Abstract
P-glycoprotein (Pgp) is an ATP-dependent hydrophobic natural product anticancer drug efflux pump whose overexpression confers multidrug resistance to tumor cells. The work reported here deals with the elucidation of the energy requirement for substrate interaction with Pgp during the catalytic cycle. We show that the Kd (412 nM) of the substrate analogue [125I]iodoarylazidoprazoin for Pgp is not altered by the presence of the nonhydrolyzable nucleotide 5′-adenylylimididiphosphate and vanadate (Kd = 403 nM). Though binding of nucleotide per se does not affect interactions with the substrate, ATP hydrolysis results in a dramatic conformational change where the affinity of [125I]iodoarylazidoprazoin for Pgp trapped in transition-state conformation (Pgp⋅ADP⋅vanadate) is reduced >30-fold. To transform Pgp from this intermediate state of low affinity for substrate to the next catalytic cycle, i.e., a conformation that binds substrate with high affinity, requires conditions that permit ATP hydrolysis. Additionally, there is an inverse correlation (R2 = 0.96) between 8AzidoADP (or ADP) release and the recovery of substrate binding. These results suggest that the release of nucleotide is necessary for reactivation but not sufficient. The hydrolysis of additional molecule(s) of ATP (or 8AzidoATP) is obligatory for the catalytic cycle to advance to completion. These data are consistent with the observed stoichiometry of two ATP molecules hydrolyzed for the transport of every substrate molecule. Our data demonstrate two distinct roles for ATP hydrolysis in a single turnover of the catalytic cycle of Pgp, one in the transport of substrate and the other in effecting conformational changes to reset the pump for the next catalytic cycle.
The best-defined form of multiple drug resistance in human cells (1–3) is caused by the overexpression of P-glycoprotein (Pgp). The multidrug resistance phenotype is elicited by the extrusion of hydrophobic chemotherapeutic drugs by Pgp using the energy of ATP (2, 4). There is now considerable evidence of drug-stimulated ATPase activity in Pgp from diverse systems (5–8) suggesting that ATP hydrolysis and drug transport are intimately linked. Additionally, Pgp has the signature of the archetype ATP binding cassette transporter, namely, two transmembrane domains composed of six transmembrane putative α-helices each and two nucleotide binding sites (NBS). Senior and colleagues (4, 9–12) have proposed a scheme for the catalytic cycle of Pgp, according to which two NBS undergo ATP hydrolysis alternately. This model is based on the fact that vanadate (Vi) trapping of the nucleotide at either catalytic site arrests ATP hydrolysis at both sites and that mutations or chemical modifications that inactivate one catalytic site also prevent catalysis at the other site (13, 14).
Several reports have used the Vi-trapped intermediate (Pgp⋅ADP⋅Vi) of Pgp to assess the catalytic cycle (9, 10, 15–18). Incubation of Pgp with ATP and Vi inhibits Pgp ATPase by trapping ADP at the catalytic site, indicating that at least one turnover is required for onset of inhibition. Moreover, given the chemical analogy between Pi and Vi, the general consensus is that Pgp⋅ADP⋅Pi and Pgp⋅ADP⋅Vi complexes are equivalent (4, 9, 15), and that the transition-state Pgp⋅ADP⋅Vi represents an intermediate state during the normal reaction pathway (4).
Studies with both cultured cells and transient expression systems suggested that substrates interact directly with Pgp, and the most compelling evidence was provided by the use of diverse substrate analogs that photoaffinity label the Pgp (19–22). We demonstrate in this study that [125I]iodoarylazidoprazosin (IAAP), an analog of the Pgp substrate prazosin, exhibits saturation binding with a Kd of 412 nM, which is not affected by the binding of nucleotides, such as the nonhydrolyzable ATP analogue 5′-adenylylimidodiphosphate (AMPPNP). Though binding of nucleotide does not affect interactions with the substrate, ATP hydrolysis has a profound effect. Thus, if Vi is used to trap Pgp in the Pgp⋅ADP⋅Vi transition-state conformation, the affinity of IAAP for this complex is reduced >30-fold. We used this reduced affinity of the transition-state conformation as an assay to understand the link between ATP hydrolysis and the interaction of substrate with Pgp. Our results demonstrate that during ATP hydrolysis Pgp undergoes a conformational change as evidenced by a drastic decrease in the affinity for substrates. The initiation of the next catalytic cycle requires that Pgp attain the conformation that binds substrate with high affinity, and ATP hydrolysis appears to be mandatory for this switch from the transition state to occur. Thus, we demonstrate a requirement for ATP hydrolysis at two distinct steps in a single turnover during the catalytic cycle of Pgp.
Materials and Methods
Chemicals.
Cyclosporin A was purchased from Calbiochem. cis(Z)-Flupentixol was from Research Biochemicals (Natick, MA), and IAAP (2,200 Ci/mmol) was obtained from NEN. 8Azido[α-32P]ATP (15–20 Ci/mmol) and 8AzidoATP were purchased from ICN. All other chemicals were obtained from Sigma.
Preparation of Crude Membranes from High Five Insect Cells Infected with Recombinant Baculovirus Carrying the Human MDR1 Gene.
High Five insect cells (Invitrogen) were infected with the recombinant baculovirus carrying the human MDR1 cDNA with a 6-histidine tag at the C-terminal end [BV-MDR1 (H6)] as described (16). Crude membranes were prepared as described (7, 16).
Photoaffinity Labeling with IAAP.
The crude membranes (10–50 μg) were incubated with the drug or modulator for 3 min at room temperature in 50 mM Tris⋅HCl, pH 7.5, and IAAP (unless otherwise stated, 3–6 nM) was added and incubated for an additional 5 min under subdued light. The samples then were illuminated with a UV lamp assembly (PGC Scientifics, Gaithersburg, MD) fitted with two black light (self-filtering) UV-A long wave-F15T8BLB tubes (365 nm) for 10 min at room temperature (21–23°C). After SDS/PAGE on a 8% Tris-glycine gel at constant voltage, gels were dried and exposed to Bio-Max MR film (Eastman Kodak) at −70°C for 12–24 h. The radioactivity incorporated into the Pgp band was quantified by using the Storm 860 PhosphorImager system (Molecular Dynamics) and the software imagequant.
Determination of the Kd for IAAP.
Increasing concentrations of IAAP were prepared by adding the appropriate volume of stock IAAP, in 50% acetonitrile, and evaporating the solution under a steady stream of N2 in the dark. An equal volume (1 μl) of DMSO was added to each of the vials to dissolve the IAAP. Crude membranes (20 μg) in 40 μl Tris⋅HCl, 50 mM, pH 7.5 were added to the vials containing the indicated concentration of IAAP (1–519 nM). In parallel experiments, the membrane preparations were preincubated in 40 mM Mes⋅Tris (pH 6.8), 50 mM KCl, 5 mM sodium azide, 2 mM EGTA, and 10 mM MgCl2 containing 5 mM AMPPNP or 5 mM ATP and 250 μM Vi for 10 min at 37°C before the addition of IAAP. The samples were photocrosslinked at 365 nm and after SDS/PAGE were transferred to nitrocellulose paper as described (6). The Pgp bands were identified by exposing the nitrocellulose paper to a Bio-Max MR film and the protein on the nitrocellulose strips determined by the method of Schaffner and Weissmann (23). After estimating the protein, the nitrocellulose paper strips were placed in Biosafe II scintillation fluid and the radioactivity associated with Pgp was determined in a scintillation counter.
Vi-Induced ADP or 8AzidoADP Trapping in Pgp.
The Pgp⋅ADP⋅Vi or Pgp⋅8AzidoADP⋅Vi transition-state conformation was generated as described (16) with minor modifications. Crude membranes (1 mg/ml) were incubated in the ATPase assay buffer (40 mM Mes⋅Tris, pH 6.8/50 mM KCl/5 mM sodium azide/2 mM EGTA/2 mM DTT/10 mM MgCl2) containing 1.2 mM ATP or 8AzidoATP and 250 μM Vi in the dark at 37°C for 5 min. The reaction was stopped by adding 12.5 mM ice-cold ATP and placing the sample immediately on ice. For some experiments, excess nucleotides and Vi were removed by centrifugation at 300,000 × g at 4°C for 10 min by using a S120-AT2 rotor in a RC-M120EX micro-ultracentrifuge (Sorvall). In studies that required tracking the trapping of 8Azido[α-32P]ADP on Pgp, the same procedure as described above was followed except that 50 μM 8Azido[α-32P]ATP (3–5 μCi/nmol) was used.
ATPase Assay.
Vi-sensitive ATPase activity of Pgp in crude membranes was measured as described (5, 7, 16). Vi stock solution was prepared as described (24).
Results
The Vi-Induced ADP Trapped Conformation of Pgp During ATP Hydrolysis Exhibits a Marked Decrease in Affinity for Substrate.
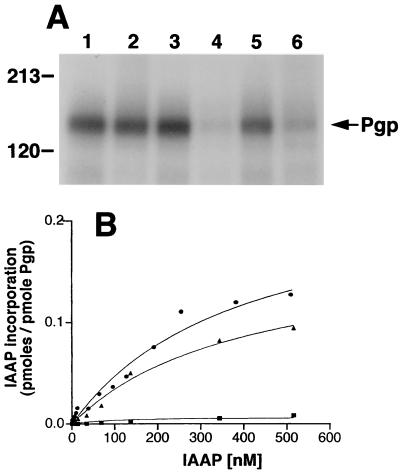
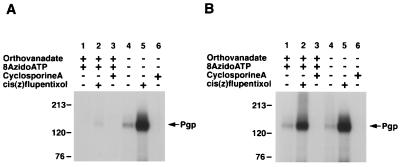
Ortho-Vi is a potent inhibitor of the ATPase activity of Pgp (6, 7), which acts by mimicking the pentacovalent phosphorus in the catalytic transition state, and traps the nucleotide tenaciously (9, 10). This transition-state conformation of Pgp is a useful tool, which synchronizes all molecules in a single conformation and, is sufficiently long-lived to study its functional characteristics. Recent studies have shown (16) that the Vi-trapped conformation of Pgp binds substrates with reduced affinity. The data in Fig. 1A shows that while nucleotides ATP and 8AzidoATP alone do not significantly affect the binding of IAAP to Pgp, the transition-state conformation of Pgp generated by Vi with either of these nucleotides results in a >80% decrease in the IAAP binding.
Figure 1.
(A) Substrate binding to Pgp in Vi-induced ADP trapped conformation. Crude membranes (20 μg protein) were labeled with 5 nM IAAP after pretreatment with ATP or 8AzidoATP in the presence or absence of Vi as described in Materials and Methods. Autoradiogram shows untreated Pgp (lane 1); Pgp pretreated at 37°C for 10 min with, 1.25 mM ATP (lane 2); 250 μM Vi (lane 3); 1.25 mM ATP and 250 μM Vi (lane 4); 1.25 mM 8AzidoATP (lane 5), or 1.25 mM 8AzidoATP and 250 μM Vi (lane 6). (B) Saturation binding of IAAP to Pgp in normal and transition-state conformation. Crude membranes (20 μg protein) were labeled with increasing concentrations (0.7–516 nM) of IAAP after pretreatment with AMPPNP or ATP in the presence of 250 μM Vi. Samples were untreated (●), treated with AMPPNP (▴), or treated with ATP (■) as described in Materials and Methods and then labeled with IAAP. The data were fitted by using the software graphpad prism 2.0 for the PowerPC MacIntosh and are representative of three independent experiments.
IAAP is a radiolabeled photoaffinity analogue of the Pgp substrate, prazosin, which specifically labels Pgp and can be competed by cyclosporin A (19, 25). IAAP incorporation into Pgp was determined as a function of substrate concentration (Fig. 1B) and it exhibited saturable binding. The single-site model of Henri-Michaelis-Menten best described the data (R2 = 0.986). This rapid equilibrium approach was deemed appropriate for determining the binding constants Kd and Bmax as the basic assumptions of the approach were satisfied: (a) The protein and ligand were allowed to interact in the dark whereby equilibrium conditions existed until the photocrosslinking step (considered analogous to termination of the reaction). (b) Sufficiently low concentrations of Pgp were used to ensure that the concentration of ligand was always in excess. (c) It is necessary to show that the binding is specific at all concentrations of the substrate. We have determined that cyclosporin A inhibits the photolabeling of Pgp with IAAP both at low and high concentrations (5 nM and 600 nM, respectively; unpublished observations).
To ensure that the decrease in affinity for IAAP (see Fig. 1B) is not a consequence of the binding of nucleotide but of ATP hydrolysis, we measured IAAP binding in the presence of AMPPNP, a nonhydrolyzable analogue of ATP, and Vi. The data for the untreated sample and that treated with the nonhydrolyzable ATP analogue AMPPNP and Vi fit well to curves with a single binding site, demonstrating similar binding constants (Kd of 412 ± 91 nM and 403 ± 117 nM, respectively). These results suggest that ATP hydrolysis and not nucleotide binding is a prerequisite for the large decrease in substrate binding. Our major aim in this study is to determine whether this decrease in affinity of Pgp in the transition-state conformation for substrate is reversible, and if so, under what conditions.
Characterization of the Pgp⋅ADP⋅Vi Transition-State Conformation of Pgp.
Earlier studies of Senior and coworkers (9–11) showed that the t1/2 for reactivation of ATP hydrolysis with the Chinese hamster Pgp in the ADP⋅Vi transition-state complex is 84 min at 37°C. We also obtained comparable results with human Pgp (data not shown). In addition, we found that incubation of Pgp at 37°C for an extended period results in greatly reduced IAAP binding (data not given). When 8AzidoATP was used instead of ATP, the t1/2 for reactivation of the Pgp⋅8AzidoADP⋅Vi complex was found to be of the order of 5–8 min for both Chinese hamster (11) and human Pgp (data not shown). The inhibition of substrate binding to the Pgp⋅ADP⋅Vi or Pgp⋅8AzidoADP⋅Vi complex is comparable (Fig. 1A), and 8AzidoATP is a good hydrolysis substrate for both Chinese hamster and human Pgp, with a Km similar to ATP (26, 27) (K. Kerr, Z.E.S, and S.V.A, unpublished work). We therefore chose to use the Pgp⋅8AzidoADP⋅Vi complex in lieu of the Pgp⋅ADP⋅Vi complex for further studies.
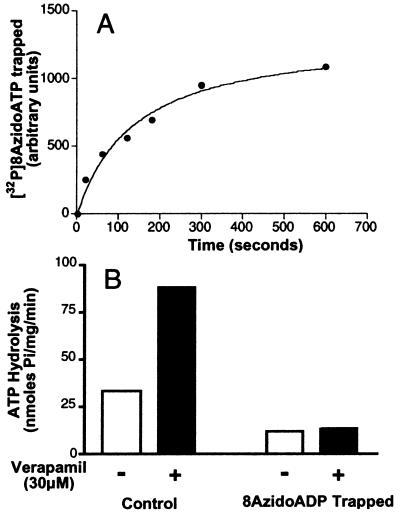
The rate of formation of the Pgp⋅8AzidoADP⋅Vi complex is shown in Fig. 2A. Incorporation of 8Azido[α-32P]ADP into Pgp is saturable (t1/2, 2.3 min). These results obtained for the recombinant human Pgp expressed in insect cells are concordant with those reported for Chinese hamster Pgp (9, 10, 28). On the basis of these results, we allowed Vi-induced 8AzidoADP trapping to occur and monitored the recovery of drug binding over 15 min after removal of excess nucleotide and Vi by centrifugation. As demonstrated above (Fig. 1A) incubation of Pgp with either ATP or 8AzidoATP in the presence of Vi results in a decrease in IAAP binding. However, it is important to demonstrate that the basal and substrate-stimulated ATPase activity of Pgp⋅8AzidoADP⋅Vi complex also is inhibited as observed previously for the Pgp⋅ADP⋅Vi complex (4, 11). Fig. 2B shows that both basal and verapamil-stimulated ATPase activity of Pgp is inhibited in the 8AzidoADP⋅Vi-trapped conformation. These results suggest that the intermediates generated by ATP or 8AzidoATP are comparable and the observation that there is decreased affinity for IAAP reflects a change in the affinity for substrate irrespective of its nature, which is a consequence of ATP hydrolysis. Moreover, it has been demonstrated recently that IAAP is transported by Pgp (S. Dey and M. M. Gottesman, personal communication).
Figure 2.
(A) Time course of Vi-induced trapping of 8Azido[α-32P]ADP on Pgp. Crude membranes (protein, 1 mg/ml) were incubated in the dark at 37°C with 50 μM 8Azido[α-32P]ATP (3–5 μCi/nmol) and 250 μM Vi in the ATPase assay buffer (see Materials and Methods). Aliquots were removed at indicated time points, reaction was terminated, and the samples were photocrosslinked by UV irradiation at 365 nm. After SDS/PAGE, radioactivity in the Pgp band was estimated by PhosphorImager analysis. (B) Basal and verapamil-stimulated ATP hydrolysis by Pgp before and after Vi trapping. ATPase activity of Pgp in crude membranes was measured by the endpoint Pi assay as described in Materials and Methods. Basal- (□) and verapamil- (30 μM) stimulated (■) ATPase was measured in control membranes and those that had been trapped into the Pgp⋅8AzidoADP⋅Vi complex, by incubation for 10 min with 1.25 mM 8AzidoATP and 250 μM Vi. Both control and Vi-trapped membranes were centrifuged to remove excess nucleotides and Vi before assaying the ATPase activity.
Correlation Between the Disassociation of 8AzidoADP and Recovery of the Binding of IAAP After Trapping of Pgp with 8AzidoADP and Vi.
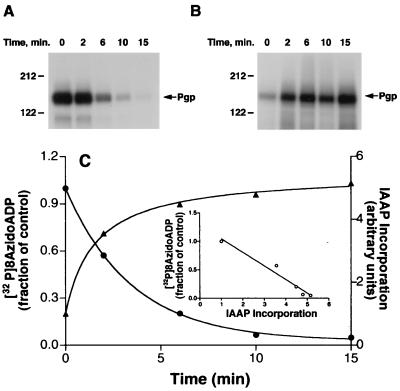
To understand how Pgp advances from the transition state (Pgp⋅8AzidoADP⋅Vi), wherein its affinity for substrate is acutely reduced, to the next catalytic cycle, we determined in parallel the rate at which 8AzidoADP disassociates from the Pgp⋅8AzidoADP⋅Vi complex and the recovery of binding of IAAP. Fig. 3A shows that the amount of 8Azido[α-32P]ADP incorporated into Pgp decreases over time when the Pgp⋅8Azido[α-32P]ADP⋅Vi complex is incubated at 37°C. There is a concomitant increase in the incorporation of IAAP into Pgp in the presence of ATP after washing off excess 8AzidoATP and Vi (Fig. 3B). Fig. 3C depicts a quantification of the data given in Fig. 3 A and B. The disassociation of 8AzidoADP shows an attendant and equivalent increase in the incorporation of IAAP. The observation that the t1/2 for the release of the trapped nucleotide and for the reactivation of IAAP incorporation are commensurate is consistent with the fact that the release of 8AzidoADP from the Pgp⋅8AzidoADP⋅Vi complex is very slow (4). Although, one or several steps between 8AzidoADP release and binding of substrate may exist, a much slower rate of 8AzidoADP release compared with other steps in the catalytic cycle would result in an overall rate comparable to the rate at which 8AzidoADP is displaced (see Fig. 3C). Although the experiments depicted in Fig. 3 indicate that the release of 8AzidoADP (or ADP) is necessary for the recovery of IAAP binding after trapping, they do not demonstrate unequivocally that the release of occluded nucleotide is sufficient.
Figure 3.
Relationship between the disassociation of 8AzidoADP and recovery of IAAP binding after Vi trapping. (A) Disassociation of 8Azido[α-32P]ADP from the Pgp⋅8Azido-[α-32P]ATP⋅Vi complex of Pgp. Crude membranes (protein, 1 mg/ml) were incubated in the dark at 37°C for 10 min with 50 μM 8Azido[α-32P]ATP (3–5 μCi/nmol) and 250 μM Vi in the ATPase assay buffer. The reaction was stopped by adding 12.5 mM ice-cold ATP and placing the tubes on ice. Untrapped nucleotides and excess Vi were removed by centrifugation at 300,000 × g for 10 min and the membranes were resuspended in the ATPase assay buffer. The resuspended membranes were placed at 37°C. Aliquots were removed at indicated intervals and placed on ice and photocrosslinked by UV irradiation at 365 nm for 5 min. (B) Recovery of IAAP binding to Pgp subsequent to formation of transition-state conformation. Crude membranes (1 mg/ml protein) were treated with 1.25 mM 8AzidoATP and 250 μM Vi for 10 min at 37°C in the ATPase assay buffer. The samples were placed on ice, and untrapped nucleotides and excess Vi were removed by centrifugation at 300,000 × g for 10 min at 4°C. The pellet was resuspended in the ATPase assay buffer containing 1.2 mM ATP and incubated at 37°C. Aliquots were transferred to ice at the indicated intervals and photolabeled with IAAP and visualized as described in Materials and Methods. Autoradiogram is shown in both A and B. (C) Relationship between the disassociation of 8AzidoADP and the recovery of IAAP binding after Vi trapping. The radioactivity associated with the Pgp bands depicted in A and B was quantified by using the Storm 860 PhosphorImager system. Disassociation of 8AzidoADP (●) and the increase in IAAP binding (▴) after Vi trapping were plotted as a function of time. (Inset) The increase in IAAP labeling, on the x-axis vs. the disassociation of 8Azido[α-32P]ADP on the y-axis, these are inversely proportional to each other with R2 = 0.959. The data are representative of three independent experiments.
Recovery of IAAP Binding to Pgp After Vi-Induced 8AzidoADP Trapping Requires ATP Hydrolysis.
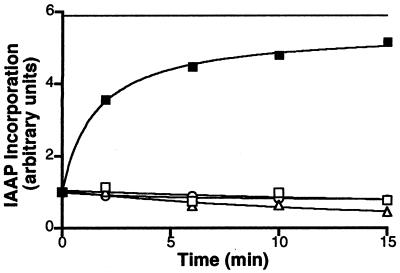
Generating the Pgp⋅8AzidoADP⋅Vi complex, washing off unbound 8AzidoATP and free Vi, and then incubating under different conditions followed by labeling with IAAP, permits us to sharply discriminate between the conditions that do and do not allow the recovery of IAAP incorporation into Pgp after formation of the 8AzidoADP-trapped intermediate. The data given in Fig. 4 demonstrate that ATP hydrolysis is essential for a recovery of IAAP incorporation into Pgp. Incubating the Pgp⋅8AzidoADP⋅Vi complex in Mg2+-free medium containing EDTA, replacing the 8AzidoATP with the nonhydrolyzable AMPPNP and the continued presence of Vi are all conditions that do not permit ATP hydrolysis, and there is no recovery of IAAP binding (Fig. 4). However, allowing the complex to hydrolyze ATP or 8AzidoATP at 37°C permits incorporation of IAAP at >80% of the levels observed in Pgp not trapped into the transition-state conformation. It is possible that ATP hydrolysis is required for disassociating 8AzidoADP (or ADP) from the complex rather than for affecting the conformational change that allows the molecule to bind substrate. Our recent observations, however, show that the rate of release of 8AzidoADP is the same in control membranes and those treated with AMPPNP or ATP in the presence of Mg+2 (data not given), demonstrating that nucleotide binding or its hydrolysis is not required for the disassociation of 8AzidoADP from the Pgp⋅8AzidoADP⋅Vi complex.
Figure 4.
Recovery of IAAP binding to the Pgp⋅8AzidoADP⋅Vi complex requires ATP hydrolysis. Crude membranes (1 mg/ml protein) were treated with 1.25 mM 8AzidoATP and 250 μM Vi for 10 min at 37°C in the ATPase assay buffer (see Materials and Methods). The reaction was stopped by placing the samples on ice. Excess 8AzidoATP and Vi were removed by centrifugation at 300,000 × g for 10 min at 4°C. The pellet was resuspended in 40 mM Mes⋅Tris, pH6.8, 50 mM KCl, 5 mM sodium azide, 2 mM EGTA, and 2 mM DTT and divided into five aliquots to which the following additions were made: 10 mM MgCl2 and 5 mM AMPPNP (▵), 10 mM MgCl2, and 1.2 mM ATP (■), 5 mM EDTA (□), and 250 μM Vi (○). The continuous horizontal line shows the extent of IAAP incorporated into an equivalent amount of untreated control membranes, which were processed in parallel. The samples were incubated at 37°C, and aliquots were removed at the indicated intervals, placed on ice, and photocrosslinked with IAAP. After SDS/PAGE, the radioactivity associated with the Pgp was estimated by PhosphorImager analyses and normalized such that the radioactivity in the untreated sample at 0 min was 1. The data are representative of three independent experiments.
Characteristics of the IAAP Binding to Pgp After Trapping with 8AzidoADP⋅Vi Are Similar to Those Observed Before Nucleotide Trapping.
The results presented above show that the Pgp⋅8AzidoADP⋅Vi complex exhibits decreased affinity for substrate, which can be restored under ATP hydrolysis condition. Fig. 5A shows that not only does the Pgp⋅8AzidoADP⋅Vi complex show reduced substrate binding but the stimulation of IAAP binding by cis(Z)-flupentixol also is abolished. It has been shown (25, 29) that cis(Z)-flupentixol stimulates the binding of IAAP to Pgp, which appears to be allosteric in nature involving interactions with Pgp at a site distinct from the substrate-binding domain (25, 30). We used this stimulation of IAAP labeling as a diagnostic test of the “native” conformation of Pgp as it is acutely sensitive to even small perturbations in the structure of Pgp (30). As stated above, with Pgp in the Pgp⋅8AzidoADP⋅Vi-trapped conformation there is no stimulation of IAAP binding by cis(Z)-flupentixol. One would, however, expect that if the reactivation of IAAP binding to Pgp represents its native conformation (i.e., pre-ATP hydrolysis state), the reactivated molecule would not only exhibit IAAP binding but this binding would be stimulated by cis(Z)-flupentixol. The data in Fig. 5B show IAAP binding to the Pgp⋅8AzidoADP⋅Vi complex that has been incubated at 37°C for 15 min to allow release of 8AzidoADP and an additional round of ATP hydrolysis. It is clear that IAAP binding is regained and it is stimulated by cis(Z)-flupentixol and inhibited by cyclosporin A. Furthermore, verapamil-stimulated ATPase activity, which is inhibited in the Pgp⋅8AzidoADP⋅Vi complex (Fig. 2B), also is recovered after incubation at 37°C for 15 min (data not given). These results suggest that Pgp, after the transition state, is in a conformation that is functionally similar to the one before being trapped with 8AzidoADP and Vi.
Figure 5.
Effect of cyclosporin A and cis(Z)-flupentixol on the IAAP labeling of Pgp in the Vi-trapped and native conformations. (A) The effect of cyclosporin A and cis(Z)-flupentixol on the IAAP labeling of control and Pgp treated with 1.2 mM 8AzidoATP and 250 μM Vi at 37°C for 10 min immediately after trapping. After incubation at 37°C, the samples were centrifuged at 300,000 × g for 10 min at 4°C and resuspended in ice-cold ATPase assay buffer. The samples then were immediately incubated with cyclosporin A (1 μM) or cis(Z)-flupentixol (25 μM) for 3 min at room temperature, with IAAP (5 nM) for an additional 3 min, and then photocrosslinked as described in Materials and Methods. (B) Same experimental conditions as A except that the samples were incubated with 1.2 mM ATP for 15 min at 37°C before treatment with cyclosporin A or cis(Z)-flupentixol and labeling with IAAP. The various treatments are given above each autoradiogram.
Discussion
Pgp is a robust drug efflux pump and its ATP hydrolytic activity, which is inhibited by Vi, is obligatorily linked to drug transport. Previous studies demonstrated reduced binding of the photoaffinity substrate analogue IAAP to Pgp in the Vi-trapped conformation (16, 25). Several reports (9, 10, 12, 15, 17) have used the transition-state (Pgp⋅8AzidoADP⋅Vi) intermediate to gain information about various steps in the ATP hydrolysis by Pgp. Although these studies have proved invaluable in understanding the ATP hydrolysis segment of the catalytic cycle of Pgp, they have not directly addressed the link between ATP hydrolysis and substrate-binding sites. There is, however, a general consensus in the literature that Pgp is an energy-driven pump, that this energy is provided by the hydrolysis of ATP, and that there is extensive communication between the substrate-binding sites and NBS of Pgp (reviewed in ref. 3). In this study, we exploited the reduced affinity of substrate for Pgp in the Vi-trapped conformation to investigate the interactions between the NBS and substrate binding sites and their energetic requirements to elucidate the catalytic cycle of Pgp.
We show that this reduced binding of IAAP occurs regardless of whether the Vi-trapped conformation is generated by using ATP or 8AzidoATP, which also is hydrolyzed by Pgp (11). Moreover, nucleotide binding alone is not sufficient to affect affinity for substrate as both ATP and 8AzidoATP in the absence of Vi do not significantly reduce the binding of IAAP to Pgp (Fig. 1A). It is thus clear that ATP hydrolysis induces a conformational change at the substrate-binding site. To quantify the magnitude of this change, we determined the binding constants for IAAP (Fig. 1B). The Kd for IAAP binding to Pgp (412 ± 91 nM), which was not affected by the presence of the nonhydrolyzable nucleotide AMPPNP and Vi (Kd = 403 ± 117). However, the IAAP binding to the Vi-trapped Pgp does not begin to show saturation even at 516 nM IAAP (because of unavailability of nonradioactive IAAP, it was not feasible to use higher concentrations). We nevertheless can estimate from this data that the affinity for IAAP is decreased at least 30-fold.
Using the Pgp⋅8AzidoADP⋅Vi conformation of Pgp, we show that the binding of IAAP can be restored to levels comparable to those obtained in the absence of Vi trapping. Fig. 5 illustrates that after incubation for 15 min at 37°C under ATP hydrolysis conditions, after trapping and removal of excess Vi and nucleotide, not only is binding of IAAP restored but so are functional characteristics such as stimulation by cis(Z)-flupentixol, inhibition by cyclosporin A and also verapamil-stimulated ATPase activity. The noteworthy features of this reversal are, however, revealed in Figs. 3 and 4. IAAP binding is not restored when the trapped Pgp is resuspended in either Mg2+ free medium containing EDTA, or in a medium containing excess AMPPNP. These data strongly suggest that not only is ATP (or 8AzidoATP) hydrolysis required for the Pgp molecule to be trapped into the Pgp⋅8AzidoADP⋅Vi conformation, but that additional ATP hydrolysis at a subsequent step is required for the molecule to be restored to its original conformation (here defined as the conformation that will bind substrate with similar properties as at the pre-ATP hydrolysis state). This observation raises the question of whether hydrolysis is required for the disassociation of the 8AzidoADP from the Pgp⋅8AzidoADP⋅Vi complex or it occurs at a subsequent step. It is unlikely that ATP hydrolysis occurs when the molecule is locked in the Pgp⋅8AzidoADP⋅Vi conformation (see Introduction). Previous work, in which ATPase activity of Pgp was restored after inhibition by Vi in the presence of ATP or ADP, does not suggest any energetic requirement at this stage (10, 11). Additionally, we found that there is a progressive disassociation of 8Azido[α-32P]ADP from the complex with a t1/2 of 2–3 min, which is not affected by the presence of nucleotides (data not given). As ATP hydrolysis is clearly not a requirement for disassociation of ADP, such a requirement must come at a step further in the cycle. The inset to Fig. 3C demonstrates a close but inverse correlation between 8AzidoADP release and the recovery of substrate binding. The data in Fig. 4 nonetheless show that while the release of nucleotide is a necessary precondition for the binding of substrate in the reactivation pathway it is not a sufficient condition. The hydrolysis of an additional molecule(s) of nucleotide is essential for the catalytic cycle to advance to completion.
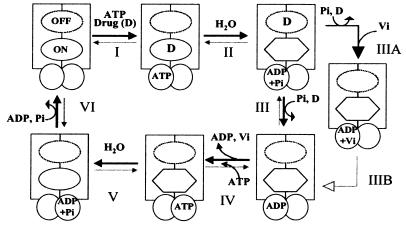
On the basis of these data and published work (4, 25), we propose a scheme for the catalytic cycle for Pgp as depicted in Fig. 6. The drug and ATP first bind to Pgp (step I), there being no energetic requirement (Fig. 1A). The studies of Liu and Sharom (31) also demonstrate, by using the fluorescent probe 2-(4-maleimidoanilino)naphthalene-6-sulfonic acid, that prior binding of ATP is not essential for drug interaction with Pgp. Thus, ATP binding could precede, follow, or accompany the binding of drug. The hydrolysis of ATP (or 8AzidoATP) and the subsequent release of Pi that generates the Pgp⋅ADP⋅Vi complex is accompanied by a conformational change that (possibly besides other effects) drastically reduces the affinity of substrate for Pgp (compare Figs. 1 and 5). It previously has been postulated that ATP hydrolysis in Pgp generates a conformation of high chemical potential. The relaxation of such a conformation could be coupled to the movement of drug binding site from an aspect of high affinity to one of low affinity (4). Experimental evidence presented here strongly supports such a notion, and plausibly the drug is extruded at this step. The extrusion of drug and the subsequent (or accompanying) disassociation of ADP (step IV) are both necessary but not sufficient for Pgp to regain its “original” conformation (Figs. 3 and 4). An additional molecule(s) of ATP (or an alternative hydrolyzable nucleotide) must be hydrolyzed (step V) to restore the conformation of Pgp to initiate the next cycle (Figs. 3–5).
Figure 6.
A proposed scheme for the catalytic cycle of Pgp. The rectangles represent the N and C halves of Pgp and the overlapping circles the two NBS. The ovals represent the substrate binding sites, the on site is depicted by the continuous perimeter, and the off site with a dotted line (see Discussion for an explanation of the on and off sites). The hexagon portrays the on site with reduced affinity for the drug. Step I: Substrate binds to the on site of Pgp, and ATP binds to one or both of the two NBS. Step II: ATP is hydrolyzed and the drug is possibly moved to the lower affinity off site. Step III: Pi is released and the drug extruded from Pgp at this step. Step IIIA: When Pi is replaced by Vi, the Pgp⋅ADP⋅Vi complex is generated, which also exhibits a reduced affinity for substrate. Step IV/step IIIB: The ADP is disassociated; however, the affinity for substrate continues to be low. Step V: After disassociation of the ADP, an additional molecule(s) of ATP is hydrolyzed and the conformation of Pgp is restored to its original state with high affinity for substrate binding. Step VI: The release of Pi, disassociation of ADP followed by the initiation of the next catalytic cycle. It is not known, however, whether release of ADP necessarily precedes the binding of the next molecule of substrate after step VI. ADP disassociation (from the previous cycle), drug binding, and ATP binding presumably could occur in any sequence in steps VI to I. Though, we depict ATP binding and hydrolysis as occurring in the N half of Pgp our data do not suggest any preference in the presence of saturating concentrations of ATP or 8AzidoATP for either of the two sites, and the process is most likely random. However consistent with the alternate site model (4), ATP hydrolysis would occur at a site different from that in step II and therefore is depicted at the NBS in the C half at step V.
This model suggests two unique and distinct roles for ATP hydrolysis in a single turnover of the catalytic cycle of Pgp. Not only is energy used in the transport of substrate but there is a clear need for ATP hydrolysis in effecting conformational changes in the molecule that make it available for the next catalytic cycle. It is worth noting that a working model for the catalytic cycle of another ATP binding cassette transporter, CFTR (cystic fibrosis transmembrane conductance regulator), has been proposed (32, 33). According to this model, a single closed-open-closed gating cycle of phosphorylated CFTR chloride channel involves hydrolysis of one ATP molecule to open it, and hydrolysis of a second ATP to close it. Thus, to complete one cycle in CFTR, similar to Pgp (see below), requires hydrolysis of two ATP molecules. Conformational changes in proteins dependent on ATP hydrolysis also have been demonstrated in other systems, e.g., the coupling of ATP hydrolysis to the unfolding of the misfolded substrate protein by the chaperonin GroEL (34, 35). Here, the hydrolysis of ATP switches the trans ring from a collapsed to an open state signaling the beginning of the next round of substrate recognition (36, 37). In recent years, the stoichiometry of number of ATP molecules hydrolyzed to substrate molecules transported has been estimated for Pgp and other ATP binding cassette transporters such as histidine permease and oligopeptide permease. It has been demonstrated that the hydrolysis of at least two molecules of ATP is required for the transport of every molecule of substrate (38–41). The stoichiometry data suggest that the hydrolysis of one ATP molecule is required to move drug from the high-affinity “on” site to the low-affinity “off” site, while the second ATP may be used either to move drug from the off site to the extracellular medium or to reset the conformation of Pgp to initiate the next cycle (18) (Fig. 6). Our data provide experimental evidence, consistent with the stoichiometry measurements, for the role for ATP hydrolysis at two distinct steps (steps II and V, Fig. 6) during the catalytic cycle. Moreover, because of common features such as substrate-stimulated ATPase activity and its inhibition by Vi, as also the stoichiometry of substrate transport and ATP hydrolysis, we suggest that this requirement for additional hydrolysis of ATP to reset the conformation during a single turnover may be common to other ATP binding cassette transporters and further work is needed to verify this.
Acknowledgments
We thank Drs. M. M. Gottesman and I. Pastan for discussions and encouragement and C. Booth, S. Dey, J. Gribar, C. Hrycyna, and K. Kerr for comments on the manuscript.
Abbreviations
- AMPPNP
5′-adenylylimidodiphosphate
- IAAP
[125I]iodoarylazidoprazosin
- NBS
nucleotide binding site
- Pgp
P-glycoprotein
- Vi
vanadate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gottesman M M, Pastan I. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman M M, Pastan I, Ambudkar S V. Curr Opin Genet Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- 3.Ambudkar S V, Dey S, Hrycyna C A, Ramachandra M, Pastan I, Gottesman M M. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 4.Senior A E, Al-Shawi M K, Urbatsch I L. FEBS Lett. 1995;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- 5.Ambudkar S V. Methods Enzymol. 1998;292:504–514. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- 6.Ambudkar S V, Lelong I H, Zhang J, Cardarelli C O, Gottesman M M, Pastan I. Proc Natl Acad Sci USA. 1992;89:8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkadi B, Price E M, Boucher R C, Germann U A, Scarborough G A. J Biol Chem. 1992;267:4854–4858. [PubMed] [Google Scholar]
- 8.Sharom F J, Yu X, Doige C A. J Biol Chem. 1993;268:24197–24202. [PubMed] [Google Scholar]
- 9.Urbatsch I L, Sankaran B, Weber J, Senior A E. J Biol Chem. 1995;270:19383–19390. doi: 10.1074/jbc.270.33.19383. [DOI] [PubMed] [Google Scholar]
- 10.Urbatsch I L, Sankaran B, Bhagat S, Senior A E. J Biol Chem. 1995;270:26956–26961. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- 11.Sankaran B, Bhagat S, Senior A E. Arch Biochem Biophys. 1997;341:160–169. doi: 10.1006/abbi.1997.9944. [DOI] [PubMed] [Google Scholar]
- 12.Senior A E, Bhagat S. Biochemistry. 1998;37:831–836. doi: 10.1021/bi9719962. [DOI] [PubMed] [Google Scholar]
- 13.Loo T W, Clarke D M. J Biol Chem. 1995;270:21449–21452. doi: 10.1074/jbc.270.37.21449. [DOI] [PubMed] [Google Scholar]
- 14.Urbatsch I L, Beaudet L, Carrier I, Gros P. Biochemistry. 1998;37:4592–4602. doi: 10.1021/bi9728001. [DOI] [PubMed] [Google Scholar]
- 15.Hrycyna C A, Ramachandra M, Ambudkar S V, Ko Y H, Pedersen P L, Pastan I, Gottesman M M. J Biol Chem. 1998;273:16631–16634. doi: 10.1074/jbc.273.27.16631. [DOI] [PubMed] [Google Scholar]
- 16.Ramachandra M, Ambudkar S V, Chen D, Hrycyna C A, Dey S, Gottesman M M, Pastan I. Biochemistry. 1998;37:5010–5019. doi: 10.1021/bi973045u. [DOI] [PubMed] [Google Scholar]
- 17.Szabo K, Welker E, Bakos, Muller M, Roninson I, Varadi A, Sarkadi B. J Biol Chem. 1998;273:10132–10138. doi: 10.1074/jbc.273.17.10132. [DOI] [PubMed] [Google Scholar]
- 18.Hrycyna C A, Ramachandra M, Germann U A, Cheng P W, Pastan I, Gottesman M M. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- 19.Greenberger L M. J Biol Chem. 1993;268:11417–11425. [PubMed] [Google Scholar]
- 20.Demmer A, Thole H, Kubesch P, Brandt T, Raida M, Fislage R, Tummler B. J Biol Chem. 1997;272:20913–20919. doi: 10.1074/jbc.272.33.20913. [DOI] [PubMed] [Google Scholar]
- 21.Bruggemann E P, Germann U A, Gottesman M M, Pastan I. J Biol Chem. 1989;264:15483–15488. [PubMed] [Google Scholar]
- 22.Morris D I, Greenberger L M, Bruggemann E P, Cardarelli C, Gottesman M M, Pastan I, Seamon K B. Mol Pharmacol. 1994;46:329–337. [PubMed] [Google Scholar]
- 23.Schaffner W, Weissmann C. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 24.Dey S, Ramachandra M, Pastan I, Gottesman M M, Ambudkar S V. Methods Enzymol. 1998;292:318–328. doi: 10.1016/s0076-6879(98)92025-0. [DOI] [PubMed] [Google Scholar]
- 25.Dey S, Ramachandra M, Pastan I, Gottesman M M, Ambudkar S V. Proc Natl Acad Sci USA. 1997;94:10594–10599. doi: 10.1073/pnas.94.20.10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urbatsch I L, Al-Shawi M K, Senior A E. Biochemistry. 1994;33:7069–7076. doi: 10.1021/bi00189a008. [DOI] [PubMed] [Google Scholar]
- 27.Al-Shawi M K, Urbatsch I L, Senior A E. J Biol Chem. 1994;269:8986–8992. [PubMed] [Google Scholar]
- 28.Sankaran B, Bhagat S, Senior A E. Biochemistry. 1997;36:6847–6853. doi: 10.1021/bi970034s. [DOI] [PubMed] [Google Scholar]
- 29.Safa A R, Agresti M, Bryk D, Tamai I. Biochemistry. 1994;33:256–265. doi: 10.1021/bi00167a034. [DOI] [PubMed] [Google Scholar]
- 30.Dey S, Hafkemeyer P, Pastan I, Gottesman M M. Biochemistry. 1999;38:6630–6639. doi: 10.1021/bi983038l. [DOI] [PubMed] [Google Scholar]
- 31.Liu R, Sharom F J. Biochemistry. 1996;35:11865–11873. doi: 10.1021/bi960823u. [DOI] [PubMed] [Google Scholar]
- 32.Senior A E, Gadsby D C. Semin Cancer Biol. 1997;8:143–150. doi: 10.1006/scbi.1997.0065. [DOI] [PubMed] [Google Scholar]
- 33.Gadsby D C, Dousmanis A G, Nairn A C. Acta Physiol Scand Suppl. 1998;643:247–256. [PubMed] [Google Scholar]
- 34.Radford S E, Dobson C M, Evans P A. Nature (London) 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 35.Sosnick T R, Mayne L, Hiller R, Englander S W. Nat Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 36.Shtilerman M, Lorimer G H, Englander S W. Science. 1999;284:822–825. doi: 10.1126/science.284.5415.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rye H S, Roseman A M, Chen S, Furtak K, Fenton W A, Saibil H R, Horwich A L. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 38.Ambudkar S V, Cardarelli C O, Pashinsky I, Stein W D. J Biol Chem. 1997;272:21160–21166. doi: 10.1074/jbc.272.34.21160. [DOI] [PubMed] [Google Scholar]
- 39.Liu C E, Liu P Q, Ames G F L. J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- 40.Mimmack M L, Gallagher M P, Pearce S R, Hyde S C, Booth I R, Higgins C F. Proc Natl Acad Sci USA. 1989;86:8257–8261. doi: 10.1073/pnas.86.21.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro A B, Ling V. Eur J Biochem. 1998;254:189–193. doi: 10.1046/j.1432-1327.1998.2540189.x. [DOI] [PubMed] [Google Scholar]