Abstract
Two major pathways for induction of apoptosis have been identified—intrinsic and extrinsic. The extrinsic pathway is represented by tumor necrosis factor family receptors, which utilize protein interaction modules known as death domains and death effector domains (DEDs) to assemble receptor signaling complexes that recruit and activate certain caspase-family cell death proteases, namely procaspases-8 and -10. The intrinsic pathway for apoptosis involves the participation of mitochondria, which release caspase-activating proteins. Bcl-2 family proteins govern this mitochondria-dependent apoptosis pathway, with proteins such as Bax functioning as inducers and proteins such as Bcl-2 and Bcl-XL serving as suppressors of cell death. An apoptosis regulator, BAR, was identified by using a yeast-based screen for inhibitors of Bax-induced cell death. The BAR protein contains a SAM domain, which is required for its interactions with Bcl-2 and Bcl-XL and for suppression of Bax-induced cell death in both mammalian cells and yeast. In addition, BAR contains a DED-like domain responsible for its interaction with DED-containing procaspases and suppression of Fas-induced apoptosis. Furthermore, BAR can bridge procaspase-8 and Bcl-2 into a protein complex. The BAR protein is anchored in intracellular membranes where Bcl-2 resides. BAR therefore may represent a scaffold protein capable of bridging two major apoptosis pathways.
Two major pathways for induction of apoptosis have been identified in recent years. One of these apoptosis pathways is represented by tumor necrosis factor (TNF)-family receptors that contain protein interaction modules known as death domains (DD) in their cytosolic regions (reviewed in refs. 1–3). On binding ligand or when overexpressed in cells, DD-containing TNF receptor family members such as Fas (CD95) aggregate, resulting in recruitment of an adaptor protein Fadd, which contains both a DD and a similar protein interaction module known as the death effector domain (DED) (4, 5). The zymogen pro-forms of certain caspase-family cell death proteases, namely procaspases-8 and -10, also contain DEDs in their N-terminal prodomains, allowing binding to Fadd/Fas complexes. This is followed by proteolytic processing and activation of the receptor-associated proteases, thereby initiating a subsequent cascade of additional processing and activation of downstream effector caspases (reviewed in refs. 1–3).
DED-containing proteins that function as antagonists of death receptor signaling have been identified in humans, mammals, and viruses (6–8). These antiapoptotic DED-containing proteins function as transdominant inhibitors, which compete for binding to the DED domains of Fadd or procaspases-8 or -10, thereby preventing assembly of a functional death-inducing complex (9).
A second major pathway for apoptosis involves the participation of mitochondria, which release cytochrome c (cyt-c), resulting in caspase activation through the effects of Apaf-1 (10, 11). Members of the Bcl-2 family play a major role in governing this mitochondria-dependent apoptosis pathway, with proteins such as Bax functioning as inducers of apoptosis and proteins such as Bcl-2 and Bcl-XL serving as suppressors of cell death (12, 13).
The Bax protein shares predicted structural similarity with the pore-forming domains of certain bacterial toxins (14) and induces release of cyt-c and triggers dissipation of the electrochemical gradient in mitochondria, even in the absence of caspases (15–17). When ectopically expressed in yeast, which have no caspases or Apaf-1 homologues, Bax targets to mitochondria, induces cyt-c release, and causes cell death (18, 19). This cytotoxic effect of Bax on yeast has permitted screens for human antiapoptosis genes that maintain cell survival despite expression of the Bax protein (20). Here we describe the cloning and characterization of human cDNAs encoding an apoptosis regulator identified through such a yeast-based screen. We have termed this protein BAR, for bifunctional apoptosis regulator, because it contains both a DED-like domain capable of suppressing apoptosis signaling through Fas (extrinsic pathway) and another domain that mediates interactions with Bcl-2 family protein and that is required for suppression of Bax-induced cell death in yeast and mammalian cells (intrinsic pathway). BAR thus represents a protein at the intersection of two major pathways controlling apoptosis.
Materials and Methods
Plasmids.
A Bgl-II fragment containing the complete ORF of BAR was isolated from a HepG2 library as described (20). cDNAs encoding full-length or fragments of BAR were generated by PCR and subcloned into various plasmids, as indicated.
Yeast Assays.
Yeast strain QX95001, containing the LEU2-marked mBax-encoding plasmid YEP51-Bax (20), was transformed with the plasmids p424, p424-BAR, p424-BAR(ΔR), and p424-BAR(ΔTM) containing the TRP marker by a lithium acetate method. Transformants were plated on SD-Leu, TRP (leucine-deficient and tryptophan-deficient SD). Protein extracts were prepared as described (18, 20). Yeast two-hybrid assays were performed as described (18, 21), by using Bcl-2 (ΔTM) proteins to avoid problems with nuclear targeting.
Cell Culture and Transfections.
293, 293T, and HT1080 cells were seeded at 5 × 105 cells per well in six-well plates and were transfected the next day with various combinations of plasmids by using SuperFect (Qiagen, Chatsworth, CA). Both floating and adherent cells (after trypsinization) were collected 24 hr after transfection and analyzed by 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) staining for assessing nuclear morphology. Transfection efficiencies were routinely >70% based on cotransfecting a green fluorescent protein (GFP)-encoding plasmid.
Caspase Assays.
Cell extracts (25 μg total protein) were prepared from transfected cells and incubated with 100 μM substrate benzyloxycarbonyl-Asp-Glu-Val-Asp-AFC (Z-DEVD-AFC) in 100 μl caspase buffer (16). Caspase activity was assayed by using a fluorometer plate reader, measuring release of fluorescent AFC.
Subcellular Fractionations.
293T cell lysates were prepared and fractionated to yield cytosolic, light-membrane, heavy-membrane, and nuclear fractions (20).
In Vitro Protein-Binding Assays.
GST-fusion proteins (≈3 μM) immobilized on glutathione-Sepharose beads were incubated with 10 μl of reticulocyte lysates (TNT-lysates, Promega) containing in vitro translated [35S] methionine-labeled proteins in 0.5 ml binding buffer (142.5 mM KCl/5 mM MgCl2/10 mM Hepes, pH 7.2/1 mM EGTA/0.2% Nonidet P-40) containing protease inhibitors for 3 hr at 4°C. Beads were washed three times in 1.5 ml binding buffer, and bound proteins were eluted by boiling in SDS-loading buffer and subjected to SDS/PAGE.
Coimmunoprecipitation Assays.
293T cells transfected with plasmids encoding Myc-BAR, Bcl-2, Bax, or other proteins were cultured with or without 20 μM MG-132 for 6 hr before lysing in HKME solution (142.5 mM KCl/5 mM MgCl2/10 mM Hepes, pH 7.2/1 mM EGTA) containing 0.4% Nonidet P-40 and protease inhibitors. Lysates were cleared by incubation with the protein G-Sepharose 4B (Zymed) and then incubated with anti-Myc antibody immobilized on agarose gel (Santa Cruz Biotechnology) at 4°C for 2 hr with constant rotation. Beads were then washed 4 times in HKME containing 0.2% Nonidet P-40 before boiling in SDS sample buffer. For analysis of interactions of BAR or BAR mutants with Flag-caspases or Flag-FADD, transfected cells were lysed in modified HKME solution, which contained 400 mM KCl and 0.8% Nonidet P-40. Immunoprecipitations were performed as above, by using either anti-Flag-M2 agarose affinity gel (Sigma) or anti-Myc immobilized agarose gel. SDS/PAGE immunoblotting was performed as described (20).
Antibody Production and Immunohistochemistry.
High-titer antisera specific for BAR were generated in rabbits by using a multiple boosting technique and recombinant GST-BAR(1–139) as immunogen, as described (22). Antibody reactivity with BAR was confirmed by immunoblot analysis of in vitro translated BAR vs. various control proteins and of lysates from 293T cells transfected with Myc-BAR or various control plasmids, revealing reactivity solely with the expected BAR protein. Specificity of antisera was determined by comparisons of immune and preimmune serum and by preadsorption of anti-BAR antisera with excess immunogen (GST-BAR) at 5 μg per 50 μl antiserum.
Computer Analysis.
BAR sequence has been analyzed with threading (23) and sensitive profile–profile alignment methods (24) developed at the authors' laboratory, as well as psi-blast (25). Three-dimensional models of identified domains were subsequently built with modeller (26) and analyzed for quality by using several structure analysis methods (27).
Results
BAR Is a Multidomain Protein.
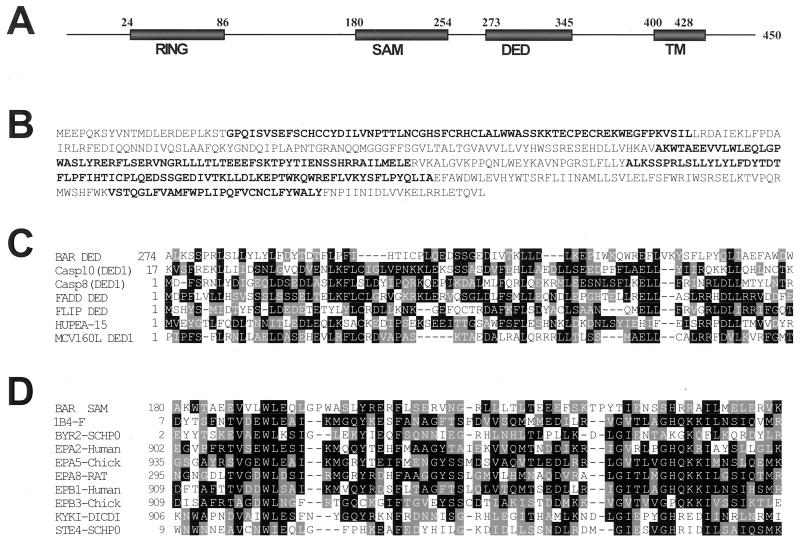
A screen for cDNAs encoding suppressors of Bax-induced cell death was performed in the yeast Saccharomyces cerevisiae, resulting in a cDNA containing an ORF encoding a human protein of 450 amino acids (≈52 kDa), to which we assigned the acronym BAR, for bifunctional apoptosis regulator (GenBank accession no. AF173003). The predicted BAR protein contains several protein domains, including: (i) an N-terminal zinc-binding RING domain at residues 24–86; (ii) a SAM domain at amino acids 180–254; (iii) a DED-like domain at 273–345; and (iv) a C-terminal hydrophobic transmembrane (TM) domain at 400–428 (Fig. 1 A and B). The first two assignments have high statistical significance, with psi-blast E-values of E−14 and E−28, respectively, indicating high confidence of the prediction. The DED-like domain is not recognized by any of the standard sequence analysis programs and instead was identified by detailed comparison to known DED proteins, as described below. The DED-like domain of BAR was most homologous to the first DED domain of procaspases-10, sharing 21% amino acid sequence identity (39% similarity), respectively, with these proteins (Fig. 1C). The BAR DED domain contains 14 of the 21 residues previously recognized to be conserved among DED-family domains based on sequence alignments (28), and 16 of the 19 hydrophobic residues known to be conserved among DED-family domains based on structural considerations (29). A consensus secondary-structure prediction based on phd (30) and nearest neighbor (31) algorithms suggested the presence of six α-helices, consistent with the known structure of other DED-family proteins (29). The SAM domain of BAR shares greatest homology with the human Ephb2 SAM domain (17% identity) and is predicted by FFAS to adopt a four α-helix bundle structure typical of SAM domains (32), with excellent conservation of the signature residues found in other members of this domain family (Fig. 1D).
Figure 1.
Sequence and domains of the BAR protein. (A) The domains of the human BAR protein are depicted, showing the RING, SAM, DED, and TM segments. (B) The predicted amino acid sequence of BAR is presented, with RING, SAM, DED, and TM domains in boldface. (C) An alignment of DED domain of BAR with DED domains from other proteins is shown, with identical and similar residues in black and gray, respectively. (D) An alignment of the SAM domain of BAR with SAM domains from other proteins is presented as above.
BAR Is a Membrane-Associated Protein That Suppresses Bax-Induced Cell Death.
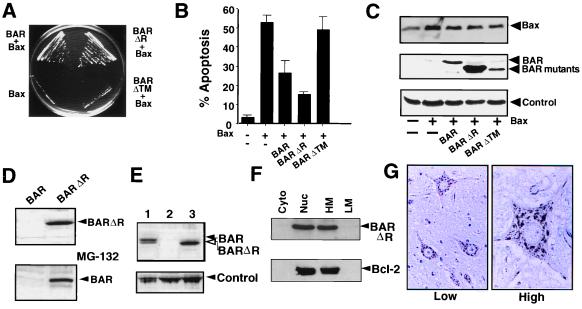
Expression of full-length BAR protein in yeast rescued cells from Bax-induced death (Fig. 2A), despite continued production of the Bax protein (not shown). Deletion of the RING domain from BAR did not interfere with its protective effect on Bax-induced cell death. In contrast, removal of the C-terminal TM domain completely abolished the ability of BAR to rescue against Bax, suggesting that membrane localization of BAR is important for its function as a Bax antagonist. Immunoblot analysis confirmed production of the BAR (ΔTM) protein at levels comparable to full-length BAR (not shown).
Figure 2.
BAR inhibits Bax-induced cell death in yeast and mammalian cells. (A) Plasmids encoding BAR, BAR(ΔR), BAR(ΔTM), or control plasmid were transformed into a yeast strain harboring YEp51-Bax. Transformants were streaked on galactose-containing synthetic medium lacking tryptophan and leucine and photographed after 4 days at 30°C. (B) 293T cells were transiently cotransfected with 0.1 μg of GFP-encoding marker plasmid and 4 μg of either pcDNA3 control (−) or 0.5 μg pcDNA3-Bax (+) plasmids together with 3.5 μg of plasmids encoding BAR, BAR(ΔR), or BAR(ΔTM) or a control plasmid (−). After 1 day, both floating and adherent cells (after trypsinization) were pooled, fixed, and stained with DAPI (18). The percentage of GFP-positive cells with fragmented nuclei or condensed chromatin (apoptotic) was determined (mean ± SD; n = 3). (C) 293T cell extracts (25 μg total protein) were prepared from the transfected cells shown in B and subjected to SDS/PAGE immunoblot analysis. (D) 293T cells were transfected with 2 μg of BAR- or BAR(ΔR)-encoding plasmids (Upper). Cell extracts (25 μg total protein) were prepared 1 day later and subjected to SDS/PAGE immunoblot analysis, by using anti-BAR antiserum. For determination of the effects of proteosome inhibition on BAR protein accumulation, 293T cells transfected with BAR-producing plasmid as above were cultured for 6 hr with (+) or without (−) 20 μM MG-132 before lysis and immunoblot analysis as above (Lower). Exposure times to x-ray film were adjusted to maximize differences. Longer exposures, however, demonstrated the presence of both full-length BAR and BAR(ΔR). (E) Immunoblot comparison of BAR levels in lysates (25 μg protein) prepared from MCF7 (1), control-transfected 293T cells (2), and BAR(ΔR)-transfected 293T cells (3), probed with anti-BAR antiserum. A band from the same blot resulting from anti-rabbit secondary antibody reactivity with an unidentified human protein is shown as a control for loading. (F) 293T cells were transiently transfected with plasmids encoding BAR(ΔR) and Bcl-2, then lysed 2 days later, and crude subcellular fractions of cytosol (Cyto), nuclei (Nuc), heavy membranes (HM), and light membranes (LM) were prepared (20). Fractions were normalized for cell equivalents and analyzed by SDS/PAGE immunoblotting, by using antisera specific for BAR (Upper) and Bcl-2 (Lower). (G) Immunohistochemistry-based analysis of BAR was performed by using paraffin-embedded tissue sections from multiple human tissues (spinal cord is shown here) and anti-BAR antiserum with DAB-colorimetric detection. Representative photomicrographs show neurons at lower (Left) and higher (Right) magnification, demonstrating punctate organelle-like distribution of BAR.
To investigate the effects of BAR on Bax-induced cell death in mammalian cells, plasmids encoding wild-type or mutant versions of BAR were cotransfected with a Bax-encoding plasmid into 293T cells, which contain low endogenous levels of BAR. Cells were recovered 24 hr later and analyzed for percentage apoptosis by staining with DAPI (18). Both BAR and BAR(ΔR) suppressed Bax-induced apoptosis (Fig. 2B), without interfering with Bax protein production (Fig. 2C). In contrast, the BAR(ΔTM) protein was ineffective at suppressing Bax-induced apoptosis, even though the protein was produced at levels similar to full-length BAR in 293T cells. We conclude therefore that BAR suppresses Bax-induced cell death in both yeast and mammalian cells, requiring the TM but not the RING domain for its Bax antagonistic function.
The BAR(ΔR) protein consistently accumulated to higher levels in cells compared with full-length BAR (Fig. 2 C and D), though no difference in plasmid-derived BAR and BAR(ΔR) mRNAs was observed (not shown). Similar to several other RING-containing proteins that are subject to proteosome-dependent degradation (33–35), the 26S proteosome protease inhibitor MG-132 markedly increased accumulation of BAR protein (Fig. 2D). Thus, steady-state levels of the BAR protein may be controlled by proteosome-dependent degradation, mediated by the N-terminal RING domain of this protein, explaining the greater suppression of apoptosis by BAR(ΔR) compared with BAR. Though deletion of the RING domain from BAR was helpful for enhancing accumulation of this protein in 293 cells, high levels of full-length BAR were found endogenously in some tumor cell lines such as MCF7 breast cancer cells (Fig. 2E), LOXIMVI and UACC-257 melanoma, and IGROV1 ovarian cancer cell lines (not shown).
The presence of a candidate TM domain in BAR suggested it could be a membrane-associated protein. Indeed, BAR protein was not extractable from cellular membrane preparations by using alkaline (pH 11.5) solution, consistent with an integral membrane protein (not shown). Subcellular fractionation experiments (Fig. 2F) revealed that BAR resides predominantly in the heavy-membrane (contains mitochondria) and nuclear fractions (presumably representing association with the nuclear envelope; see below), with little BAR present in cytosol or low-density membrane factions. Reprobing the same blots with antibodies to mitochondrial (COX-IV), cytosolic (caspase-3), and nuclear (poly-ADP ribosyl polymerase) proteins validated the fractionation procedure (not shown). BAR also cofractionated with Bcl-2 (Fig. 2F). When examined by immunohistochemistry by using normal human tissue sections and monospecific anti-BAR antisera, BAR was present in an organellar pattern within the cytosol of most cells (Fig. 2G and data not shown). Preimmune serum and anti-BAR antiserum preadsorbed with recombinant BAR protein resulted in negligible immunostaining, confirming the specificity of these results (not presented). Microscopy examination of cells expressing GFP-tagged BAR confirmed these findings (not shown). BAR mRNA and protein were widely expressed in human tissues, as determined by Northern and immunoblot analyses (unpublished observations).
BAR Modulates Bax-Induced Apoptosis Through a SAM Domain-Dependent Mechanism.
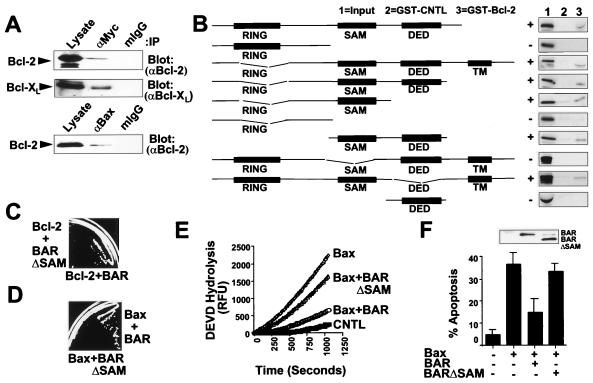
To explore how BAR might modulate Bax-induced cell death pathways, we examined whether the BAR protein can associate with members of the Bcl-2 family. For these experiments, Myc-epitope-tagged or untagged BAR or BAR(ΔR) proteins were coexpressed by transient transfection in 293T cells together with Bcl-2, Bcl-XL, Bak, or Bax. Coimmunoprecipitation assays determined that BAR can specifically associate with antiapoptotic Bcl-2 and Bcl-XL proteins (Fig. 3A), but not with Bax or Bak (not shown). The efficiency with which Bcl-2 coimmunoprecipitated with BAR was comparable to Bax (Fig. 3A) under the conditions of these assays.
Figure 3.
SAM Domain of BAR Is Required for Interaction with Bcl-2 and Inhibition of Bax-Induced Apoptosis. (A) 293T cells were transiently transfected with plasmids encoding Myc-BAR(ΔR) and either Bcl-2 or Bcl-XL (Upper) or with plasmids encoding Bax and Bcl-2 (Lower). Cells were lysed 2 days later in buffer containing 0.4% Nonidet P-40, and immunoprecipitations were performed by using anti-Myc or anti-Bax antibodies or by using mouse IgG1 as a control. Immune complexes and lysates (representing ≈5% of input) were subjected to SDS/PAGE and immunoblot analysis by using antisera for detection of Bcl-2 or Bcl-XL. (B) For in vitro binding studies, in vitro translated 35S-labeled BAR mutant proteins were incubated with 10 μg of either GST-Bcl-2 or GST (control; CNTL) recombinant proteins, and protein complexes were recovered on glutathione-Sepharose and analyzed by SDS/PAGE followed by autoradiography. In vitro translation mixes (10% input) were run directly in gels as a control. A wide variety of control GST proteins were tested, thus confirming specificity of BAR interactions with Bcl-2 (not shown). All BAR fragments shown were >50% intact when produced by in vitro translation in reticulocyte lysates, but others attempted could not be produced without extensive degradation, such as SAM only. (C) For yeast two-hybrid assay, 1 μg pGilda-Bcl-2(ΔTM) was cotransformed into EGY191 cells with 1 μg pJG4–5-BAR or pJG4–5-BAR(ΔSAM). Transformants were initially selected on glucose-containing medium lacking histidine and tryptophan. Two-hybrid interactions were assayed by streaking onto galactose plates lacking leucine, histidine, and tryptophan. Use of various positive and negative control proteins in yeast two-hybrid assays confirmed the validity of these observations (not shown). (D) For cytotoxicity assays in yeast, plasmids encoding BAR or BAR(ΔSAM) were transformed into a yeast strain harboring YEp51-Bax. Transformants were then streaked on galactose-containing synthetic medium lacking tryptophan and leucine. Photograph was taken after 4 days at 30°C. (E) For caspase assays, 293T cells were transfected with either 4 μg of vector control plasmid (CNTL) or with 0.5 μg Bax-encoding plasmid together with 3.5 μg of either vector control plasmid or plasmids encoding BAR or BAR(ΔSAM). Cell extracts were prepared 24 hr after transfection, normalized for protein content (25 μg), and incubated with 100 μM DEVD-AFC. Enzyme activity was determined by the release of the AFC fluorophore (expressed as relative fluorescence units, RFU). (F) A portion of the transfected 293T cells described in E were stained with DAPI, and the percentage of GFP-positive cells with fragmented or condensed nuclei (apoptotic) was determined (mean ± SD; n = 3). Inset shows immunoblot analysis of lysates from transfected cells using anti-BAR antiserum with enhanced chemiluminescence-based detection.
The region within BAR required for its interaction with Bcl-2 was mapped by using a series of N- and C-terminal truncation mutants and internal fragments of BAR for in vitro protein-binding assays (Fig. 3B). Fragments of BAR that lacked the RING, DED, or TM domains retained Bcl-2-binding ability, indicating that none of these domains is necessary. In contrast, all fragments of BAR that retained the SAM domain bound Bcl-2 in vitro, whereas all fragments lacking this domain failed to bind Bcl-2. The dependence of the SAM domain for interactions with Bcl-2 was also confirmed by yeast two-hybrid assays (Fig. 3C).
We next explored the relevance of the SAM domain of BAR for suppression of Bax-induced cell death in yeast and for Bax-induced caspase activation and apoptosis in human cells. When coexpressed in yeast with Bax, the full-length BAR protein suppressed Bax-induced cell death and permitted growth on selective medium, whereas the BAR (ΔSAM) protein failed to rescue yeast from Bax-induced cell death (Fig. 3D). Immunoblot analysis confirmed production of the BAR and BAR (ΔSAM) proteins at comparable levels in yeast (not shown). For mammalian cell experiments, 293T (Fig. 3) cells were cotransfected with a Bax-producing plasmid together with plasmids encoding either BAR or BAR (ΔSAM) proteins. Cells were recovered 1 day later, and either cell lysates were prepared for assaying caspase activity by using the fluorigenic substrate Asp-Glu-Val-Asp-aminofluorocoumarin (DEVD-AFC) (Fig. 3E) or the percentage of apoptotic cells was quantified based on DAPI staining (Fig. 3F). Overexpression of Bax induced caspase activation and apoptosis, which were both markedly suppressed by coexpression of full-length BAR but not by BAR (ΔSAM) (Fig. 3 E and F). Immunoblot analysis confirmed that neither BAR nor BAR(ΔSAM) interfered with Bax protein production and demonstrated that the BAR and BAR (ΔSAM) proteins were produced at roughly equivalent levels in these transfected cells (Fig. 3F and data not presented). Taken together, the results demonstrate that the SAM domain of BAR is required both for its interactions with Bcl-2 and its ability to suppress Bax-induced cell death.
BAR Binds DED-Containing Caspases and Suppresses Fas-Induced Apoptosis.
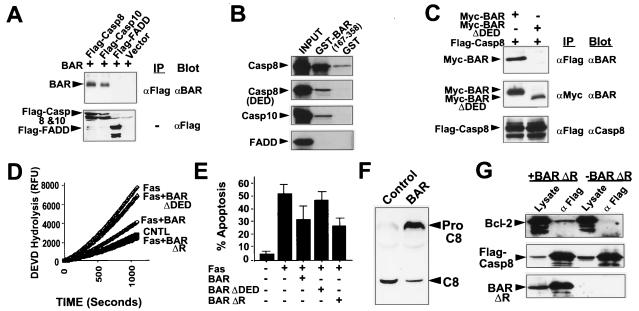
The presence of a DED-like domain in the BAR protein suggested that it might interact with other DED-containing proteins. Indeed, BAR and BAR(ΔR) specifically interacted with procaspase-8 and -10 in coimmunoprecipitation assays by using lysates from transfected 293T cells (Fig. 4A and data not shown). In contrast, BAR and BAR(ΔR) did not coimmunoprecipitate with Fadd (Fig. 4A), even though interactions of Fadd with procaspases-8 and 10 were detectable under the same experimental conditions (not shown). Thus, BAR associates with some but not all DED-family proteins. Mutagenesis studies confirmed a role for the DED domains of BAR, procaspase-8 and -10 for mediating their interactions (Fig. 4 B and C).
Figure 4.
DED Domain of BAR Is Required for Interactions with Procaspases-8 and -10 and for Inhibition of Fas-induced Apoptosis. (A) Coimmunoprecipitation assays were performed as described above by using 293T cells transfected with plasmids encoding Myc-BAR and either Flag-procaspase-8 (cys/ala), procaspase-10 (cys/ala), or Fadd. (B) For in vitro binding studies, in vitro translated 35S-labeled procaspase-8, procaspase-8 domain (DED), procaspase-10, and FADD proteins were incubated with 10 μg of either GST or GST-BAR-DED (residues 167–358) recombinant proteins. Protein complexes were recovered on glutathione-Sepharose beads, followed by analysis by SDS/PAGE autoradiography. In vitro translation mixes (10% input) were run directly in gels as a control. A wide variety of control GST proteins were tested, thus confirming specificity of BAR interactions with procaspases-8 and -10 (not shown). (C) 293T cells were cotransfected with 5 μg of either BAR- or BAR(ΔDED)-encoding plasmids and 5 μg of a plasmid encoding Flag-procaspase-8. Cell extracts were prepared 2 days later, and immunoprecipitates were prepared by using either anti-Flag or anti-Myc monoclonal antibodies. Immune complexes were subjected to SDS/PAGE immunoblot analysis, by using anti-BAR and anti-procaspase-8 antibodies. (D) For caspase activity assays, 293T cells were transfected with either control plasmid or cotransfected with 0.3 μg of Fas-encoding plasmid and 3.7 μg of either control plasmid or plasmids encoding BAR, BAR(ΔDED) or BAR(ΔR). Cell extracts were prepared 1 day later, normalized for total protein (25 μg), and incubated with 100 μM DEVD-AFC. Enzyme activity was determined by the release of the AFC fluorophore. (E) A portion of the transfected 293T cells described above were stained with DAPI, and the percentage of GFP-positive cells with fragmented or condensed nuclei (apoptotic) was determined (mean ± SD; n = 3). (F) 293T cells were transfected with plasmids encoding procaspase-8 and either pcDNA3 (control) or pcDNA3-Myc-BAR. Cell lysates were prepared 1 day later, normalized for protein content (50 μg), and analyzed by SDS/PAGE immunoblotting by using anti-caspase-8 antiserum. The positions of the unprocessed procaspase-8 and the large subunit of processed caspase-8 are indicated by arrows. (G) 293T cells were cotransfected with 5 μg each of plasmids encoding Bcl-2, Flag-Procaspase-8 and either 10 μg of BAR encoding plasmid (+BAR) or control plasmid (−BAR). Cell extracts were prepared 2 days later and immunoprecipitates were prepared by using anti-Flag antibody. Immune complexes were analyzed by SDS/PAGE immunoblotting, probing the same blot sequentially with anti-Bcl-2 antiserum (Top), anti-Flag monoclonal antibody (Middle), and anti-BAR antiserum (Bottom), with antibody stripping between each detection. Data are representative of several experiments and include a wide variety of control transfections and immunoprecipitations that further confirmed specificity of the observed interactions.
Because caspase-8 is essential for Fas-induced apoptosis (36, 37), we sought evidence that BAR could modulate this apoptotic pathway. For these experiments, 293 (Fig. 4) or HT1080 (not shown) cells were transfected with plasmids encoding Fas in combination with either a control plasmid or plasmids producing the BAR, BAR(ΔR) and BAR(ΔDED) proteins. Caspase activity and apoptosis were then assayed after 1 day. 293 and HT1080 cells were chosen for these studies because overexpression of Fas triggers apoptosis in these cells through a Bcl-2-independent mechanism (20, 38), thus avoiding any contributions that BAR might make with respect to modulation of Bcl-2 family proteins. As shown, Fas stimulated activation of DEVD-cleaving caspases and triggered apoptosis (Fig. 4 D and E), both of which were partially blocked by coexpression of BAR or BAR(ΔR). In contrast, BAR(ΔDED) and BAR (ΔR/ΔDED), which failed to bind procaspase-8, were ineffective at blocking Fas-induced activation of caspases and apoptosis (Fig. 4 D and E). Thus, BAR requires the DED domain to interact with procaspase-8 and to suppress Fas-induced apoptosis. Immunoblot analysis also revealed correlations with Fas-induced processing of procaspase-8, with BAR reducing the amount of processed procaspase-8 produced as a result of Fas overexpression (Fig. 4F).
BAR Can Mediate Association of Bcl-2 and Procaspase-8.
Recognizing that BAR is capable of interacting with both Bcl-2 and procaspase-8, we determined whether BAR bridges these two proteins together in a complex. Accordingly, Bcl-2 and Flag-procaspase-8 were coexpressed in 293T cells by transient transfection, in the presence or absence of cotransfected BAR. Coimmunoprecipitation assays revealed that Bcl-2 was readily detected in caspase-8-containing immune complexes when using lysates from cells overexpressing BAR but not in lysates of control transfected 293T cells (Fig. 4G), which contain relatively little endogenous BAR protein. Coexpression of Bax prevented BAR-mediated coimmunoprecipitation of Bcl-2 and procaspase-8 (not shown), suggesting that Bcl-2 cannot simultaneously bind Bax and BAR. We therefore conclude that BAR can bridge Bcl-2 and procaspase-8, at least when overexpressed, thus bringing together members of two important families of proteins involved in apoptosis regulation.
Discussion
BAR represents an apoptosis regulator having unique features heretofore not seen in other modulators of cell death pathways. This protein has a multidomain structure, which includes RING, SAM, DED, and TM domains, thus suggesting it may serve as a scaffold protein that integrates interactions and communication between two types of apoptosis-regulatory proteins—namely, the DED-containing initiator caspases and Bcl-2 family proteins. Though the extrinsic (e.g., Fas/death receptor) and intrinsic (e.g., Bax/mitochondrial) pathways for apoptosis are capable of operating independently, crosstalk between these two major pathways for apoptosis also occurs (2, 3, 12, 13). In this regard, overexpression of Bcl-2 or Bcl-XL can protect some cell lines in vitro and some tissues in vivo from Fas-induced apoptosis (39–43), but not others. Sensitivity to Bcl-2 protection from Fas-induced apoptosis correlates with reduced activation of procaspase-8 after Fas crosslinking (44), requiring a mitochondrial amplification step to achieve sufficient activation of downstream caspases for triggering apoptosis (45). Antiapoptotic DED-containing proteins such as BAR, Bap31 (46), and Flip (7) may compete with adaptor proteins such as Fadd for binding to procaspases-8 and -10, thus reducing the amount of caspase processing and activation (9).
At present, it remains unclear why BAR suppressed Bax-induced cell death in yeast, given that we have been unable to demonstrate interaction of BAR with Bax by coimmunoprecipitation or yeast two-hybrid assays. However, the observation that the SAM and TM domains of BAR are required for suppression of Bax-induced cell death in both yeast and mammalian cells suggests a conserved mechanism.
Though gene transfer-mediated overexpression of BAR was used here for assaying its effects on apoptosis pathways, the levels of BAR produced by transfection of 293T and HT1080 cells were similar to the endogenous levels of BAR found in a variety of human tumor cell lines, based on comparisons by immunoblotting (unpublished observations). Thus, levels of BAR sufficient to impact apoptosis pathways can occur at least within the context of cancer. Further analysis of BAR in other cellular contexts and ultimately by gene ablation experiments in mice is required to understand the overall importance of this protein for the in vivo regulation of apoptosis.
Acknowledgments
We thank E. Smith, R. Cornell, and S. Farrar for manuscript preparation; F. Pio for computer analysis; S. Matsuzawa (The Burnham Institute), D. Sykes (Buffalo General Hospital, Buffalo, NY), and S. Torii (Gunma University, Gunma, Japan), for plasmids; and the National Institutes of Health (NIH)/National Institutes on Aging (AG15393), A.G. and K.P. under NIH Grant GM60049, CaP-CURE, and the Department of Defense Breast Cancer Research Program (Q.X. and H.Z.) for generous support.
Abbreviations
- TNF
tumor necrosis factor
- DD
death domain
- DED
death effector domain
- DAPI
4′,6-diamidine-2′-phenylindole dihydrochloride
- GFP
green fluorescent protein
- TM
transmembrane
References
- 1.Wallach D, Boldin M, Varfolomeev E, Beyaert R, Vandenabeele P, Fiers W. FEBS Lett. 1997;410:96–106. doi: 10.1016/s0014-5793(97)00553-x. [DOI] [PubMed] [Google Scholar]
- 2.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi A, Dixit V. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 4.Chinnaiyan A M, O'Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 5.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 6.Bertin J, Armstrong R, Ottilie S, Martin D, Wang Y, Banks S, Wang G, Senkevich T, Alnemri E, Moss B, et al. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J-L, Schröter M, Burns K, Mattmann C, et al. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 8.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schroter M, et al. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 9.Tschopp J, Irmler M, Thome M. Curr Opin Immunol. 1998;10:552–558. doi: 10.1016/s0952-7915(98)80223-9. [DOI] [PubMed] [Google Scholar]
- 10.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 11.Li P, Nijhawan D, Budihardjo I, Srinivasula S, Ahmad M, Alnemri E, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 12.Adams J, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 13.Green D, Reed J. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 14.Schendel S, Montal M, Reed J C. Cell Death Differ. 1998;5:372–380. doi: 10.1038/sj.cdd.4400365. [DOI] [PubMed] [Google Scholar]
- 15.Narita M, Shimizu S, Ito T, Chittenden T, Lutz R J, Matsuda H, Tsujimoto Y. Proc Natl Acad Sci USA. 1998;95:14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurgensmeier J M, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Proc Natl Acad Sci USA. 1998;5:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finucane D M, Bossy-Wetzel E, Cotter T G, Green D R. J Biol Chem. 1998;274:2225–2233. doi: 10.1074/jbc.274.4.2225. [DOI] [PubMed] [Google Scholar]
- 18.Zha H, Fisk H A, Yaffe M P, Mahajan N, Herman B, Reed J C. Mol Cell Biol. 1996;16:6494–6508. doi: 10.1128/mcb.16.11.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manon S, Chaudhuri B, Buérin M. FEBS Lett. 1997;415:29–32. doi: 10.1016/s0014-5793(97)01087-9. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, Reed J C. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang H G, Rapp U R, Reed J C. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 22.Krajewski S, Krajewska M, Shabaik A, Wang H-G, Irie S, Fong L, Reed J C. Cancer Res. 1994;54:5501–5507. [PubMed] [Google Scholar]
- 23.Jaroszewski L, Rychlewski B, Zhang B, Godzik A. Protein Sci. 1998;7:1431–1440. doi: 10.1002/pro.5560070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rychlewski L, Jaroszewski L, Li W, Godzik A. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A, Blundell T L. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 27.Jaroszewski L, Pawłowski K, Godzik A. J Mol Model. 1998;4:294–309. [Google Scholar]
- 28.Peter M, Medema J, Krammer P. Cell Death Differ. 1997;4:523–525. doi: 10.1038/sj.cdd.4400285. [DOI] [PubMed] [Google Scholar]
- 29.Eberstadt M, Huang B, Chen Z, Meadows R, Ng S, Zheng L, Lenardo M, Fesik S. Nature (London) 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 30.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 31.Rychlewski L, Godzik A. Protein Eng. 1997;10:1143–1153. doi: 10.1093/protein/10.10.1143. [DOI] [PubMed] [Google Scholar]
- 32.Thanos C, Goodwill K, Bowie J. Science. 1999;283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 33.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 34.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamura T, Koepp D, Conrad M, Skowyra D, Moreland R, Iliopoulos O, Lane W, Kaelin W J, Elledge S, Conaway R, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 36.Muzio M, Chinnaiyan A M, Kischkel F C, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 37.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 38.Frisch S M, Vuori K, Kelaita D, Sicks S. J Cell Biol. 1996;135:1377–1382. doi: 10.1083/jcb.135.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan A, Li F, Wong A, Kodandapani L, Smidt R, Krebs J, Fritz L, Wu J, Tomaselli K. J Biol Chem. 1998;273:4523–4529. doi: 10.1074/jbc.273.8.4523. [DOI] [PubMed] [Google Scholar]
- 40.Medema J, Scaffidi C, Krammer P, Peter M. J Biol Chem. 1998;273:3388–3393. doi: 10.1074/jbc.273.6.3388. [DOI] [PubMed] [Google Scholar]
- 41.Torigoe T, Millan J A, Takayama S, Taichman R, Miyashita T, Reed J C. Cancer Res. 1994;54:4851–4854. [PubMed] [Google Scholar]
- 42.Lacronique V, Mignon A, Fabre M, Viollet B, Rouquet N, Molina T, Porteu A, Henrion A, Bouscary D, Varlet P, et al. Nat Med. 1996;2:80–86. doi: 10.1038/nm0196-80. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez I, Matsuura K, Khatib K, Reed J C, Nagata S, Vassalli P. J Exp Med. 1996;183:1031–1036. doi: 10.1084/jem.183.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli K, Debatin K-M, Krammer P, Peter M. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuwana T, Smith J J, Muzio M, Dixit V, Newmeyer D D, Kornbluth S. J Biol Chem. 1998;273:16589–16594. doi: 10.1074/jbc.273.26.16589. [DOI] [PubMed] [Google Scholar]
- 46.Ng F W H, Nguyen M, Kwan T, Branton P E, Nicholson D W, Cromlish J A, Shore G C. J Cell Biol. 1997;39:327–338. doi: 10.1083/jcb.139.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]