Abstract
Using a strategy consisting of (i) the isolation of cell walls from synchronously differentiating cells of Zinnia, (ii) the generation of mAbs with an antibody phage display method, and (iii) screening with a subtraction method, we isolated mAbs recognizing vascular development-specific cell wall components without prior antigen identification. One of the isolated mAbs, designated CN 8, recognized a cell wall component contained in the hemicellulosic fraction. Immunohistochemical analyses showed that the CN 8 epitope was localized to the cell wall of immature tracheary elements and xylem parenchyma cells. In immature tracheary elements, the CN 8 epitope had a polarized localization pattern regardless of whether the cells are formed as parts of vessels in situ or as single tracheary elements in vitro, suggesting that cell polarity autonomously formed on the cell wall may function in tracheary element differentiation.
The walls of plant cells are supermolecular structures, comprising polysaccharides, proteins, and phenolic compounds (1, 2). Recent studies have suggested that structure and composition of the cell wall change dynamically during plant development, which makes it likely that these changes play essential roles in development (3). However, it is difficult to monitor a change in a specific component of the cell wall at the molecular level during development. Wall components are embedded in the matrix of the wall and difficult to isolate. Therefore, the interrelationship between changes in specific wall components and developmental events in plant has not been fully understood.
Use of antibodies raised against some wall components may overcome the difficulty of monitoring the wall components during development (4). Indeed, mAbs raised against some wall components have revealed dynamic changes in the wall components coupled with developmental events (5–9). However, it is still hard to isolate antibodies against ill-defined wall components that mark specific developmental stages of cells. To isolate such mAbs, we devised a strategy consisting of the following procedures: (i) using a cell culture system that allows us to define the specific stages of cell differentiation and to prepare the homogeneous walls from differentiating cells, (ii) making an antibody library to cell walls by using phage display technology, and (iii) screening differentiating cell-specific mAbs by using a subtractive procedure.
Single mesophyll cells isolated from Zinnia elegans transdifferentiate into tracheary elements (TEs) when cultured in a medium containing auxin and cytokinin (10). This in vitro differentiation is found to mimic vascular differentiation in plant (11). Because it occurs synchronously and at high frequency, we can isolate a relatively large amount of cells that are at the same stage of differentiation. We tried to isolate cell wall fractions from differentiating cells that had not yet developed morphological features unique to TEs and then to prepare mAbs against the cell wall at the early stage of differentiation.
To isolate the mAbs, we adopted an antibody phage display method (12) coupled with a subtractive procedure. The reason is that the number of developmental stage-specific epitopes in the cell walls may be limited; however, the number of nonspecific epitopes in the cell walls may be much larger, and therefore we needed a method allowing large scale generation of mAbs and an efficient screening. Indeed, the phage display subtraction method has been used successfully for the isolation of blood cell-specific (13, 14) and tumor-specific (15) mAbs. Using this procedure, we successfully isolated differentiation-specific phage antibodies (ΦAbs), one of which recognized a cell wall component localized in a tip of immature TEs.
Materials and Methods
Cell Wall Preparation.
Zinnia mesophyll cell culture was performed with the method detailed in ref. 10, modified as described in ref. 16. The culture medium containing 0.1 mg/liter 1-naphthaleneacetic acid (NAA) and 0.2 mg/liter benzyladenine (BA) was used for TE induction (17). Mesophyll cells isolated from Zinnia elegans seedlings differentiated into TEs in which secondary walls were thickened when cultured for more than 48 h in the TE-inductive medium (Fig. 1). To isolate antibodies that recognize the cell wall components that appear before secondary walls are thickened, the cell wall was prepared after 42 h of culture. The cell wall was designated “pre-TE wall” and used as antigen for later procedures. Not only differentiation but also cell division is induced in a liquid medium containing NAA and BA (Fig. 1A). The control cell wall (control wall) was therefore prepared from the cell that had been cultured for 42 h in a control medium containing only 0.1 mg/liter NAA in which only cell division was induced (Fig. 1A). Cultured cells were collected by centrifugation and resuspended in equal volumes of ice-cold 0.5 M Hepes buffer (pH 7.5) containing 10 mM DTT. The cells were sonicated until their chloroplasts were flaked away and then centrifuged at 500 × g for 1 min. The precipitate was washed several times with 10 volumes of ice-cold 50 mM Hepes buffer (pH 7.5) containing 1 mM DTT and 1% (wt/vol) Triton X-100 and then with ice-cold 50 mM Hepes buffer (pH 7.5). Final precipitate was used as a cell wall fraction for later experiments.
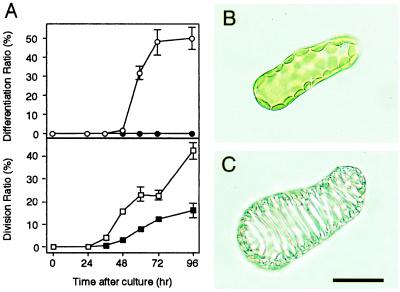
Figure 1.
(A) Time courses of TE-differentiation and division of Zinnia mesophyll cells cultured in the TE-inductive (○ and □; NAA + BA) or a control medium (● and ■; NAA). Each point represents the mean result from three samples (n = 500 in a sample), and vertical lines show standard deviations. (B) A single mesophyll cell just after the isolation. (C) A TE on which sculptured secondary wall is thickened. (Bar = 30 μm.)
Generation of mAbs.
A female BALB/c mouse was hyperimmunized by intraabdominal injections with pre-TE wall. An injection was performed with pre-TE wall (prepared from 3 × 106 cells) suspended in 0.3 ml of PBS at 2-week intervals three times. The immunized mouse showed about 1:100,000 of antibody titer, whereas preimmune serum showed about 1:500 when tested by ELISA against pre-TE wall. Its spleen was harvested 3 days after the final injection. Poly(A)+ RNA of the murine spleen was purified by using a FastTrack mRNA isolation kit (Invitrogen). A single-chain antibody variable domain (scFv) phage display library (2.1 × 106 members) was constructed by using the Recombinant Phage Antibody System (Amersham Pharmacia). The scFv phage display library was enriched for the pre-TE wall binding clones by four rounds of panning against pre-TE wall. Panning procedure was based on the manufacturer's protocol. In brief, approximately 1011 plaque-forming units of ΦAbs derived from the library of transformed TG1 cells was added to suspension of pre-TE wall from 5 × 106 cells, coprecipitated with pre-TE wall by centrifugation at 2,000 × g for 30 s. The precipitated ΦAbs were infected to TG1 cells and propagated for further enrichment. The ratio of binding phage (output) to applied phage (input) was determined after each panning. Four rounds of panning resulted in 2,500-fold enrichment.
Subtraction.
ΦAbs (106 plaque-forming units) from the library enriched by four rounds of panning were suspended in PBS containing 1% (wt/vol) BSA and 0.05% Triton X-100. The suspension was incubated with control wall from 5 × 106 cells overnight at 4°C, and the ΦAbs that bound to control wall were removed by centrifugation at 2,000 × g for 30 s. The supernatant was incubated with pre-TE wall from 106 cells overnight at 4°C, and ΦAbs binding to pre-TE wall were collected and infected to TG1 cells.
ELISA.
ΦAbs were prepared from supernatant of culture of single bacterial colonies, purified by precipitation with polyethylene glycol, and resuspended in 1/10 volume of 2× YT medium, which contained 1.6% (wt/vol) bacto-tryptone, 1% (wt/vol) bacto-yeast extract, and 0.5% (wt/vol) NaCl (18). The ΦAb suspension was diluted 2-fold in PBS containing 2% (wt/vol) BSA (PBS-B), and 100 μl of the diluted ΦAb was incubated overnight at 4°C with the cell wall (0.5 × 106 cells) that had been blocked with PBS-B and that had been treated with 1% (wt/vol) H2O2 for 3 h at 4°C to inactivate intrinsic peroxidase activity. After several washes with PBS containing 0.05% Triton X-100 (PBS-T), anti-M13 antibody horseradish peroxidase conjugates (Amersham Pharmacia) diluted in PBS-B (1:5,000) were then reacted to the cell wall overnight at 4°C. After several washes with PBS-T, 100 μl of 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) substrate solution was added to the cell wall. After 20 min, 100 μl of 2% (wt/vol) oxalic acid was added to stop the color development, and the absorbance (A) of the supernatant was measured at 415 nm with a spectrophotometer. An A415 of solution reacted without ΦAb treatment was subtracted from an A415 of each sample as background. M13 phage instead of ΦAb was used as a negative control.
To perform ELISA against whole cells, cultured cells were fixed for 30 min in 0.2% (wt/vol) glutaraldehyde in 50 mM phosphate buffer (pH 7.5) and then for 1 h in a mixture of 0.2% glutaraldehyde and 3.6% (wt/vol) formaldehyde in 50 mM phosphate buffer (pH 7.5). CN 8 scFv protein was produced in Escherichia coli and purified by anti-peptide-tag (anti-E-tag) affinity column (Amersham Pharmacia). The fixed cells were dehydrated, rehydrated through graded ethanol series, and then immersed in PBS. After the intrinsic peroxidase activity was inactivated by treatment with 3% H2O2 in PBS for 10 min at room temperature twice, the rehydrated cells were blocked with a blocking regent containing casein (Dako). The pretreated cells were reacted with 1 μg/ml of CN 8 scFv in PBS-B and then with horseradish peroxidase-labeled anti-E-tag antibody (Amersham Pharmacia) diluted in PBS-B (1:8,000), each for 1 h at room temperature. The reaction with the substrate and the measurement of absorbance were performed as described above. An A415 of cells reacted without primary antibody treatment was subtracted from an A415 of each sample as background.
Immunodetection of CN 8 Epitope.
Cultured Zinnia cells were fixed, dehydrated, rehydrated, blocked as mentioned above, and used for immunodetection of CN 8 epitope. The shoot apex of 14-day-old seedlings and stems of 2-month-old plants of Zinnia were fixed and embedded in paraffin as described (19). The sections of the Zinnia plants were also used for immunodetection of CN 8 epitope after being deparaffinized. Plant materials were probed in order with 1–5 μg/ml of CN 8 scFv, 1 μg/ml of anti-E-tag antibody (Amersham Pharmacia), and then 10- to 50-fold diluted alkaline phosphatase-labeled dextran polymer that was conjugated with anti-mouse Ig antibody (EnVision AP; Dako), each for 1 h at room temperature. After several washes with PBS-T, alkaline phosphatase-dependent color was developed for 10–30 min at room temperature by using Enzyme-Labeled-Fluorescence-97 (Molecular Probes) as a phosphatase substrate. After staining with 0.05% safranin O for 2 min, samples were observed under a fluorescence microscope. The green fluorescent signal of the Enzyme-Labeled-Fluorescence-97 was detected with a UV laser. Light-field micrographs were converted into pseudocolors and layered with the fluorescent micrographs with photoshop 5.0 (Adobe Systems, Mountain View, CA).
Cell Wall Solubilization.
The cell wall from 107 cells that had been cultured for 72 h in the TE-inductive medium was extracted sequentially with 3 ml of 2 M CaCl2, 50 mM cyclohexane-trans-1,2-diamine-NNN′N′-tetra-acetate (CDTA), 4% (wt/vol) KOH, and then 24% KOH at 4°C each for 12 h with shaking. These extracts were blotted on the nitrocellulose membrane. After blocking with PBS-B, antibody treatments were performed as described above. The color development was performed by using the Immun-Blot Assay Kit (Bio-Rad).
Results
Isolation of TE-Differentiation Specific mAbs.
We produced the scFv phage display library enriched for the clones that bound preferentially to pre-TE wall that was prepared from cells that had been cultured for 42 h in the TE-inductive medium. To check the quality of the enriched library, 26 monoclonal ΦAbs were prepared from randomly picked single bacterial colonies. All ΦAbs bound to pre-TE wall as determined by ELISA. Sequencing of scFv genes indicated that the 26 ΦAbs originated from nine individual clones with frequencies of 11, 5, 2, 2, 2, 1, 1, 1, and 1. Then, it was concluded that the enriched library consisted of a large repertoire of ΦAbs binding to pre-TE wall.
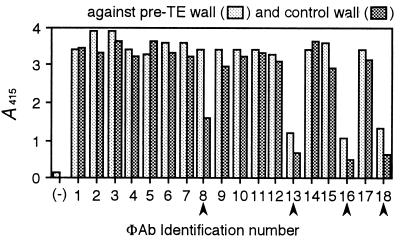
To isolate mAbs specific to TE differentiation, the enriched library was subtracted by using control wall. ΦAb suspension of the enriched library was adsorbed with an excess of control wall, and then the ΦAbs that bound to pre-TE wall were selected out of the suspension. When 18 of the selected ΦAbs were tested by ELISA against pre-TE and control wall, 4 ΦAbs had clear preferential binding to pre-TE wall (Fig. 2). Sequencing of scFv genes indicated that the four ΦAbs had been derived from four individual clones. CN 8, a clone that showed the most distinct binding among the four was used for further analysis. Monoclonal scFv protein was produced from CN 8, purified by anti peptide-tag affinity column, and designated CN 8 scFv.
Figure 2.
Screening of pre-TE wall-specific ΦAbs from subtracted library by ELISA. Arrowheads indicate ΦAbs that bound preferentially to pre-TE wall. (−), the negative control in which the M13 phage that did not display scFv was used instead of ΦAb.
Involvement of CN 8 Epitope in Vascular Differentiation.
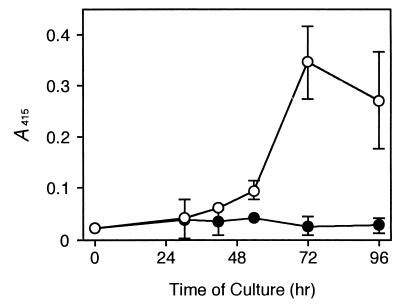
Whole-cell ELISA against cultured Zinnia cells indicated that the CN 8 epitope increased specifically in cells cultured in the TE-inductive medium but not in those in a control medium (Fig. 3). The increase occurred at 42 h of culture, which was 6 h before secondary wall thickenings started, and peaked at 72 h of culture at which time TE differentiation reached maximum.
Figure 3.
Changes in CN 8 epitope in isolated Zinnia mesophyll cells during culture in the TE-inductive (○) or a control (●) medium. Each point represents the mean result from three samples, and vertical lines show standard deviations.
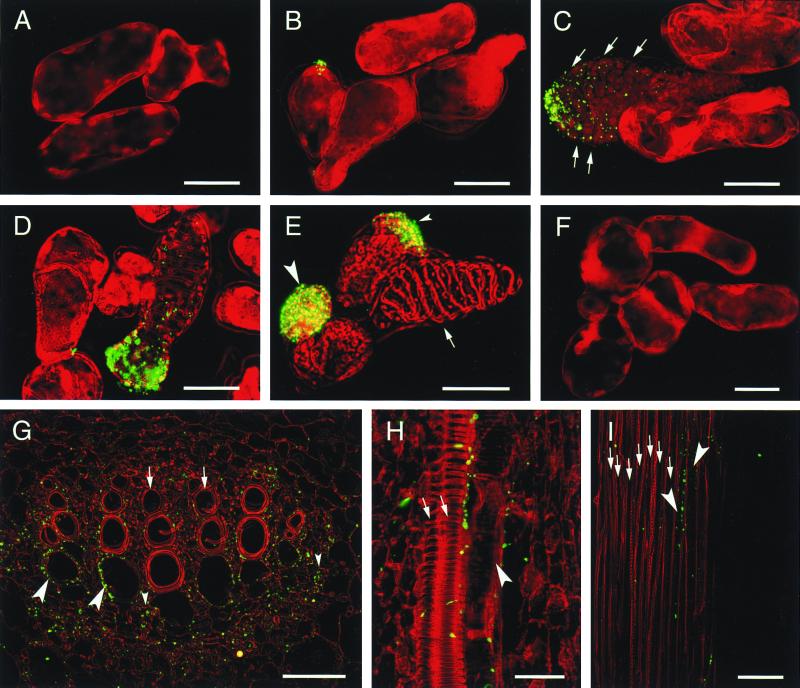
To know in which type of cells the CN 8 epitope localizes, immunohistochemical analyses were performed for cultured cells and plants of Zinnia (Fig. 4). No signal was detected in cells cultured in a control medium throughout the culture period (Fig. 4F). In cells cultured in the TE-inductive medium, the CN 8 epitope was not detected before 30 h of culture (Fig. 4A). At 42 h of culture when there are no cells having a visible sign of secondary wall thickenings, a subpopulation of cells had the CN 8 epitope localized at a tip of the cell (Fig. 4B). Observation of a plasmolyzed cell indicated that the CN 8 epitope was localized in the cell wall (Fig. 4B). Stronger CN 8 epitope signals with a gradient from a tip of the cell were detected in immature TEs that were recognized by predicted secondary wall thickening area from which chloroplasts were kept away (Fig. 4C). The CN 8 epitope increased further without changing its location in maturing TEs on which the secondary wall was slightly thickened (Fig. 4D). However, a mature TE with fully thickened secondary wall often had no signal (Fig. 4E, arrow). The CN 8-positive TEs appeared at 54 h of culture, peaked at 72 h (14% of total TEs), and then decreased. Non-TE cells with strong signals were also observed when cultured for 72 h in the TE-inductive medium (Fig. 4E, large and small arrowheads). Most of the CN 8-positive non-TE cells were round and cytoplasm-rich and had the CN 8 epitope over the cell (Fig. 4E, large arrowhead). Some non-TE cells showed the localization of the CN 8 epitope in a tip of the cell (Fig. 4E, small arrowhead), like an immature TE (Fig. 4C). Those CN 8-positive non-TE cells with strong signals comprised approximately 20% of total cells at 72 h and 96 h of culture but zero up to 54 h.
Figure 4.
Immunolocalization of the CN 8 antigen in cultured Zinnia cells (A–F), in the shoot apex of 14-day-old Zinnia seedlings (G and H), and in the stem of 2-month-old Zinnia plants (I). The yellowish green color indicates CN 8 epitope localization in the cell pseudocolored in red. Cells were cultured in the TE-inductive (A–E) or a control medium (F) for 30 h (A), 42 h (B), and 72 h (C–F) and treated with CN 8 scFv. (G) Cross section of midvein in a first foliage leaf. (H) Longitudinal section of midvein in a second foliage leaf. (I) Longitudinal section of stem vein. Arrows in C indicate the chloroplast-depleted area in an immature TE. The arrow and large and small arrowheads in E indicate a mature TE, a round CN 8-positive cell, and a semielongated CN 8-positive cell in which the CN 8 antigen localized in a tip, respectively. Arrows and large arrowheads in G–I and small arrowheads in G indicate mature TEs, immature TEs, and xylem parenchyma cells, respectively. (Bars in A–F = 30 μm; bars in G and H = 50 μm; bar in I = 100 μm.)
In the shoot apex of young seedlings and stems of adult plants of Zinnia, the CN 8 epitope was localized preferentially in the cell wall in immature TEs but not in mature TEs (Fig. 4 G–I). Immunohistochemistry with longitudinal sections clearly indicated that the CN 8 epitope was localized preferentially on one side along the long axis of immature TEs (Fig. 4 H and I). This polarized location of the CN 8 epitope on cell wall of immature TEs in Zinnia plants is consistent with the result obtained in vitro. In addition to immature TEs, xylem parenchyma cells (Fig. 4G, small arrowheads) and immature xylem cells, which are to differentiate into TEs or xylem parenchyma cells, were also labeled with the CN 8 scFv.
Characterization of CN 8 Antigen.
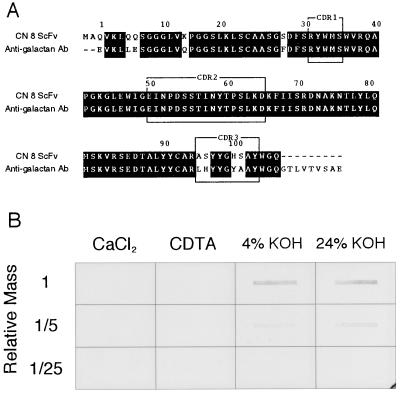
A search of the DNA database indicated that the deduced amino acid sequence of variable domain in an immunoglobulin heavy chain (VH) of CN 8 scFv had a significant (92%) homology with VH of anti-β-(1 → 6)-d-galactan mAb (Fig. 5A). The great similarity over the VH region suggested that the two VHs are derived from the same preimmune repertoire. Although the amino acid sequences of two complementarity determining regions, CDR1 and CDR2, were identical in VHs of CN 8 scFv and anti-galactan mAb, those of CDR3 were different, suggesting that the two mAbs may recognize similar but not the same set of epitopes. Indeed, CN 8 scFv did not recognize a commercial galactan or an arabinogalactan standard (Aldrich; data not shown). To know the nature of the antigen recognized by CN 8 scFv, cell wall components were extracted with CaCl2, CDTA, 4% (wt/vol) KOH, and 24% KOH from the wall of cells that had been cultured for 72 h in the TE-inductive medium. Immunoblot analysis of the extracts showed that the CN 8 antigen was present preferentially in 4% KOH and 24% KOH fractions (Fig. 5B). These results suggest that CN 8 antigen may be a component of hemicellulose.
Figure 5.
(A) Comparison of amino acid sequences of VH between CN 8 scFv and anti-(1 → 6)-β-d-galactan mAb (referred to in ref. 20). Complementarity determining regions (CDR1–3) and numbers of amino acid residues were assigned according to the method described in ref. 21. Identical amino acid residues are shown against black backgrounds. (B) Immunoblot assay of cell wall extracts with CN 8 scFv. The cell wall fraction was prepared from the cells that had been cultured for 72 h in the TE-inductive medium. Cell wall components were extracted sequentially with CaCl2, CDTA, 4% (wt/vol) KOH, and 24% KOH and then blotted on a nitrocellulose membrane.
Discussion
We succeeded in isolating mAbs that recognize wall components of vascular cells. The success justifies our strategy for the preparation of developmental stage-specific mAbs consisting of (i) the isolation of wall fractions from synchronously differentiating cells with an in vitro system, (ii) the generation of mAbs with the phage display method, and (iii) screening with a subtraction method. The first application of antibody phage display technology to plant cell wall research was made for the isolation of CCRC-R1, which is a mAb against deesterified rhamnogalacturonan II (8). Anti-homogalacturonan mAbs were also isolated by an interesting application of antibody phage display technology, which allowed the production of mAbs against nonimmunogenic substances by a large library consisting of preimmune repertoire (22). In addition, the classical mAb-generating methods have contributed to preparing mAbs against specific components isolated from cell walls such as arabinogalactan proteins (5). Some of the mAbs against arabinogalactan proteins mark vascular cells preferentially at specific developmental stages (23–25). However, vascular cell-specific antibodies were still few. To our knowledge, our approach is the first that has led to the success in the isolation of vascular cell wall-specific mAbs without the prior isolation or identification of antigens and opens the way for the generation of mAbs recognizing cell wall components specific to vascular cells on a large scale.
Of mAbs isolated as vascular cell wall-specific, the CN 8 scFv was found to be a unique type of antibody. The immunohistochemical analyses for the section of the shoot apex of Zinnia seedlings clearly indicated that the CN 8 scFv labeled immature TEs and xylem parenchyma cells. In immature TEs, the CN 8 epitope was localized on the cell walls with a polarity. This localization indicates the presence of a polarized cell wall component in immature TEs. Interestingly, single TEs formed in vitro also had this polarity. Therefore, this polarity must be formed in differentiating TEs autonomously. In the shoot apex, TEs are continuously formed from the distal to basal side of the vein, and the perforation occurs only on the distal end wall to release degraded cell contents. Preliminary analysis of vessel formation in the Zinnia shoot apex suggested that the CN 8 epitope was localized at the side where the perforation may occur (data not shown). Scanning microscopic observation revealed that even in single TEs formed in vitro, the pore opens on one end of the cell (J. Nakashima, personal communication). The CN 8 epitope does not seem to be involved directly in the pore formation at the given end, because there are non-TEs with similar polarity. However, polarized location of a wall component may be important in the determination of a developmental axis of cells. In fucoid zygotes, some cell wall components locating with a polarity are postulated to anchor the cytoskeleton via plasma membrane proteins and then fix a developmental axis of zygote (26). Further studies of the CN 8 epitope, specifically of its localization mechanism and interrelationship between the plasma membrane and the cytoskeleton, will shade a new light on the formation of polarity in developing single cells.
In cultured cells, the CN 8 scFv labeled not only immature TEs but also two other types of non-TE cells, round cells mainly and semielongated cells partly. The round cells had strong signals all over the primary walls. The fact that the CN 8 epitope is present on the cell wall of xylem parenchyma cells as well as immature TEs in situ suggests that cultured non-TE cells with strong signals of the CN 8 epitope are xylem parenchyma-like cells. In plants, there is a strong functional relation between xylem parenchyma cells and differentiating TEs. Glycine-rich protein 1.8 is known to be produced by xylem parenchyma cells that export the protein to the walls of differentiating TEs (27). Xylem parenchyma cells may also provide lignin precursors to differentiating TEs, as suggested by the fact that a gene encoding cinnamyl alcohol dehydrogenase, which catalyzes a late step of biosynthesis of lignin precursors, is expressed in xylem parenchyma cells as well as differentiating TEs (28). It is reported for cultured Zinnia cells that lignification in TEs continues even after TEs lose their cell contents (29). In fact, the medium in which differentiating cells are cultured contains a relatively large amount of lignin precursors (Y. Sato, personal communication). The accumulation of such precursors was suppressed by an inhibition of secretion, even after most of the TEs died (Y. Ito and H.F., unpublished results). These results indicate that a subpopulation of non-TE cells produces lignin precursors. Taken together with our observations, it is likely that some cultured cells differentiate into xylem parenchyma cells that provide lignin precursors to differentiating TEs even in vitro. This idea raises an important question as to how xylem parenchyma cells and TEs are controlled to differentiate in association with each other. The in vitro Zinnia system will also be useful system for such analysis.
Our preliminary characterization suggests that the CN 8 epitope is a cell wall component contained in the hemicellulosic fractions and not directly related to lignification. The identification of CN 8 epitope is important for understanding the function and localization mechanism of CN 8 antigen.
Acknowledgments
The authors thank Drs. M. McCann and K. Roberts (John Innes Centre) for critical reading of the manuscript. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (nos. 10304063, 10219201, and 10182101 to H.F.; nos. 10158204 and 09740587 to T.D.), by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS-RFTF96L00605 to H.F.), and by Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to N.S.).
Abbreviations
- TE
tracheary element
- ΦAb
phage antibody
- NAA
1-naphthaleneacetic acid
- BA
benzyladenine
- scFv
single-chain antibody variable domain
- CDTA
cyclohexane-trans-1,2-diamine-NNN′N′-tetra-acetate
- VH
variable domain in an immunoglobulin heavy chain
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the DDBJ/EMBL/GenBank databases (accession no. AB036341).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050582197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050582197
References
- 1.Varner E J, Lin L S. Cell. 1989;56:231–239. doi: 10.1016/0092-8674(89)90896-9. [DOI] [PubMed] [Google Scholar]
- 2.Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 3.Pennell R I. Curr Opin Plant Biol. 1998;1:504–510. doi: 10.1016/s1369-5266(98)80043-5. [DOI] [PubMed] [Google Scholar]
- 4.Knox J P. Int Rev Cytol. 1997;171:79–120. doi: 10.1016/s0074-7696(08)62586-3. [DOI] [PubMed] [Google Scholar]
- 5.Knox J P, Linstead P J, Cooper P C, Roberts K. Plant J. 1991;1:317–326. doi: 10.1046/j.1365-313X.1991.t01-9-00999.x. [DOI] [PubMed] [Google Scholar]
- 6.Smallwood M, Beven A, Donovan N, Neill S J, Peart J, Roberts K, Knox J P. Plant J. 1994;5:237–246. [Google Scholar]
- 7.Kreuger M, van Holst G J. Planta. 1995;197:135–141. [Google Scholar]
- 8.Williams M N V, Freshour G, Darvill A G, Albersheim P, Hahn M G. Plant Cell. 1996;8:673–685. doi: 10.1105/tpc.8.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCabe P F, Valentine T A, Forsberg L S, Pennell R I. Plant Cell. 1997;9:2225–2241. doi: 10.1105/tpc.9.12.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda H, Komamine A. Plant Physiol. 1980;65:57–60. doi: 10.1104/pp.65.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda H. Plant Cell. 1997;9:1147–1156. doi: 10.1105/tpc.9.7.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoogenboom H R. Trends Biotechnol. 1997;15:62–70. doi: 10.1016/S0167-7799(97)84205-9. [DOI] [PubMed] [Google Scholar]
- 13.de Kruif J, Terstappen L, Boel E, Logtenberg T. Proc Natl Acad Sci USA. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel D L, Chang T Y, Russell S L, Bunya V Y. J Immunol Methods. 1997;206:73–85. doi: 10.1016/s0022-1759(97)00087-2. [DOI] [PubMed] [Google Scholar]
- 15.Cai X, Garen A. Proc Natl Acad Sci USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiyama M, Fukuda H. Plant Tissue Culture Manual. Dordrecht, the Netherlands: Kluwer; 1995. [Google Scholar]
- 17.Sugiyama M, Fukuda H, Komamine A. Plant Cell Physiol. 1986;27:601–606. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Demura T, Fukuda H. Plant Cell. 1994;6:967–981. doi: 10.1105/tpc.6.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao D N, Rudikoff S, Krutzsch H, Potter M. Proc Natl Acad Sci USA. 1979;76:2890–2894. doi: 10.1073/pnas.76.6.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed. Bethesda, MD: National Institutes of Health; 1991. [Google Scholar]
- 22.Willats W G, Gilmartin P M, Mikkelsen J D, Knox J P. Plant J. 1999;18:57–65. doi: 10.1046/j.1365-313x.1999.00427.x. [DOI] [PubMed] [Google Scholar]
- 23.Schindler T, Bergfeld R, Schopfer P. Plant J. 1995;7:25–36. doi: 10.1046/j.1365-313x.1995.07010025.x. [DOI] [PubMed] [Google Scholar]
- 24.Knox J P. FASEB J. 1995;9:1004–1012. doi: 10.1096/fasebj.9.11.7544308. [DOI] [PubMed] [Google Scholar]
- 25.Stacey N J, Roberts K, Carpita N C, Wells B, McCann M C. Plant J. 1995;8:891–906. [Google Scholar]
- 26.Kropf D L. Plant Cell. 1997;9:1011–1020. doi: 10.1105/tpc.9.7.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryser U, Keller B. Plant Cell. 1992;4:773–783. doi: 10.1105/tpc.4.7.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feuillet C, Lauvergeat V, Deswarte C, Pilate G, Boudet A, Grima-Pettenati J. Plant Mol Biol. 1995;27:651–667. doi: 10.1007/BF00020220. [DOI] [PubMed] [Google Scholar]
- 29.Sato Y, Watanabe T, Komamine A, Hibino T, Shibata D, Sugiyama M, Fukuda H. Plant Physiol. 1997;113:425–430. doi: 10.1104/pp.113.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]