Abstract
Notch signaling is involved in cell differentiation and patterning during morphogenesis. In the Drosophila wing, Notch activity regulates the expression of several genes at the dorsal/ventral boundary, and this is thought to elicit wing-cell proliferation. In this work, we show the effect of clones of cells expressing different forms of several members of the Notch signaling pathway, which result in an alteration of Notch activity. The ectopic expression in clones of activated forms of Notch or of its ligands (Delta or Serrate) in the wing causes outgrowths associated with the appearance of ectopic wing margins. These outgrowths consist of mutant territories and of surrounding wild-type cells. However, the ectopic expression of Delta, at low levels in ventral clones, causes large outgrowths that are associated neither with the generation of wing margin structures nor with the expression of genes characteristic of the dorsal/ventral boundary. These results suggest that Notch activity is directly involved in cell proliferation, independently of its role in the formation of the dorsal/ventral boundary. We propose that the nonautonomous effects (induction of extraproliferation and vein differentiation in the surrounding wild-type cells) result from pattern accommodation to positional values caused by the ectopic expression of Notch.
Cell–cell interactions play a crucial role during the development of multicellular organisms, mainly in the coordination of cell proliferation and differentiation. Among the best known signaling pathways of intercellular communication is that of Notch (N), which is involved in multiple processes during Drosophila development. Notch and members of its signaling pathway are conserved in evolution (1). The different elements of the pathway are the transmembrane proteins encoded by Delta (Dl) and Serrate (Ser), which are ligands for the Notch receptor, and several intracellular components including Suppressor of Hairless [Su (H)] (which encodes a transcription factor), the Enhancer of split complex [E(spl)] [encoding basic helix—loop–helix (bHLH) proteins] and Hairless (H), which encodes a negative regulator of the pathway (1–4). The function of Notch appears to control local cell interactions related to cell determination. Thus for instance during neurogenesis Notch signaling functions to single out neural precursor cells from a field of undifferentiated cells of the neuroectoderm (5).
One of the best characterized morphogenetic processes in which Notch is involved is the definition of the dorsal/ventral (d/v) boundary and wing margin patterning during the development of the imaginal wing disc (revised in ref. 6). The wing develops from a group of cells segregated from the embryonic ectoderm in early embryogenesis, which after proliferation in the larval stages will form the imaginal wing disc (7). Clonal analysis reveals that the wing is divided into four compartments (anterior, posterior, dorsal, and ventral). The subdivision of the wing into anterior/posterior (a/p) compartments occurs before the segregation of the disc from the epidermis in the embryo (8). The d/v boundary is established later, during the proliferation of disc cells (9). The formation and maintenance of the d/v boundary requires the locally restricted activation of Notch signaling (6, 10). This boundary is formed as consequence of the confrontation of two cell populations, dorsal cells that express the selector gene apterous (ap) (11) and cells that do not express it. The gene ap activates the expression of both the Notch ligand Serrate and of fringed, a gene involved in the regulation of Ser and Dl activity (12–15). Although Ser is effective only at activating N in ventral cells, it has been proposed that Dl is required in ventral cells along the boundary to active N in the dorsal compartment (16, 17). Notch activation at the d/v boundary is, in turn, required for the localized expression of different genes involved in the formation of the d/v boundary and wing margin patterning, such as wingless (wg), vestigial (vg), Distal less (Dll), and cut (ct) (15, 18–24). It has been proposed that it is the function of vg and wg that leads to the proliferation of the wing disc by using the d/v boundary as the organizing center (revised in ref. 6).
Indirect evidence suggests that the effect of Notch signaling on cell proliferation is not the simple consequence of the regulation of vg or wg expression. Thus, the ectopic expression of either vg or wg does not reproduce the phenotype of the ectopic expression of an activated form of Notch (23, 25, 26). Moreover, clones of cells homozygous for loss-of-function alleles of N have poor viability, even clones that do not touch the d/v border, suggesting that the function of this gene is necessary throughout the wing for cell proliferation (27). The results of the present work suggest a direct function of Notch signaling on cell proliferation, independently of the generation of the d/v boundary.
Materials and Methods
Genetic Strains.
We used the gain-of-function alleles AxM3 and Ax16172 (28), the vg d/v boundary [vg(B)] and quadrant [vg(Q)] enhancers lacZ constructs (21, 29) (kindly provided by S. Carroll, Laboratory of Molecular Biology, University of Wisconsin), the UAS lines UAS-Nintra, UAS-Ser (23) (a gift from J. F. de Celis, Department of Genetics, Cambridge University), UAS-Dl (30), UAS-wg (31) (kindly provided by S. S. Huppert and F. Diaz-Benjumea, CBMSO, Universidad Autónoma de Madrid, respectively), and the GAL4 line GAL4-MS1096 (32).
Generation of Mosaics.
Clones of cells expressing GAL4 were induced 24–48, 48–72, or 72- 96 hr after egg laying (AEL) by 7-min heat shocks at 37°C in flies of the following genotypes: (i) f36a FLP1.22; P [abx/Ubx<FRT f+FRT>GAL4-lacZ]/UAS-Nintra or UAS-wg or UAS-Dl. The flip-out of the <FRT f+FRT> cassette results in the expression of a GAL4-lacZ fusion gene under the control of the abx/Ubx promoter (23). (ii) y w FLP1.22; Act5C<FRT yellow+ FRT> GAL4 UAS-GFP/UAS-Nintra or UAS-Dl or UAS-Ser. The flip-out of the <FRT yellow+ FRT> cassette results in the expression of the transcriptional activator GAL4 gene under the control of the Act5C promoter (33). Clones were detected by expression of GFP or LacZ expression and were analyzed in third instar larvae or adult flies.
Cell Lineage in Ax Mutant Background.
AxM3/Ax16172;Act5C>Draf+<>nuc-lacZ/+;hsp70-flp/+ mutant or control larvae Act5C>Draf+<>nuc-lacZ/+;hsp70-flp/+ (34) were heat shocked 6 min at 37°C at 36 hr AEL. β-Galactosidase detection was performed in third instar imaginal wing discs as described (35).
In Situ Hybridization and Immunocytochemistry.
Whole-mount in situ hybridization with digoxigenin-labeled DNA probes in imaginal discs was performed as described in ref. 35. For immunocytochemistry, we used rabbit anti-β-galactosidase (Cappel) and anti-VG (36) (kindly provided by S. Carroll), mouse monoclonal anti-WG (37) (kindly provided by S. Cohen), anti-CT (38), and anti-Dll (19). Secondary antibodies were from Jackson ImmunoResearch (used at 1/200 dilution). Hoechst 33258 was performed as described (39).
Estimation of Number of Mitotic Cells.
We have studied the number of mitotic cells in the wing region close to the d/v boundary of nine mutant and control third instar imaginal wing discs by using a light microscope and a lens of ×60.
Results
Abruptex Cell Lineage.
Alleles of Notch, Abruptex (Ax) cause ectopic and increased activity of Notch (28, 40). Certain heteroallelic combinations between different alleles give rise to discs much larger that wild-type discs (28). In these discs, Notch signaling is greatly enhanced, as suggested by the enlarged expression of wg and ct close the d/v border (16, 26). To see whether Ax mutations lead to a preferential growth in cells close to the d/v boundary, we have carried out a cell lineage analysis in wing discs of the heteroallelic combination AxM3/Ax16172 by generating clones of β-galactosidase-expressing cells (see Materials and Methods). This analysis consists of the study of the behavior and proliferation parameters of clones of cells marked (β-galactosidase in this case) in a wild-type or mutant background. The average size of the clones initiated at 36-hr AEL in the Ax mutant discs is larger all over the wing than in wild-type controls (wt: 138 ± 141; Ax: 313 ± 469 number of cells). In addition, whereas we never find clones of more than 600 cells on control discs, in mutant discs clones can be of more than 2,000 cells. The topographical distribution of different clone sizes in Ax mutant discs is, like in wild-type discs, heterogeneous, i.e., not restricted to cells in the d/v border or any particular region of the disc. These results suggest that in Ax mutant discs, the cell proliferation rate is higher than in control wings.
Ectopic Expression of Member Genes of the Notch Pathway.
To study the effect of Notch signaling directly on cell proliferation, we generated by flip-out recombination clones of cells expressing different gene constructs that modify the activity of Notch signaling. In this analysis, we have used two different promoters for inducing GAL4-expressing clones: one, the strong (abx/Ubx) promoter, which is associated to the adult cell marker forked (23), and the weaker promoter (Actin), which drives the expression of GAL4 associated with the cell marker yellow (33). This allows us to compare high or low levels of expression of the UAS-associated gene.
We first studied the effects of overexpression of the intracellular part of Notch (Nintra), which corresponds to an activated form of the receptor. By using the strong promoter, we observed the phenotypes previously described (23). Thus, clones of Nintra-expressing cells in the wing pouch of both wing surfaces cause wing outgrowths. They contain small territories of mutant cells in the tip of the outgrowth that differentiate wing margin pattern elements and wild-type cells in the base of the outgrowth that differentiate wing-blade tissue with identifiable vein patterns (Fig. 1 A and B) (23). In these outgrowths, the nonautonomous induction of proliferation of wild-type cells has been explained as a consequence of the appearance of a new d/v boundary that drives, as in the normal wing d/v boundary, cell proliferation (6, 10). Interestingly, the extent of the outgrowth formed by wild-type cells depends on the position where the clones were initiated more than on the size of the mutant clone. The largest outgrowths appear in clones close to the a/p boundary, away from the d/v border, whereas the smaller outgrowths appear close to the d/v boundary (Fig. 1 A and B). This finding suggests that the nonautonomous proliferation of wild-type cells associated to the mutant clone is independent of the putative amount of d/v signal. Rather, it seems that outgrowths are generated by intercalary cell proliferation, as seen by pattern duplication, to accommodate for discrepancies in positional values along the a/p axis of the wing (see Discussion). Clones of Nintra-expressing cells generated by using the weak (actin) promoter grow normally, without causing outgrowths, indicating that in these clones, Notch activity is low.
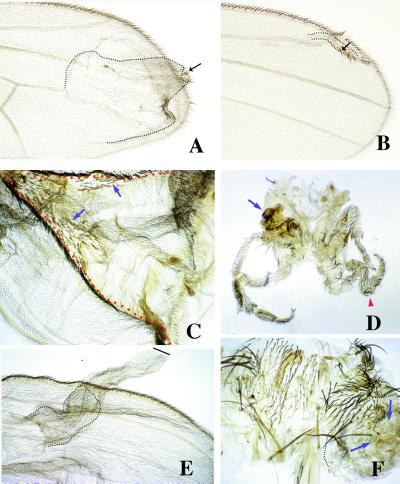
Figure 1.
Adult phenotypes caused by clones of ectopic expression of Nintra using the abx/Ubx promoter (A and B) and Dl by using the actin promoter (C–F). In A and B, clones of Nintra-expressing cells (forked) in the wing blade cause the ectopic differentiation of wing margin structures (arrows) and induce the proliferation of wild-type cells surrounding the clone (dotted line). The clones generated close to the a/p boundary and away from the d/v boundary cause larger outgrowths (A) than clones close to the d/v border (B). In C, a clone (red dotted line) of Dl-expressing cells (yellow) in the dorsal compartment causes wing outgrowths that contain wing margin structures at the clone boundaries (arrows indicate mutant elements). (D) Large outgrowths in the legs associated with clones of Dl-expressing cells (arrow indicates a large outgrowth in the coxal region and arrowhead, an enlarged leg). In E, clones of Dl-expressing cell in the ventral compartment of the wing cause large outgrowth that is not associated with the differentiation of wing margin structures. The wild-type cells constitute only a small fraction of the outgrowth (dotted line). (F) Large outgrowth (dotted line) in the notum caused by a clone (arrows) of Dl-expressing cells.
Clones of Ser overexpression by using the strong promoter give similar phenotypes to that of Nintra clones when they appear in the ventral surface of the wing. Dorsal clones, however, show no outgrowths, as already described (23). By using the weak promoter, the clonal phenotypes are like those with the strong promoter, namely outgrowths in ventral clones only (data not shown).
Clones of Dl-expressing cells using the strong promoter cause outgrowths in dorsal wing surfaces, as described (23). Interestingly, clones of Dl-expressing cells, using the weak promoter, cause large outgrowths in both wing surfaces. However, whereas dorsal outgrowths are associated with the differentiation of wing margin structures (Fig. 1C), the outgrowths induced in the ventral compartment almost never differentiate wing margin elements (Fig. 1E). The rare (4/90) cases where margin structures differentiate ventrally were cases where all clones were close to the endogenous margin. In these ventral outgrowths, mutant cells form most of the tissue. Large outgrowths also appear in the proximal regions of the legs and in the notum (Fig. 1 D and F). In these regions, the clones differentiate ectopic sensory elements characteristic of the position where they appear, features suggestive of N insufficiency.
Ventral Clones of Dl-Expressing Cells Do Not Generate a d/v Boundary.
The phenotype of ventral Dl-expressing clones without margin elements suggests that the ectopic activation of Notch signaling could induce cell proliferation independently of the differentiation of a d/v wing margin. We therefore analyzed the effect of clones of Dl-expressing cells generated by flip-out recombination by using the weak promoter (low Dl expression) on the expression of different genes known to be involved in the specification of patterning of the d/v boundary, such as; cut (ct), Distalles (Dll), vestigal (vg), and wingless (wg) (6). Notch signaling at the d/v boundary induces the localized expression of these genes in both the wing margin of normal wings and the ectopic wing margin associated to clones of N-, Ser-, and Dl-expressing cells (6, 23). Clones in the dorsal surface of low Dl-expressing cells cause ectopic expression of ct in both the cells expressing the Dl ligand and the adjacent wild-type cells outside the clone (Fig. 2 B and C). The ectopic activation of ct within the dorsal clones suggests that Notch signaling is greatly enhanced in the same Dl-expressing cells, in contrast to clones of Dl-expressing cells induced by using a strong promoter, where ct is expressed only outside the clone. It has been proposed that ectopic expression of the ligands Dl or Ser at high levels has a dominant-negative effect (23, 25). By contrast, in the ventral wing surface, clones of Dl-expressing cells express ct neither in the clone nor outside it (Fig. 2 A and C).
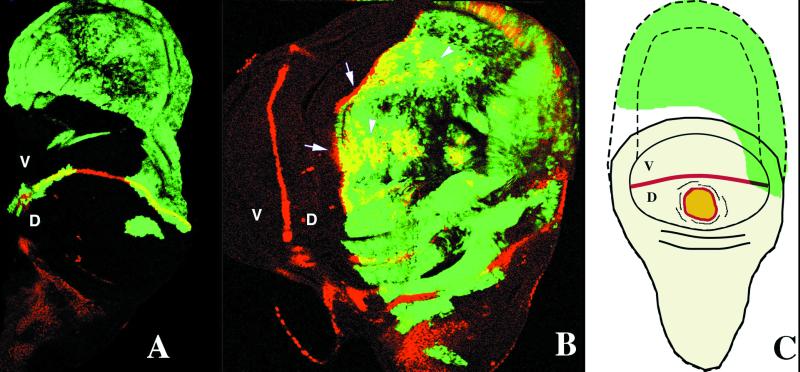
Figure 2.
Effects of clones of Dl-expressing cells (green, GFP labeled) on the expression of CT (antibody against CT, staining in red) in third instar imaginal wing discs in the dorsal (D) and ventral (V) compartments. In A, ventral clones cause large outgrowths where CT is not ectopically expressed. In B, dorsal clones of Dl-expressing cells induce the ectopic expression of CT in some cells within the clone (arrowheads) and in the wild-type cells surrounding the clone (arrows). In C, schematic representation of the effects caused by clones of Dl-expressing cells. Ventral clones that do not express CT cause large outgrowth, and dorsal clones cause the ectopic expression of CT within the clone (yellow) and in adjacent wild-type cells (red).
The same differences in clonal behavior apply to the expression of the gene Distalless (Dll), which is expressed in a wide region of the wing pouch. Dorsal clones of Dl-expressing cells ectopically express Dll in both mutant and wild-type cells, reproducing the same pattern as that in the normal d/v boundary, whereas ventral clones do not express it.
The Drosophila vestigial (vg) gene is required for wing-cell proliferation, and its expression in the wing is driven by at least two enhancers (21, 29). The vg boundary enhancer [vg(B)] is activated by Notch at the d/v boundary early in the development of the wing discs (29). The vg quadrant enhancer [vg(Q)], which acts later, drives vg expression in the developing wing blade (21). Both enhancers are activated in the outgrowth formed by clones of low Dl-expressing cells in the dorsal compartment in Dl-expressing cells as well as in the wild-type cells surrounding the clone (Fig. 3 A, C, E, and G). However, neither of the two enhancers are activated in clones of Dl-expressing cells in the ventral compartment (Fig. 3 B, C, F, and G). Thus the extraproliferation observed in ventral clones cannot be consequence of the activation of these vg enhancers. Surprisingly, an antibody against VG protein shows expression of the protein in most cells that constitute the ventral outgrowths (Fig. 3 D and I). This suggests that there are other regulatory regions of vg expression in addition to the known vg(Q) and vg(B) enhancers. Outgrowths also appear in clones of the notum and legs, but in these cases VG is not expressed (data not shown). Thus, expression of vg, which occurs throughout the mosaic wing, cannot account for the extra growth because it appears in clones in the notum and leg where vg is not expressed.
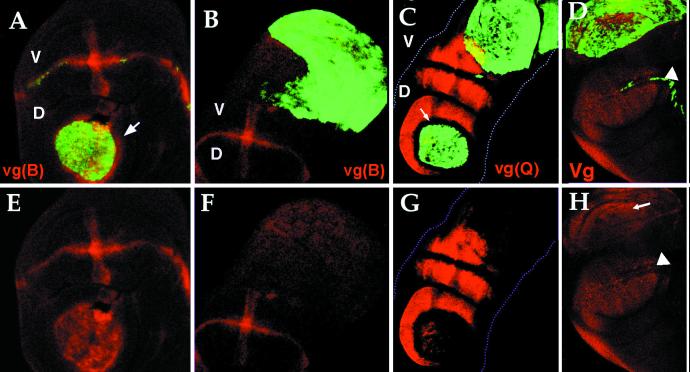
Figure 3.
Effect of clones of Dl-expressing cells (green, GFP) on the expression of vestigial boundary enhancer [vg(B)] red in (A, B, E, and F), quadrant enhancer [vg(Q)] red in (C and G), and pattern of VG protein monitored by using an antibody against VG (D and H). In A and E, the vestigial boundary enhancer [vg(B)] is activated in the outgrowth caused by a dorsal clone in both mutant and wild-type cells surrounding the clone (arrow), but not in ventral clones (B and F). In C and G, the Quadrant Enhancer [vg(Q)] is activated in the outgrowth caused by Dl dorsal clones but not in ventral. In dorsal clones the vg(Q) enhancer is repressed in wild-type cells adjacent to the clone but ectopically activated in both mutant cells and wild-type cells [arrow indicates the wild-type cells adjacent to the clone that not express vg(Q) enhancer]. In D and H, the entire wing pouch, including the outgrowth, expresses the VG protein (arrow) (arrowheads point to the d/v boundary). The cells in the outgrowth that do not show VG expression are in a different focal plane.
In the d/v boundary, the activation of the expression of wingless also depends on Notch signaling activity. This gene has been proposed to play an important role in the development of the wing, promoting growth (19). Dorsal clones of low Dl-expressing cells express high levels of wg in both the clone and in the wild-type cells adjacent to it (Fig. 4 A, C, D, and F). We also observe occasional clones of Dl-expressing cells that induce the ectopic expression of a new ring of wg expression between the two rings of cells with high levels of wg expression (Fig. 4 A and D).
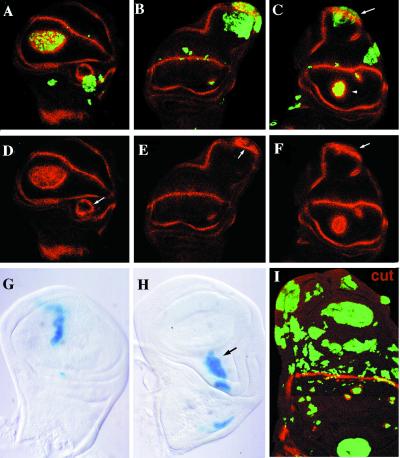
Figure 4.
Expression pattern of WG (A–F) and CT (I) (red) in wing discs with clones of Dl-expressing cells (green, GFP); clones of wg-expressing cells (G and H) (blue, β-galactosidase). In A–F, different examples of clones of Dl-expressing cells. In A, C, D, and F, dorsal clones express high levels of WG within the clone as well as in the wild-type cells adjacent to the clone (arrowhead in C). In B, C, E, and F, the clones in the ventral compartment that abut the external and internal rings of WG expression cause large outgrowths with elongation of the internal ring of WG expression (arrows in C). High levels of WG in some cells within the clone can be observed (arrows in E and F). In G and H, clones of wg-expressing cells in the wing blade never cause outgrowths, whereas they do in notum and wing base (H). In I, compare the small size of clones of Dl-expressing cells closer to the d/v boundary with the large ones in the proximal region of the wing.
In the ventral compartment, however, clones of Dl-expressing cells that touch the external and internal rings of wg expression in the base of the wing cause large outgrowths and, as a consequence, the internal and external rings of wg expression appear elongated. These outgrowths are again formed by both mutant and wild-type cells (Fig. 4 B, C, E, and F). The ectopic growth observed in Dl-expressing cells could be a direct consequence of the ectopic expression of wg induced by Notch. We have therefore studied the effects of ectopic expression of wg in clones. The ectopic expression of wg using the weaker promoter never causes any phenotype (19). However, when we use the strong (abx/Ubx) promoter, we observe that these clones induce large outgrowths in the hinge and notum regions with wing histotype (data not shown). The same large clones in the wing blade never cause outgrowths (Fig. 4 G and H) (23). In adult wings, these wg clones are associated with the differentiation of wing margin structures but not with outgrowths (data not shown) (25, 41).
As with Nintra-expressing clones, the size and effect on cell proliferation caused by Dl-expressing clones depend on the position where the clones were initiated. In contrast to clones expressing Nintra that are always small, clones of Dl-expressing cells are in general much larger. However, clones close to the d/v boundary are smaller than clones in proximal regions of the wing blade or close to the a/p boundary (Fig. 4I). This is an indication that cell proliferation in outgrowths is negatively correlated with the distance to the d/v boundary. In addition, proximal clones cause lesser nonautonomous effects in the surrounding wild-type cells than distal clones. We can clearly establish a topological relationship between the size of the outgrowth and its position in the wing blade more clearly in adult wings. There, the final size of the outgrowth (autonomous or nonautonomous effects) depends on the distance between the place where the clones were generated and the tip of the wing rather than on the size of the mutant clone.
Cell Proliferation Dynamics in Dl Ectopic Expression.
The results presented above suggest that the activity of Notch signaling could have a direct effect on the control of cell proliferation. We have analyzed the cell-cycle dynamics in clones of Dl-expressing cells. In wild-type wing discs, cells in the different phases of the cell cycle appear mainly in small synchronous clusters that are not clonally derived (19, 34, 42). The G2/M transition was here monitored by the accumulation of the string (stg) gene product. Using the UAS/GAL4 system, we have overexpressed Dl throughout the dorsal compartment of the wing (using the GAL4-MS1096 line). We found that in the dorsal compartment, the number of cells labeled with string is higher than in the ventral compartment (data not shown). Because the high number of clusters makes it difficult to identify individual clusters, we have further analyzed mitotic cells corresponding to a shorter phase of the cycle by using Hoechst 33258. The numbers of cells in mitosis (meta and anaphases) is higher in Dl ectopic expression regions (average 30 ± 7) than in controls (average 22 ± 6).
Discussion
In Drosophila, the Notch signaling pathway is involved in processes of cell specification, such as the determination of neuroblasts (43) and wing vein cells (44). In these processes, Notch activity prevents cell determination. The Notch pathway also operates in cell patterning and morphogenesis, such as in leg joints (45) and the wing margin (6). In these processes, reduction of Notch activity leads to both decreased proliferation and cell fate changes. The effects of Notch on proliferation, however, are thought to be mediated by the activation of downstream genes that promote cell proliferation. It has been proposed that Notch may contribute to wing proliferation and patterning by the activating vg and wg in the wing margin (6). In this view, the enlarged wing discs seen in Ax mutants (N gain-of-function allele) could result from the ectopic activity of N close to the d/v boundary, which is associated with an expanded expression of wing margin genes over the wing blade (26, 46).
The results presented here indicate that Notch activation leads to cell proliferation directly, independently of the generation of a d/v boundary as a reference for growth. The overexpression in the wing discs of an activated form of N (Nintra) or its ligands (Dl and Ser) in clones causes extra growth. These outgrowths display wing margin structures and ectopic expression of vg and wg in the tip of the outgrowth (23) and are formed by mutant cells in the tip and wild-type cells in its stem, the latter frequently constituting the majority of the outgrowth. The extraproliferation of the wild-type cells is considered to result from a nonautonomous response to proliferation signals emanating from the ectopic d/v boundary (6). A topological analysis of these outgrowths reveals that Nintra or Ser clones close to the wing margin contain mutant cells and few wild-type cells, whereas clones away from the wing margin (in the wing base or close to the a/p boundary) contain large outgrowths formed mainly by wild-type cells. These outgrowths display pattern landmarks, such as veins, which allow the identification of pattern duplications resulting from intercalary proliferation. This gives a continuous pattern of positional values of the mosaic wing. Similar intercalary growth occurs in clones that overexpress en+ (the selector gene for posterior compartment) in the anterior compartment or in en− or ap− clones in the posterior or dorsal compartments, respectively (11). The size of the outgrowth varies depending on the distance from the a/p boundary in the first case or the d/v boundary in the second. We have interpreted these findings as resulting from positional value “accommodation” of mutant and wild-type cells in mosaics (47). Accommodation reflects merely local control of cell proliferation to attain all differential positional values, as opposed to growth driven by morphogen signals emanating from reference boundaries acting at long distances.
Clones that overexpress Dl under a strong promoter are like those of Nintra: margin elements and gene expression pattern characteristic of the wing margin abut the clones from the outside. The outgrowths caused by clones of low Dl overexpression also appear in both the dorsal and ventral wing surface (as in clones of activated forms of Notch), but clones in the ventral surface show ectopic expression of d/v boundary markers (ct, Dll, wg) neither within the clone nor outside it. Both dorsal and ventral outgrowths contain Dl mutant and nonmutant cells. Significantly, the mutant territory is larger in ventral than in dorsal clones. Thus, ventral outgrowths are contributed mainly by Dl-expressing cells that in the adult are not associated with pattern duplications. This is an indication of lack of accommodation to high positional values and reflects merely more cell proliferation.
The different clonal responses to Dl depending on the amount of its overexpression could reflect degrees of Notch activation. Thus, high levels of Dl are associated with dominant-negative effects on the Notch protein (revised in ref. 1). It is likely that the lower levels of Dl obtained with the weak GAL4 promoter, compared with those with the strong promoter, are sufficient to activate Notch but insufficient to produce a dominant-negative effect.
The observation that outgrowths caused by weak Dl overexpression in the ventral wing surface are not accompanied by the expression of wing margin markers begs the question of how activated Notch signaling causes extra cell proliferation. We have shown that the overexpression of Dl does not lead to the expression of wg, a Notch downstream gene considered directly responsible for extra cell proliferation. In addition, the overexpression of wg by itself in the wing blade of wild-type wings does not lead to extra growth (25). The overexpression of Dl induces the expression of neither the vg(B) (boundary) nor the vg(Q) (quadrant) enhancers of vg. The fact that VG protein appears in the outgrowth caused by Dl-expressing cells suggests that vg has other enhancers. Importantly, the extra growth cannot be explained by vg expression because it is ubiquitous in the wing blade. Moreover, the Dl-expressing clonal outgrowths in the notum and legs are not accompanied by VG expression.
Thus, the outgrowths caused by Dl overexpression do not seem to result from secondary consequences of the generation of a wing margin or from the activity of genes active in the wing margin that supposedly promote growth. This conclusion leads us to consider the possibility that Notch activity is directly involved in cell proliferation. In fact, N− recombinant cells anywhere in the wing fail or proliferate much less than control clones when they are induced early or late, respectively (27). It remains open which genes involved in cell proliferation are activated by Notch activity. The epidermal growth factor receptor (Egfr) pathway is one candidate (48). It has been shown that clones doubly mutant for N and members of the Egfr pathway fail to proliferate or proliferate less that control N, suggesting synergistic interactions (27). A role of N− in cell proliferation has been proposed for the cell cycle phase transitions in the wing margin (49). Similarly, there is suggested involvement of the N homologue in Caenorhabditis elegans (GLP-1) in the proliferation of germ cells, because a constitutively active form of this gene prevents exit of the cells from the mitotic cycle (50). In the same line are our findings that Ax mutant discs grow faster and continue to grow longer than normal discs, while failing to differentiate wing pattern elements. Thus, the role of N on cell proliferation and cell differentiation in different morphogenetic processes may be reduced to its function in maintaining cell proliferation as opposed to cell differentiation.
Acknowledgments
We thank Drs. J. Campos-Ortega, J. F. de Celis, T. Casci, M. Freeman, and A. M. Martínez-Arias for constructive criticism of the manuscript. We also thank S. M. Cohen, S. B. Carroll, F. Díaz-Benjumea, J. F. de Celis, and M. A. T. Muskavitch for generously providing flies and antibodies. P. Martín, R. Hernández, A. López, and A. Hernando contributed with skillful technical assistance. A.B. is a postdoctoral fellow of the Comunidad Autonoma de Madrid. This work was supported by grant PB92–0036 of the Comision Interministerial de Ciencia y Tecnologia (Spain) and by an institutional grant from the Fundación Ramón Areces to the Centro de Biologia Molecular “Severo Ochoa.”
Abbreviations
- d/v
dorsal/ventral
- a/p
anterior/posterior
- AEL
after egg laying
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040576497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040576497
References
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Knust E, Schrons H, Grawe F, Campos-Ortega J A. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrons H, Knust E, Campos-Ortega J A. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecourtois M, Schweisguth F. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 6.Irvine K D, Vogt T F. Curr Opin Cell Biol. 1997;9:867–876. doi: 10.1016/s0955-0674(97)80090-7. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S M. In: The Development of Drosophila melanogaster. Bate M, Martínez Arias A, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 8.García-Bellido A, Ripoll P, Morata G. Nat New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Bellido A. In: Cell Patterning. Porter R, Rivers J, editors. Vol. 29. Boston: CIBA Symposium; 1975. pp. 161–183. [Google Scholar]
- 10.Blair S S. BioEssays. 1995;17:299–309. doi: 10.1002/bies.950170406. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Benjumea F, Cohen S M. Cell. 1993;75:741–752. doi: 10.1016/0092-8674(93)90494-b. [DOI] [PubMed] [Google Scholar]
- 12.Cohen B, MacGuffin M E, Pfeifle C, Segal D, Cohen S M. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- 13.Blair S S, Brower D L, Thomas J B, Zabortink M. Development (Cambridge, UK) 1994;120:1805–1815. doi: 10.1242/dev.120.7.1805. [DOI] [PubMed] [Google Scholar]
- 14.Irvine K D, Wieschaus E. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Irvine K D, Carroll S B. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- 16.de Celis J F, García-Bellido A, Bray S J. Development (Cambridge, UK) 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 17.Doherty D, Feger G, Younger-Shepherd S, Jan L Y, Jan Y N. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- 18.Couso J P, Knust E, Martínez-Arias A. Curr Biol. 1995;5:1437–1448. doi: 10.1016/s0960-9822(95)00281-8. [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Benjumea F J, Cohen S M. Development (Cambridge, UK) 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- 20.Rulifson E J, Blair S S. Development (Cambridge, UK) 1995;121:2813–2824. doi: 10.1242/dev.121.9.2813. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Sebring A, Esch J J, Kraus M E, Vorwerk K, Magee J, Carroll S B. Nature (London) 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 22.Neumann C J, Cohen S M. Development (Cambridge, UK) 1996;122:3477–3485. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- 23.de Celis J F, Bray S. Development (Cambridge, UK) 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 24.Micchelli C A, Rulifson E J, Blair S S. Development (Cambridge, UK) 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 25.Klein T, Martinez-Arias A. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- 26.Go J M, Eastman D S, Artavanis-Tsakonas S. Development (Cambridge, UK) 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- 27.de Celis J F, Garcia-Bellido A. Mech Dev. 1994;46:109–122. doi: 10.1016/0925-4773(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 28.de Celis J F, Garcia-Bellido A. Genetics. 1994;136:183–194. doi: 10.1093/genetics/136.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams J A, Paddock S W, Vorwek K, Carroll S B. Nature (London) 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- 30.Huppert S S, Jacobsen T L, Muskavitch M A T. Development (Cambridge, UK) 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence P A, Bodmer R, Vincent J P. Development (Cambridge, UK) 1995;121:4303–4308. doi: 10.1242/dev.121.12.4303. [DOI] [PubMed] [Google Scholar]
- 32.Capdevila J, Guerrero I. EMBO J. 1994a;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. Development (Cambridge, UK) 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 34.Milán M, Campuzano S, García-Bellido A. Proc Natl Acad Sci USA. 1996;93:640–645. doi: 10.1073/pnas.93.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domínguez M, Campuzano S. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams J A, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 37.Brook W J, Cohen S M. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- 38.Blochlinger K, Jan L Y, Jan Y N. Genes Dev. 1991;5:1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- 39.Edgar B A, O'Farrell P H. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welshons W J. Genetics. 1971;68:259–268. doi: 10.1093/genetics/68.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein T, Martinez-Arias A. Dev Biol. 1998;194:196–212. doi: 10.1006/dbio.1997.8829. [DOI] [PubMed] [Google Scholar]
- 42.Milán M, Campuzano S, García-Bellido A. Proc Natl Acad Sci USA. 1996;93:11687–11692. doi: 10.1073/pnas.93.21.11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campos-Ortega J A, Knust E. Annu Rev Gen. 1990;24:387–407. doi: 10.1146/annurev.ge.24.120190.002131. [DOI] [PubMed] [Google Scholar]
- 44.de Celis J F, Bray S, García-Bellido A. Development (Cambridge, UK) 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 45.de Celis J F, Tyler D M, de Celis J, Bray S J. Development (Cambridge, UK) 1998;125:4617–4626. doi: 10.1242/dev.125.23.4617. [DOI] [PubMed] [Google Scholar]
- 46.de Celis J F, de Celis J, Ligoxiars P, Preiss A, Delidakis C, Bray S. Development (Cambridge, UK) 1996;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- 47.García-Bellido A, Cortés F, Milán M. Proc Natl Acad Sci USA. 1994;91:10222–10226. doi: 10.1073/pnas.91.21.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diaz-Benjumea F, Hafen E. Development (Cambridge, UK) 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- 49.Johnston L A, Edgar B A. Nature (London) 1998;394:82–84. doi: 10.1038/27925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry L W, Westlund B, Schedl T. Development (Cambridge, UK) 1997;124:925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]