Abstract
The direct developing anuran, Eleutherodactylus coqui, lacks a tadpole, hatching as a tiny frog. We investigated the role of the metamorphic trigger, thyroid hormone (TH), in this unusual ontogeny. Expression patterns of the thyroid hormone receptors, TRα and TRβ, were similar to those of indirect developers. TRβ mRNA levels increased dramatically around the time of thyroid maturation, when remodeling events reminiscent of metamorphosis occur. Treatment with the goitrogen methimazole inhibited this remodeling, which was reinitiated on cotreatment with TH. Despite their radically altered ontogeny, direct developers still undergo a TH-dependent metamorphosis, which occurs before hatching. We propose a new model for the evolution of anuran direct development.
The term “amphibian” conveys the image of a dual life history, split between water and land. However, 29 different anuran reproductive modes have been defined, on the basis of the site of egg development (1). Included in this developmental smorgasbord are species in which the larvae develop inside a foam nest (Chiromantis), those that brood live embryos in their stomach (Rheobatrachus), “marsupial frogs” in which the tadpoles develop within a parental pouch (Gastrotheca), and the most extreme modification, direct development, where the free-living larva has been deleted from the life history (Eleutherodactylus).
Direct development in Eleutherodactylus is a derived characteristic (2), yet embryogenesis in these frogs is so radically modified that little hint of their biphasic metamorphic ancestry is visible. Development is entirely terrestrial, with tiny froglets hatching from their jelly capsules 3 wk after fertilization. Many larval characteristics are lacking, including cement gland, lateral line organs, coiled gut, larval mouth parts, and some cranial cartilages (3). The only obviously larval feature is the tail, which shrinks before hatching.
The ontogenetic modifications responsible for direct development are unknown. Because direct developers are thought to have evolved from biphasic frogs (1, 2), the role of thyroid hormone (TH) in direct development is of particular interest. TH, shown in other systems to signal through thyroid hormone receptors (TRs), controls the larval-to-adult transition in biphasic frogs (4). TRβ expression in both Xenopus laevis and Rana catesbeiana is temporally correlated with the onset of metamorphosis, implicating this gene as a likely metamorphic mediator (5–7). The role of TH in direct development is unclear. Both chemical and surgical ablation of the thyroid axis in Eleutherodactylus martinicensis inhibited tail regression and pronephric degeneration (8, 9). These events were precociously induced by 3,3′,5-triiodothyronine (T3) treatment, but other organ systems appeared unaffected (8). Although the similarity between such degenerative changes in Eleutherodactylus and those occurring in metamorphosis was noted, the specificity of effects caused by drug treatment was questioned. It was thought likely by these authors that direct development was largely independent of thyroid functioning (8, 9).
Treatment of embryos with T3 resulted in the precocious induction of the urea-cycle enzyme, arginase, in the liver (10), paralleling the metamorphic response of the liver in other frogs (11). Outgrowth of limb explants was not stimulated by T3 (12). The thyroid gland is first detectable about 1.5 wk after fertilization, at Townsend–Stewart stage 10 (TS10) (13) and appears most active during the last third of development [TS12–15 (14)]. Because thyroid maturation occurs well after hatching in biphasic frogs, yet before hatching in Eleutherodactylus, it was suggested that the thyroid axis has been heterochronically accelerated in direct developers (14, 15).
Here, we report the temporal expression pattern of TRs in Eleutherodactylus coqui and the developmental effects of methimazole treatment, which inhibits TH synthesis. Our results demonstrate that TH is necessary for the completion of direct development in E. coqui and lead us to propose a new model for the evolution of direct development.
Materials and Methods
Embryo Culture and Drug Treatment.
Adult E. coqui were collected at the El Verde Field Station, Puerto Rico, under permits issued by the Departamento de Recursos Naturales, Puerto Rico. Mating pairs were housed at 26°C with a 12-hr light/dark cycle. Embryos were obtained from spontaneous matings, staged according to Townsend and Stewart (13), and cultured in 20% Steinberg's solution [12 mM NaCl/130 μM KCl/70 μM Ca(NO3)2/170 μM MgSO4/920 μM Tris, pH 7.4]. T3 was dissolved in 1 M NaOH to produce a 2.5 mM stock solution. A 50-nM working solution was prepared by dilution of the stock with 20% Steinberg's solution and adjustment of the pH to 7.4. Methimazole was dissolved in 20% Steinberg's solution, to produce a 1 mM working solution. Embryos were anesthetized in 0.2% 3-aminobenzoic acid ethyl ester (MS222) before sacrifice.
TR Cloning.
Total RNA was isolated by using the Trizol method (BRL) from TS13 embryos that had been immersed in 1.5 μM T3 for 3 days. Reverse transcription and PCR were performed with oligonucleotides previously used to clone R. catesbeiana TRα and TRβ (16). PCR conditions were 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, with a 10-sec extension per cycle. The 254-bp TRα and 258-bp TRβ PCR products were gel purified and cloned into pBluescript II SK+ (Stratagene). Sequencing indicated that E. coqui TRα and TRβ shared 94% and 91% identity, respectively, with their R. catesbeiana orthologues, and the plasmids were named EcTRα and EcTRβ.
Ribosomal Protein L8 Cloning.
Ribosomal protein L8 (L8) was amplified from TS12 liver cDNA that had been primed with random hexamers. PCR primers were designed by using pc/gene based on the orthologous X. laevis and Homo sapiens sequences. Primer sequences were (5′-3′): CAGGAATTCGACATTATCCATGATCCAGGCCG (+); GCAAAGCTTCAGTCTTTGTACCGCGCAGACG (−). Underlined regions indicate restriction sites added to facilitate subcloning. Amplification conditions were 26 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a 10-sec extension per cycle. A 616-bp product was purified and subcloned as for the TRs. The plasmid was partially sequenced and named EcL8.
Northern Blotting.
Four micrograms of total embryo RNA was separated on a formaldehyde gel and blotted onto a Hybond N+ membrane (Amersham) overnight. Blots were baked at 80°C for 1.5 hr and prehybridized for 30 min at 65°C in a hybridization oven in Rapid Hyb buffer (Amersham). L8 probe was random primed by using the NEBlot labeling kit (NEB). Overnight hybridization at 65°C was followed by two 15-min low-stringency washes at 55°C and two 15-min high-stringency washes, also at 55°C. The low-stringency washes were in 2× SSC, 0.1% SDS, and the high-stringency washes were in 0.1% SSC, 0.1% SDS.
Reverse Transcription (RT)–PCR Assay.
The assay was based on the method devised for the relative quantitation of the multidrug resistance gene, mdr-1 (17). RT was performed by using random hexamers, with 1 μg total embryo RNA as template. PCR conditions were 26 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. Amplifications from different primer sets were performed in separate reactions. The E. coqui primers used were (5′-3′), TRα: CACCCGAAATCAGTGCCAGC (+), GATCATCTCCTCCTTCCGACGC (−); TRβ: TTTGGATGACAGCAAGCGTTTGGC (+), CCCACTCTTCTGATGTTGGCTCAGG (−); L8: GCTCCTCTTGCCAAGGTTGC (+), TCATAGCCACACCACGGACACG (−). The molecular sizes of the amplified products of TRα, TRβ, and L8 were 148, 122, and 448 bp, respectively. PCR products were analyzed on a 5% acrylamide/1× Tris–borate/EDTA gel and electroblotted onto Hybond N+ membrane, which was alkali denatured and baked. PCR products were detected by hybridization with end-labeled gene-specific oligos: CCTGGATGATTCCAAGCGGGTAGCC (TRα); CGCAAGCACGAACTGCAGAAAACAG (TRβ); CGTGATCCCTACAGGTTTAAGAAG (L8). Overnight hybridization at 40°C was followed by 1× low-stringency wash and 2× high-stringency washes, each for 15 min at 40°C. The low-stringency wash was in 5× SSC, 0.1% SDS. The α and β high-stringency washes were in 0.2× SSC, 0.1% SDS; the L8 low-stringency wash was in 1× SSC, 0.1% SDS. Signals were quantitated by using the Storm 860 PhosphorImager (Molecular Dynamics) with imagequant software. The linear amplification range for each primer set was determined by amplification of a dilution series of cDNA. In subsequent assays, the input cDNA amount was within the linear range. Input cDNA amounts (in RNA equivalents) for the different genes analyzed were: ≤25 ng for TRα, ≤75 ng for TRβ, and ≤25 ng for L8. All cDNA amplifications were duplicated. The input cDNA quantity/quality between different samples within a series was controlled by expressing the TRα and TRβ signals relative to the L8 signal from that sample. L8 is a ribosomal protein expressed at constant levels throughout development (data not shown). The values quoted in Results for TRα and TRβ levels are normalized values representing the TR/L8 ratios of the samples.
Morphometry.
Embryo dimensions were measured by using an ocular micrometer in a Nikon dissecting microscope. The Kruskal–Wallis and Tukey multiple comparisons tests were performed by using the kwikstat 4 program (TexaSoft, Dallas).
Whole-Mount Immunochemistry.
Embryos were fixed in Dent's fixative (80% methanol/20% DMSO), skinned, stored at −20°C, and examined by whole-mount immunochemistry (18). The 12/101 monoclonal mouse antibody against skeletal muscle, obtained as a hybridoma culture supernatant from the Developmental Studies Hybridoma Bank, Iowa City, IA, was used at a 1:20 dilution. A goat anti-mouse alkaline phosphatase-conjugated secondary antibody was used at a 1:500 dilution.
Cartilage Staining.
Embryos were processed for cartilage staining with Alcian blue according to ref. 18.
Results
Establishment of Assay Conditions.
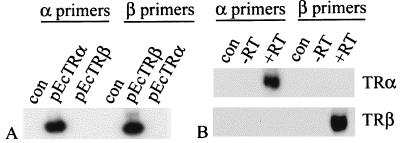
Expression of E. coqui TRs was examined by using a relative-quantitative reverse transcription–PCR assay in which expression of the TRs was normalized relative to the levels of ribosomal protein L8, which was found by Northern blotting to be expressed at constant levels throughout the developmental stages examined (data not shown). Sequence comparison showed that the E. coqui TR paralogues shared 74% identity, so gene specificity of TR amplification was confirmed by performing PCR by using 0.1 ng of either EcTRα or EcTRβ vector as template, with TRα and TRβ assay primers added in separate reactions. These primers did not amplify the paralogous gene product (Fig. 1A). The end-labeled TR oligos did not crossreact (Fig. 1B), providing further evidence of the assay's specificity.
Figure 1.
PCR specificity. (A) The TRα- and TRβ-specific primers do not amplify the paralogous receptors. Plasmids were used as templates. dCTP (1 μCi) was added to each reaction, and the gel was exposed to film. (B) The TRα- and TRβ-specific oligo probes do not crosshybridize with the paralogous receptor PCR products. TS12 embryo cDNA was used as template. Controls are water blanks.
Developmental Expression of TR mRNAs.
The developmental expression patterns of TRα and TRβ mRNAs are shown in Fig. 2 Left. TRβ mRNA levels changed dramatically during embryogenesis. In this developmental series, TRβ mRNA was barely detectable at TS5. Although no band was visible in this TS7 sample, TRβ expression was detected as faint bands in nine pre-TS9 embryo samples, demonstrating that TRβ mRNA is present at very low levels before development of the thyroid gland. By TS10, when the thyroid gland becomes histologically apparent (14), TRβ mRNA was clearly detectable, and levels continued to rise during the last third of embryogenesis. Surprisingly, TRβ mRNA levels remained high even after hatching. In three of four developmental series, the posthatching levels were higher than at any stage during embryogenesis. Mean TRβ mRNA levels from 1- to 2-wk posthatching froglets were 10% greater than those found in TS14/TS15 animals. This contrasts with the situation in X. laevis, where TRβ mRNA levels decrease markedly on completion of tail resorption (5). Substantial TRα expression occurred early in embryogenesis (Fig. 2). In contrast with TRβ levels, TRα expression in the same embryos was much more constant. Although some variation was observed between individual samples, no clearly discernible developmental trends were evident.
Figure 2.
Developmental expression of TRs, analyzed by RT-PCR (Left). 2wk ph, 2 wk posthatching. Drawings of selected stages are depicted over the relevant lanes. No products were detected in the absence of RT (data not shown). TR expression in full grown unmatured oocytes relative to TS12 levels (Right). L8 is the loading control.
To quantitate the changes in TR expression levels during embryogenesis, mRNA levels between early and late stage embryos were compared (Table 1). TRα mRNA levels showed at most a modest increase, whereas TRβ mRNA expression underwent a massive developmental induction in the same embryos, which was over an order of magnitude in four of six cases.
Table 1.
Developmental up-regulation of TR RNAs
| Early stage | Late stage | Fold increase
|
|
|---|---|---|---|
| TRα | TRβ | ||
| TS4 | 1 wk posthatch | 2 | 12 |
| TS5 | Hatching | 1 | 10 |
| TS6 | 1 wk posthatch | 1 | 3 |
| TS7 | Hatching | 1 | 6 |
| TS7 | Hatching | 3 | 25 |
| TS7 | TS14 | ND | 14 |
Six experiments quantitating the fold increase in TR levels between early and late stages of embryogenesis. A value of 1 indicates equivalent levels at the two stages. All values were normalized relative to L8 expression. TRα was not measured in one sample set (ND). ND, not determined.
To address the possibility that maternal TR mRNA stores may facilitate direct development, TR mRNA expression in the E. coqui full-grown but unmatured oocyte was examined. The oocyte was loaded with mRNAs of both receptors (Fig. 2 Right), and TRβ mRNA levels were substantially higher than those found in TS12 embryos, a stage when zygotic induction of this gene is well underway.
Thyroid Dependency of Direct Development.
TS8–10 embryos (n > 50) were treated with 1 mM methimazole, an inhibitor of TH synthesis that prevents metamorphosis in X. laevis (19). Development was normal until TS12, at which stage embryogenesis arrested (Fig. 3). The ontogenetic effects of methimazole were pleiotropic. Effects on head, limb, muscle, tail, and skin development are described below. Treatment of early embryos (TS4–6, n = 24) did not enhance the effect of methimazole, presumably because zygotic hormone synthesis had not yet begun.
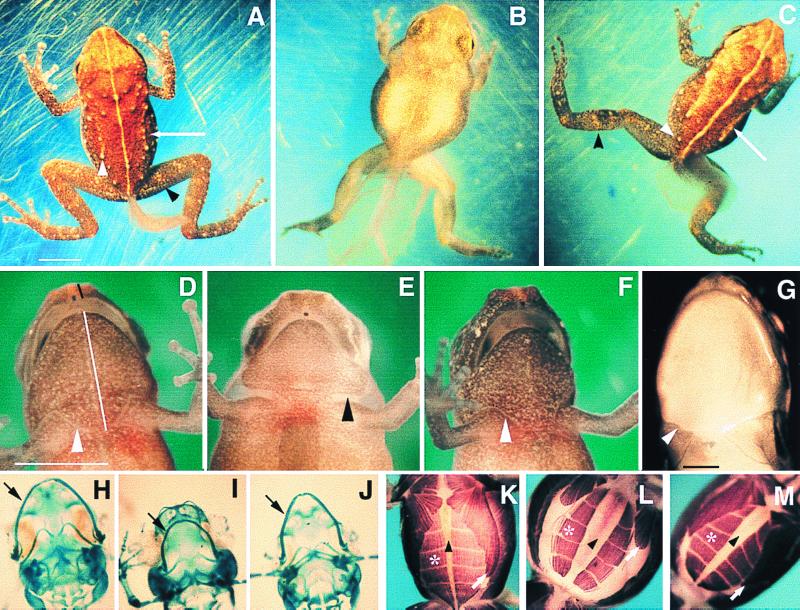
Figure 3.
Developmental effects of methimazole treatment. All E. coqui specimens are early posthatching stage. Scale bars represent 2 mm. (A) Control froglet with regressing tail. Adult skin characters include leg bands (black arrowhead), tubercles (white arrowhead), and raised dorsolateral ridges (white arrow). (B) Methimazole-treated sibling. (C) T3-rescued froglet. (D) Control froglet, ventral view. Skin has sutured (arrowhead). Black line indicates “snout–upper jaw” length, and white line represents the “lower jaw–suture line” length, measured for morphometric analysis (Table 2). (E) Methimazole-treated embryo, with a gap between head and trunk skin (arrowhead). (F) T3-rescued embryo. (G) Suture marks of newly metamorphosed Rana pipiens (arrowhead). (H–J) Meckel's cartilage (arrow) in control (H), methimazole-treated (I), and T3-rescued animals (J). (K) Control froglet, exhibiting convergence of the rectus abdominis (*) on the midline (arrowhead) and posterior elongation of the pectoral muscles (white arrow), events inhibited by methimazole (L) and rescued by T3 (M).
Although development was arrested, the embryos appeared healthy, indicating that the effects of methimazole were not caused by drug toxicity. Embryos cultured in methimazole for 3 wk after the controls reached hatching stage remained morphologically inhibited. Methimazole-treated embryos developed toepads and eyelids, which normally appear after TS13, indicating that the effects of methimazole were caused by specific inhibition of the embryonic thyroid axis and not merely by general developmental retardation. This was conclusively demonstrated by rescuing ontogenesis with exogenous TH. TS8–10 embryos (n > 50) treated with 1 mM methimazole were subsequently cotreated with 50 nM T3 for 8 days, starting at TS13. Development was substantially rescued (Fig. 3C). Some rescued embryos were morphologically indistinguishable from controls, although in many cases lower-jaw remodeling was incomplete. Whereas methimazole-arrested embryos remained aquatic, both control and rescued froglets adopted a terrestrial habitat, hopping out of the culture solution and suspending themselves from the Petri-dish lid. Treatment with T3 for longer periods generally resulted in death.
Morphometric measurements were made to determine quantitatively the extent and significance of differences between control, methimazole-inhibited, and T3-rescued embryos. Characters selected for morphometric analysis were hindlimb and tail length, jaw shape, and ventral body width. In all cases, the methimazole-treated embryos were significantly different from their untreated siblings (Table 2). With the exception of thigh length, rescue appeared complete, because no significant differences were found between control and T3-rescued embryos.
Table 2.
Morphometric analysis of methimazole-treated embryos
| Morphological character | Mean value ± 1 SD
|
||
|---|---|---|---|
| Control | Meth | Meth/T3 | |
| Snout–upper jaw* | 0.3 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.1 |
| Lower jaw–suture* | 2.5 ± 0.2 | 2.0 ± 0.2 | 2.3 ± 0.2 |
| Calf† | 2.7 ± 0.2 | 2.2 ± 0.2 | 2.7 ± 0.2 |
| Thigh† | 2.8 ± 0.2 | 2.0 ± 0.1 | 2.4 ± 0.1 |
| Tail | 3.2 ± 2.2 | 5.6 ± 0.5 | 2.5 ± 0.8 |
| Ventral width | 2.8 ± 0.2 | 3.5 ± 0.2 | 2.9 ± 0.2 |
Statistically significant differences (P < 0.05) between means of different groups within a row are indicated by bold font. Meth, methimazole-treated; Meth/T3, methimazole/T3 cotreatment.
*Measurement points indicated on Fig. 3D.
†Length:width ratios; all other values are in millimeters.
In metamorphosing anurans, the larval head is radically remodeled during metamorphosis. The mouth parts of methimazole-treated E. coqui were indistinguishable from controls until TS12, but elongation of the lower jaw during TS13 was inhibited. The prominent snout typical of froglets did not form; instead, the methimazole-treated embryos had a rounded head, characteristic of the embryonic head shape (Fig. 3 D–F). Morphometric measurements of head shape (snout–upper jaw, lower jaw–suture line) demonstrated that remodeling was significantly inhibited by methimazole treatment and restored to control levels by cotreatment with T3 (Table 2). Examination of Meckel's cartilage (Fig. 3 H–J), which extends dramatically during anuran metamorphosis (20), indicated that elongation of this element in E. coqui was inhibited by methimazole treatment, an effect that was partially rescued by T3 treatment (n = 14 per treatment group).
Limb buds appear during TS4 of E. coqui development, shortly after neural fold closure. Although limb bud formation in E. coqui is temporally comparable with amniotes, it is heterochronically accelerated relative to metamorphosing anurans, where limbs form after hatching (21). Methimazole treatment, even from early stages (TS4–6, n = 24), had no effect on foot paddle (TS7) or limb differentiation, and digits formed normally at TS9. During TS12, limbs normally undergo rapid elongation. This growth was inhibited by methimazole, and the hindlimbs appeared short and stumpy (Fig. 3B). Inhibition of limb elongation was confirmed by the finding of significant differences in both the length/width ratios of both the lower leg and thigh regions of the hindlimb (Table 2). T3 cotreatment completely rescued the dimensions of the lower leg segment and partially rescued those of the thigh.
Morphometric analysis of the ventral width of embryos indicated that the methimazole-treated embryos were significantly wider than their control and T3-rescued counterparts (Table 2). During normal development, embryonic ventral width decreases. This is coincident with the repositioning of the rectus abdominis muscle bands from their initial lateral position to a more ventral one and also with the growth of the body musculature (22). Because in tadpoles this ventral convergence of the rectus abdominis occurs during metamorphosis (23, 24), the axial musculature was examined to determine whether this event is TH dependent in E. coqui (Fig. 3 K–M). Movement of the rectus abdominis toward the ventral midline was inhibited by methimazole, and a large amount of yolk was visible on the ventrum. Additionally, the pectoral muscle did not extend posteriorly but remained small and similar in appearance to a TS12–13 embryo. The lateral muscle bands, composed of the musculus obliquus externus and the musculus transversus, normally extend both dorsally and ventrally to cover the yolk between the dorsal axial musculature and the rectus abdominis. In methimazole-treated embryos, these lateral muscles remained as a relatively narrow band, so a significant amount of yolk was visible when the embryos were viewed from a lateral aspect. Cotreatment with T3 reinitiated remodeling of all affected muscle groups.
The tail is one of the few organs in which TH has been developmentally implicated in Eleutherodactylus. T3 treatment of TS7–9 embryos caused precocious tail regression in E. coqui (12), and treatment of E. martinicensis with the goitrogen phenylthiourea (PTU) inhibited tail resorption (8). Methimazole, like PTU, inhibited tail regression. Cotreatment with exogenous T3 restored tail resorption to control levels (Fig. 3C; Table 2).
In contrast with the highly differentiated skin of early posthatching embryos, the skin of methimazole-treated embryos had a uniform appearance (Fig. 3 A and B), lacking tubercles, complex pigmentation, dorsolateral ridges, and leg bands typical of hatched froglets. The predominant pigment cell was the melanophore, but the concentration of this cell type was insufficient to obscure the underlying yolk, which was visible through the dorsal surface. The skin of T3-rescued embryos was indistinguishable from that of hatchling frogs (Fig. 3C). Methimazole also inhibited suturing of the ventral epidermis at the junction of the head and the trunk that normally occurs during TS13 (Fig. 3 D and E). T3 treatment rescued these suturing events (Fig. 3F), which have an intriguing parallel in biphasic frogs, where ventral suturing occurs in the corresponding region after the limbs burst through the operculum during metamorphic climax (25, 26).
Temporal Sensitivity to Methimazole.
Because methimazole affects the synthesis of TH, the effects of methimazole should diminish once zygotic hormone synthesis begins. This was tested by commencing methimazole treatment at progressively later stages. Methimazole treatment at TS9 (n = 24) caused inhibition of development, as described above. Embryos treated at TS11 showed varying degrees of further development, including partial limb growth and partial skin transformation (n = 20). All embryos treated after TS11 (n = 25) exhibited extensive remodeling, including skin transformation and suturing, limb elongation, and lower-jaw extension. Many of the TS13-treated embryos looked like normal froglets, although several retained large tails.
Discussion
E. coqui developmental TR mRNA expression patterns are similar to those in X. laevis. TRα mRNA is detectable within a few days of fertilization (5), at Nieuwkoop and Faber (NF) stage 35 in X. laevis (27). Substantial levels are found well before thyroid gland maturation at NF54. Likewise, E. coqui TRα mRNA levels were fairly constant, showing no appreciable developmental increase. We found significant levels at TS4–6, before thyroid gland differentiation at TS10. (14). TRβ mRNA is low in premetamorphic X. laevis, but increases dramatically at NF54, just before the onset of metamorphic remodeling (5, 28). Similarly, only small amounts of E. coqui TRβ mRNA are present before thyroid gland differentiation. During TS12–15, when the thyroid gland appears most active, TRβ mRNA levels are massively elevated relative to early embryogenesis. At this time, remodeling events reminiscent of metamorphosis occur, including tail regression (13), limb elongation (12), and cranial remodeling (29, 30). The correlation of high TRβ mRNA levels with both peak thyroid activity and remodeling provides a tempting parallel with the metamorphic period in biphasic frogs.
We have demonstrated the necessity for TH signaling in direct development by inhibiting the thyroid axis by using the metamorphic inhibitor methimazole, which resulted in developmental arrest at approximately TS12. Affected organ systems included the skin, limbs, jaw musculature, and cartilages, tail, and axial musculature. The apparently smooth and continuous development of E. coqui embryos into the adult morphology is revealed to be a biphasic process on methimazole treatment: the first phase occurs independently of zygotic thyroid production, and the second phase requires embryonic TH synthesis. The division between these two phases occurs around TS11, when a slight reduction in the efficacy of methimazole is detected. Cotreatment of methimazole-inhibited embryos with T3 allows them to reinitiate ontogenesis and attain the frog morphology. This inhibition and rescue experiment demonstrates that many remodeling events in E. coqui are TH dependent. The hypothesis that direct development results from emancipation of ontogeny from TH control (8, 9) is not supported by our results.
Hanken et al. (29) noted that the developmental remodeling of some cranial cartilages, occurring during TS12–14 of E. coqui development, had similarities with metamorphic transformations. However, previous studies failed to implicate TH in such remodeling. Our investigation shows that, as in metamorphosing anurans, elongation of Meckel's cartilage, the most dramatic event of cranial metamorphosis, has retained TH dependency in E. coqui.
Likewise, previous attempts to implicate TH in E. coqui limb development were unsuccessful (12). Here, we found that, whereas limb differentiation was unaffected by methimazole, the late phase of limb elongation, which occurs at TS12–14, requires TH. Similarly, in biphasic anurans, completion of limb development is thyroid dependent, but a reasonable degree of differentiation, including some cartilage development, occurs in the absence of a thyroid gland (12). This suggests that even though early limb development in Eleutherodactylus is highly derived relative to metamorphosing frogs, mechanistically both morphogenetic processes converge at a similar developmental stage, requiring TH to complete their growth.
Although remodeling of the hypaxial musculature occurs during metamorphosis in biphasic anurans (23, 24), there is no direct evidence in the literature for the role of TH in this event in any frog. In our investigation of E. coqui, methimazole prevented ventral convergence of the rectus abdominis, posterior extension of the pectoral muscles, and extension of the lateral muscle bands. All three events were rescued by T3, indicating that TH signaling is responsible for the extensive remodeling of the hypaxial musculature that occurs in the later phases (TS12–15) of E. coqui embryogenesis.
Another character in E. coqui documented here is the ventral suturing of the head and body epidermis at TS13. Its TH dependence was demonstrated by methimazole inhibition and T3 rescue. In biphasic frogs, the formation of a contiguous ventral epidermis also appears TH dependent, because hypophysectomized tadpoles treated with low doses of thyroxine do not undergo proper epidermal remodeling in the opercular region (31). Despite the radically different modes of early limb development and the highly modified opercular morphogenesis in E. coqui (32), this direct developer still recapitulates the ventral suturing seen in biphasic frogs. Similarly, the TH-dependent transformation of E. coqui skin appears to recapitulate the transition found in other frogs (33, 34) and can be regarded as another metamorphic remodeling event that has been retained in direct developers. In all these examples, initially divergent early embryogenesis converges on a common morphogenetic pathway utilizing TH signaling.
Methimazole will not interfere with the action of preexisting maternally derived hormone, and TH has been found in fish eggs and frog gastrulae (35, 36). Therefore, a role for TH in early embryogenesis cannot be discounted. The possibility that precocious embryonic accumulation of TRβ mRNA may mediate the early events of direct development is not supported by these data, because high levels of TRβ mRNA are not found before development of the thyroid gland. However, it is conceivable that maternally derived TRβ protein, perhaps translated from the TR mRNAs found in full-grown unmatured oocytes, could be involved in early development. TR messages are also present in X. laevis primary oocytes (6, 37), but their function is unknown.
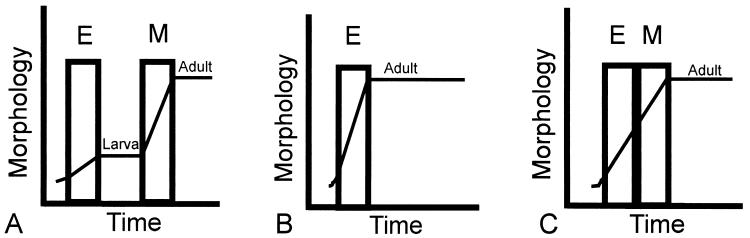
How might direct development have evolved? A way to visualize the evolution of direct development, suggested by Hanken (38), is shown in Fig. 4 A and B. In the ancestral life history, the embryogenic and metamorphic periods are temporally separated by the developmentally static larval phase (Fig. 4A). Direct development is viewed as proceeding directly to the adult morphology during embryogenesis, the metamorphic phase of the life history having been deleted (Fig. 4B). We propose that this metamorphic phase was retained during the evolution of direct development but was directly juxtaposed with the embryonic phase (Fig. 4C). The intervening larval phase was deleted. This model is supported empirically by the observation that TRβ mRNA is low in the embryonic period and up-regulates during the “metamorphic” remodeling period. The existence of this cryptic metamorphosis in Eleutherodactylus is proven by the inhibition of metamorphic-type remodeling when TH signaling is inhibited.
Figure 4.
Models of direct development. A and B based on ref. 38. E, embryogenesis; M, metamorphosis.
Hanken et al. (15) suggested that key aspects of the thyroid axis may have been conserved and advanced earlier into E. coqui development, because thyroid axis activation occurs prehatching in E. coqui and posthatching in biphasic frogs such as X. laevis. However, as noted by these authors, hatching takes 3 wk in E. coqui rather than 50 hr, as in X. laevis. Therefore, hatching is not a reliable character from which to judge developmental heterochronies. Although the thyroid axis and concomitant metamorphic events can be viewed as heterochronically accelerated relative to the development of early embryonic characters when compared with biphasic anurans, this can be explained by deletion of the larval period, rather than by the acceleration of thyroid signaling into early E. coqui embryogenesis.
The importance of TH for the completion of direct development indicates that even in anurans with radically altered ontogenies, the key metamorphic signaling pathway has been conserved. Although early development in Eleutherodactylus shows profound evolutionary innovation, such as loss of cement gland and dramatic reorganization of cranial cartilages and muscles (3, 29, 30), later development appears similar to biphasic anurans, with many remodeling events retaining TH dependence. We suggest that the complex and highly coordinated TH signaling cascade that orchestrates metamorphosis may constrain the adult morphology, explaining the absence of morphological innovation in adult frogs, even in species which have undergone extensive larval repatterning.
Acknowledgments
We thank J. Aubin, D. Brown, T. Candeliere, H. Fang, and Y. Marikawa for their helpful suggestions, Y. Marikawa for drawings, and the staff of the El Verde Field Station, Puerto Rico, for use of their facilities. The field station is part of the Long Term Ecological Research project funded by the National Science Foundation. E. coqui were collected under permits issued by the Departamento de Recursos Naturales, Puerto Rico. The 12/101 antibody developed by J. Brockes was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was funded by a grant from Natural Sciences and Engineering Research Council, Canada, to R.P.E. E.M.C. was supported by University of Toronto fellowships and an Ontario Government Scholarship.
Abbreviations
- T3
3,3′,5-triiodothyronine
- TH
thyroid hormone
- TR
thyroid hormone receptor
- TSn
Townsend–Stewart stage n
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF201957 (EcTRa) and AF201958 (EcTRb)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050501097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050501097
References
- 1.Duellman W E, Trueb L. Biology of Amphibians. New York: McGraw–Hill; 1986. pp. 13–50. [Google Scholar]
- 2.Heyer W R. Smithsonian Contrib Zool. 1975;199:1–55. [Google Scholar]
- 3.Elinson R P. Semin Dev Biol. 1990;1:263–270. [Google Scholar]
- 4.Tata J R. In: Metamorphosis: Postembryonic Reprogramming of Gene Expression in Amphibian and Insect Cells. Gilbert L I, Tata J R, Atkinson B G, editors. New York: Academic; 1996. pp. 465–503. [Google Scholar]
- 5.Yaoita Y, Brown D D. Genes Dev. 1990;4:1917–1924. doi: 10.1101/gad.4.11.1917. [DOI] [PubMed] [Google Scholar]
- 6.Kawahara A, Baker B S, Tata J R. Development (Cambridge, UK) 1991;112:933–943. doi: 10.1242/dev.112.4.933. [DOI] [PubMed] [Google Scholar]
- 7.Helbing C, Gergely G, Atkinson B G. Dev Gen. 1992;13:289–301. doi: 10.1002/dvg.1020130406. [DOI] [PubMed] [Google Scholar]
- 8.Lynn W G, Peadon A M. Growth. 1955;19:263–286. [PubMed] [Google Scholar]
- 9.Hughes A. J Embryol Exp Morphol. 1966;16:401–430. [PubMed] [Google Scholar]
- 10.Callery E M, Elinson R P. J Exp Zool. 1996;275:61–66. doi: 10.1002/(SICI)1097-010X(19960501)275:1<61::AID-JEZ9>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Cohen P P. Science. 1970;168:533–543. doi: 10.1126/science.168.3931.533. [DOI] [PubMed] [Google Scholar]
- 12.Elinson R P. J Exp Zool. 1994;270:202–210. doi: 10.1002/jez.1402700209. [DOI] [PubMed] [Google Scholar]
- 13.Townsend D S, Stewart M M. Copeia. 1985. 423–436. [Google Scholar]
- 14.Jennings D H, Hanken J. Gen Comp Endocrinol. 1998;111:225–232. doi: 10.1006/gcen.1998.7111. [DOI] [PubMed] [Google Scholar]
- 15.Hanken J, Jennings D H, Olsson L. Am Zool. 1997;37:160–171. [Google Scholar]
- 16.Schneider M J, Davey J C, Galton V A. Endocrinology. 1993;133:2488–2495. doi: 10.1210/endo.133.6.8243269. [DOI] [PubMed] [Google Scholar]
- 17.Murphy L D, Herzog C E, Rudick J B, Fojo A T, Bates S E. Biochemistry. 1990;29:10351–10356. doi: 10.1021/bi00497a009. [DOI] [PubMed] [Google Scholar]
- 18.Klymkowsky M W, Hanken J. Methods Cell Biol. 1991;36:419–441. doi: 10.1016/s0091-679x(08)60290-3. [DOI] [PubMed] [Google Scholar]
- 19.Brown D D. Proc Natl Acad Sci USA. 1997;94:13011–13016. doi: 10.1073/pnas.94.24.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose C S, Reiss J O. In: The Skull. Hanken J, Hall B K, editors. Chicago: Univ. of Chicago Press; 1993. pp. 289–346. [Google Scholar]
- 21.Gosner, K. L. Herpetologica16, 183–190.
- 22.Elinson R P, Fang H. Dev Genes Evol. 1998;208:457–466. doi: 10.1007/s004270050203. [DOI] [PubMed] [Google Scholar]
- 23.Ryke P A J. Acta Zoo. 1953;34:1–70. [Google Scholar]
- 24.Lynch K. J Morph. 1984;182:317–337. doi: 10.1002/jmor.1051820307. [DOI] [PubMed] [Google Scholar]
- 25.Helff O M. J Exp Zool. 1926;45:1–67. [Google Scholar]
- 26.Taylor A C, Kollros J J. Anat Rec. 1946;94:2–23. doi: 10.1002/ar.1090940103. [DOI] [PubMed] [Google Scholar]
- 27.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis (Daudin) New York: Garland; 1994. [Google Scholar]
- 28.Kanamori A, Brown D D. J Biol Chem. 1992;267:739–745. [PubMed] [Google Scholar]
- 29.Hanken J, Klymkowsky M W, Summers C H, Seufert D W, Ingebrigtsen N. J Morphol. 1992;211:95–118. doi: 10.1002/jmor.1052110111. [DOI] [PubMed] [Google Scholar]
- 30.Hanken J, Klymkowsky M W, Alley K E, Jennings D H. Proc R Soc London B. 1997;264:1349–1354. doi: 10.1098/rspb.1997.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollros J J. In: Comparative Endocrinology. Gorbman A, editor. New York: Wiley; 1959. pp. 340–350. [Google Scholar]
- 32.Callery, E. M. & Elinson, R. P. (2000) Dev. Genes Evol., in press. [DOI] [PubMed]
- 33.Fox H. In: Metamorphosis. Balls M, Bownes M, editors. Oxford: Clarendon; 1985. pp. 59–87. [Google Scholar]
- 34.Furlow J D, Berry D L, Wang Z, Brown D D. Dev Biol. 1997;182:284–298. doi: 10.1006/dbio.1996.8478. [DOI] [PubMed] [Google Scholar]
- 35.Weber G M, Okimoto D K, Richman N H, Grau E G. Gen Comp Endocrinol. 1992;85:392–404. doi: 10.1016/0016-6480(92)90084-w. [DOI] [PubMed] [Google Scholar]
- 36.Weber G M, Farrar E S, Tom C K F, Grau E G. Gen Comp Endocrinol. 1994;94:62–71. doi: 10.1006/gcen.1994.1060. [DOI] [PubMed] [Google Scholar]
- 37.Eliceiri B P, Brown D D. J Biol Chem. 1994;269:14459–65. [PubMed] [Google Scholar]
- 38.Hanken J. J Evol Biol. 1992;5:549–57. [Google Scholar]