce the enterobactin siderophore at rates of 100–200 min−1 (11). Second, it is a relatively simple NRPS, with the EntE, B, and F proteins providing one, one, and four domains, respectively, to provide a two-module assembly line. There are two A domains that can be compared for in trans recognition of cognate and noncognate carrier protein domains: the A domain of the single-domain 59-kDa EntE and the A domain of the four-domain 142-kDa EntF (Fig. 2). The EntE A domain is specific for aryl carboxylic acids, functioning as a chain-initiating aryl-N-capping catalyst, whereas the embedded A domain in EntF is a serine-specific amino acid-activating catalyst. The partner for EntE is the holo form of the EntB carrier protein domain in a physiological in trans interaction, whereas the acceptor for the EntF A domain is the holo form of the PCP domain, fused in cis, the typical situation for internal chain-elongating modules of NRPS.
First attempts to excise and express the EntF A domain as a typical 50- to 60-kDa A domain fragment failed to give soluble folded protein with any detectable activity (23), highlighting the ongoing unpredictability of stable folding of internal NRPS domains (26, 32–34). Therefore, we retained the native N terminus of EntF and expressed the two domain C-A fragment of EntF as a 108-kDa protein (residues 1–974), which proved to be soluble and contain an active A domain when purified as a C-terminal His6-tagged fragment. The first assay of A domain function was the classic serine-dependent [32P]PPi-ATP exchange assay, which yielded kcat of 990 min−1 and Km for serine of 14 mM. The catalytic efficiency ratio (kcat/Km) was 70 mM−1min−1, compared with 1,120 mM−1min−1 for the full-length four-domain EntF protein, suggesting some loss of catalytic power in the truncated C-A fragment for the first step in A domain catalysis, seryl-AMP formation.
A second assay for A domain integrity was the net ATPase rate, dependent on the amino acid serine. In contrast to the PPi-ATP exchange, which measures the ability to form enzyme-bound seryl-AMP reversibly and release the unlabeled PPi product into solution to exchange with [32P]PPi, the serine-dependent ATPase activity assay measures net release of PPi into solution. Catalytic turnover of ATP to PPi, as opposed to stoichiometric accumulation of seryl-AMP in the A domain active site, must be accompanied by release of seryl-AMP to regenerate the free A domain to catalyze another cycle of net hydrolysis. Thus, this assay is a measure of the leak rate, the frequency of loss of the thermodynamically activated aminoacyl-AMP from the A domain active site.
The leak rates of both the EntF A domain for seryl-AMP (0.2 min−1) and of the EntE A domain for 2,3-dihydroxybenzoyl-AMP (0.7 min−1) are indeed low, indicating efficient sequestration. With a kcat for enterobactin production of 100–200 min−1, these leak rates represent a loss of less than 0.1 to 0.4% of the activated intermediate at these two stages of the enterobactin assembly line. The double-domain EntF C-A displayed a 10-fold higher leak rate (2.3 min−1) than intact EntF, implying a less tightly bound seryl-AMP intermediate, perhaps indicating some slight loss of architectural integrity of the A domain of this 108-kDa protein fragment. A Km for serine for EntF C-A 20-fold higher than intact EntF supports this notion. In addition, we note that the continuous spectrophotometric assay to measure PPi facilitates assessment of aminoacyl-AMP leak rate, a valuable indicator of A domain integrity and a necessary baseline parameter for evaluating PCP aminoacylation.
A third attribute of the A domain of the EntF C-A fragment assessed was its ability to transfer the thermodynamically activated seryl moiety onto the terminal thiol of the Ppant chains of various holo PCP fragments. The initial assay was TCA precipitation of radioactivity from labeled serine, followed with a second assay, validation of covalent incorporation of radioactive serine into the PCP fragment by SDS/PAGE gel electrophoresis and autoradiography. The third assay, successful with SrfB1 PCP, was direct MS determination of the molecular weight increase in the PCP fragment as a consequence of seryl-S-PCP thioesterification. A scan of several holo PCP fragments available to us (20, 27, 28, 35) showed two interesting trends. First, under no experimental conditions could we detect seryl transfer to the cognate PCP-TE fragment of EntF (the other half of the protein). Second, there was detectable serylation of both the holo SrfB1 PCP and the Ybt PCP1. These latter two were selected for kinetic characterization and yielded saturation kinetics, yet very slow rates of aminoacylation, 0.01 to 0.5 min−1. These transfer rates are slower than the release rate of seryl-AMP (2.3 min−1) from the EntF C-A fragment A domain noted above. Therefore, a conservative interpretation is that covalent capture of the serinyl moiety by the holo PCPs' Ppant thiol could arise from prior aminoacyl-AMP release and nonspecific chemical aminoacylation of the thiol in solution by the mixed acyl-AMP anhydride. This should be competable by other thiols, and indeed DTT (data not shown) prevents detectable covalent labeling of the PCPs.
The EntE A domain serves as an interesting control and counterpart to the behavior of the EntF A domain. EntE shows a similarly slow substrate-mediated net ATPase activity (0.7 min−1) for its natural substrate, DHB, whereas the substrate analogue salicylate displays a markedly faster leak rate (9 min−1), suggesting weaker binding or less efficient sequestration. Nevertheless, introduction of EntE's specific partner holo carrier protein, the ArCP domain of EntB, induces a dramatic increase in net ATPase rate (to 95 min−1), consistent with our earlier complementary measurement of [14C]-salicylation rates of holo EntB ArCP (130 min−1) (10). On the other hand, the noncognate SrfB1 PCP, although salicylated by EntE, acquires label at the slow rate of 0.5 min−1, explicable as nonenzymatic capture via the leak rate of salicyl-AMP into solution before reaction. This result argues for high selectivity in this A/ArCP domain functional pairing.
As a final evaluation of the two enterobactin A domains, thiol-dependent stimulation of PPi release was examined. For the A domains of several aminoacyl-tRNA synthetases (aaRS) such activity has been studied (38), and various small thiol nucleophiles, including CoASH and pantetheine, could be aminoacylated, albeit slowly (kcat/Km 0.1 mM−1 min−1). The rate of release of aminoacyl-AMP from these aaRS has not been reported, and whether the observed thioesterification resulted from thiol binding at the aaRS active site or instead nonenzymatic scavenging from solution remains unclear. Toward EntE, thiols CoASH, pantetheine, and DTT triggered significant increases in PPi release and not surprisingly, pantetheine, the uncharged prosthetic arm of the natural cosubstrate carrier proteins, was the most efficient nucleophile (kcat/Km 1.6 mM−1 min−1). EntF exhibited a more restricted access to thiols, as did the EntF C-A fragment. It is conceivable that for an A-PCP pair fused naturally in cis, protein–protein interactions between holo PCP and the A catalytic site control access to potential nucleophiles, but this conjecture awaits structural confirmation of the architecture of A-PCP domain pairs.
There is one case known of an internal amino acid (cysteine)-activating A domain, the high molecular-weight protein 2 component of yersiniabactin synthetase, which services not one but three PCP domains, two in cis (PCP1, PCP2) and one in trans (PCP3) in the second, high molecular-weight protein 1 subunit (36). As expected, in the absence of holo PCP, this unusual A domain tightly retains cysteinyl-AMP (leak rate of 0.14 min−1) (37), but in the presence of holo PCP3 turns over at a rate of 195 min−1. In addition, cysteine analogue alternate substrates are transferred to PCP3 at rates comparable to cysteine, suggesting a lack of specificity of the holo PCP for this A domain-bound activated amino acid (Fig. 1, Step 2). Rapid loading of the EntB ArCP with the DHB analogue salicylate also supports this notion.
The in trans valination of the free-standing BlmI holo PCP domain of the bleomycin cluster (30) may present a related biologic context. The free-standing A domains in the chloroeremomycin biosynthesis cluster (39) and in the syringomycin cluster (40) may analogously function in trans to provide branch entry points into NRPS assembly lines. The holo BlmI valination, recently reported, is worth reexamination for both kinetics of valyl transfer and correlation to leak rate of the valyl-AMP from the valine-activating A domain. If that observed valination occurs at a rate slower than a measured leak rate of valyl-AMP, then the possibility is open that the qualitative valination represents merely a scavenging reaction from valyl-AMP in solution and therefore would be unlikely to be predictive of in vivo paired function of those domains.
Acknowledgments
We are grateful to Luis Quadri (Weill Medical College of Cornell University, New York, NY) and Debbie Miller (Harvard Medical School, Boston, MA) for providing SrfB1 PCP and Ybt PCP1, respectively, for initial experiments, and to Tom Keating (Harvard Medical School, Boston, MA) for synthesis of MesG. This work was supported in part by funds from National Institutes of Health Grant GM 20011 to C.T.W. D.E.E. and H.C.L. are National Science Foundation Graduate (NSF)Research Fellows. C.A.S.-R. was an NSF Minority Postdoctoral Research Fellow (DBI-9707847).
Abbreviations
- NRPS
nonribosomal peptide synthetase
- PCP
peptidyl carrier protein
- PPi
pyrophosphate
- Ppant
phosphopantetheine
- A
adenylation
- C
condensation
- TE
thioesterase
- DHB
2,3-dihydroxybenzoic acid
- CoASH
CoA
- MBP
maltose-binding protein
- TCA
trichloroacetic acid
- MesG
2-amino-6-mercapto-7-methylpurine ribonucleoside
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040572897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040572897
References
- 1.Kleinkauf H, von Dohren H. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 2.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997b;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 3.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 4.Walsh C T, Gehring A M, Weinreb P H, Quadri L E N, Flugel R S. Curr Opin Chem Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 5.Stachelhaus T, Mootz H D, Marahiel M. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 6.Jakubowski H, Goldman E. Microbiol Rev. 1992;56:412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. p. 212. [Google Scholar]
- 8.Cane D, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 9.Stachelhaus T, Schneider A, Marahiel M A. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 10.Gehring A M, Bradley K A, Walsh C T. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 11.Gehring A M, Mori I, Walsh C T. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 12.Neilands J B. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 13.Shaw-Reid C A, Kelleher N L, Losey H C, Gehring A M, Berg C, Walsh C T. Chem Biol. 1999;6:385–400. doi: 10.1016/S1074-5521(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning. A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C T. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.McCafferty D G, Lessard I A D, Walsh C T. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 18.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 19.Quadri L E N, Weinreb P H, Lei M, Nakano M M, Zuber P, Walsh C T. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 20.Suo Z, Walsh C T, Miller D A. Biochemistry. 1999;38:14023–14035. doi: 10.1021/bi991574n. [DOI] [PubMed] [Google Scholar]
- 21.Rusnak F, Faraci W S, Walsh C T. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 22.Webb M R. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring A M. Ph.D. thesis. Boston, MA: Harvard University; 1998. p. 330. [Google Scholar]
- 24.Conti E, Stachelhaus T, Marahiel M, Brick P. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieckmann R, Pavela-Vrancic M, von Dohren H, Kleinkauf H. J Mol Biol. 1999;288:129–140. doi: 10.1006/jmbi.1999.2671. [DOI] [PubMed] [Google Scholar]
- 26.Mootz H, Marahiel M. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreb P H, Quadri L E N, Walsh C T, Zuber P. Biochemistry. 1998;37:1575–1584. doi: 10.1021/bi9719859. [DOI] [PubMed] [Google Scholar]
- 28.Stachelhaus T, Huser A, Marahiel M A. Chem Biol. 1996;3:913–921. doi: 10.1016/s1074-5521(96)90180-5. [DOI] [PubMed] [Google Scholar]
- 29.Mootz H D, Marahiel M A. Curr Opin Biotechnol. 1999;10:341–348. doi: 10.1016/S0958-1669(99)80062-7. [DOI] [PubMed] [Google Scholar]
- 30.Du L, Shen B. Chem Biol. 1999;6:507–517. doi: 10.1016/S1074-5521(99)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson C R. Chem Rev. 1997;97:2525–2535. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]
- 32.Konz D, Klens A, Schorgendorfer K, Marahiel M A. Chem Biol. 1997;4:927–937. doi: 10.1016/s1074-5521(97)90301-x. [DOI] [PubMed] [Google Scholar]
- 33.Quadri L E, Sello J, Keating T A, Weinreb P H, Walsh C T. Chem Biol. 1998a;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 34.Konz D, Doekel S, Marahiel M A. J Bacteriol. 1999;181:133–140. doi: 10.1128/jb.181.1.133-140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehmann D E, Gehring A M, Walsh C T. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 36.Gehring A M, Mori I, Perry R, Walsh C T. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 37.Keating, T. A., Suo, Z., Ehmann, D. E. & Walsh, C. T. (2000) Biochemistry, in press. [DOI] [PubMed]
- 38.Jakubowski H. Biochemistry. 1998;37:5147–5153. doi: 10.1021/bi972528v. [DOI] [PubMed] [Google Scholar]
- 39.van Wageningen A M A, Kirkpatrick P N, Williams D H, Harris B R, Kershaw J K, Lennard N J, Jones S J, Solenberg P J. Chem Biol. 1998;5:155–162. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- 40.Guenzi E, Galli G, Grgurina I, Gross D, Grandi G. J Biol Chem. 1998;273:32857–32863. doi: 10.1074/jbc.273.49.32857. [DOI] [PubMed] [Google Scholar]