Abstract
Mammalian nonhomologous DNA end joining employs Ku70, Ku80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4, and DNA ligase IV (Lig4). Herein, we show that Ku70 and Ku80 deficiency but not DNA-PKcs deficiency results in dramatically increased death of developing embryonic neurons in mice. The Ku-deficient phenotype is qualitatively similar to, but less severe than, that associated with XRCC4 and Lig4 deficiency. The lack of a neuronal death phenotype in DNA-PKcs-deficient embryos and the milder phenotype of Ku-deficient versus XRCC4- or Lig4-deficient embryos correlate with relative leakiness of residual end joining in these mutant backgrounds as assayed by a V(D)J recombination end joining assay. We conclude that normal development of the nervous system depends on the four evolutionarily conserved nonhomologous DNA end joining factors.
DNA double-strand breaks (DSBs) in eukaryotic cells can be repaired either by homologous recombination or by nonhomologous end joining (NHEJ) mechanisms. Both pathways are conserved in organisms from yeast to humans (1). Although homologous recombination is the predominant DSB repair (DSBR) pathway in yeast, NHEJ seems more important in mammalian cells. The five identified mammalian NHEJ proteins are generally expressed (1). These include Ku70, Ku80, and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which are subunits of the DNA-dependent protein kinase, as well as XRCC4 and DNA Ligase IV (Lig4), which function together in a ligation reaction (2). Gene-targeted mutation confirmed roles for all five mammalian proteins in aspects of NHEJ and V(D)J recombination (3–14). However, the apparent requirement of XRCC4 and Lig4 but not Ku70, Ku80, or DNA-PKcs for murine embryonic development and generation of a normal nervous system was unanticipated (8, 12, 15).
V(D)J recombination is the process by which variable region exons of T cell receptor and Ig genes are assembled from component gene segments (16). The recombination activating gene (RAG) proteins RAG1 and RAG2, which are expressed together specifically in lymphoid cells, initiate V(D)J recombination by introducing DSBs between recombination signal sequences (RS) and potential V, D, or J coding segments (17). RAG-mediated DNA cleavage generates blunt, 5′ phosphorylated RS ends and covalently sealed hairpin coding ends. Subsequently, RS ends are joined precisely, whereas coding ends are joined in a reaction that involves hairpin opening and potential loss/addition of nucleotides. However, both of these joining reactions employ generally expressed NHEJ components (1).
Mammalian cells deficient in Ku70, Ku80, XRCC4, or Lig4 share common phenotypic features, including ionizing radiation sensitivity, growth defects, premature senescence, and inability to join RAG-generated coding or RS ends (3–7, 9, 10, 12). Therefore, all of the defective processes likely result from a defective NHEJ pathway that employs these four factors. In contrast, DNA-PKcs deficiency resulted in a subset of the cellular phenotypes associated with deficiencies in the other four NHEJ proteins, including defective coding joining, variable ionizing radiation sensitivity, and (at most) modest RS joining defects (11, 13, 14). The less dramatic effects of DNA-PKcs deficiency might be due to a redundant factor or a more limited role for DNA-PKcs in NHEJ. In the latter context, it is notable that Ku70, Ku80, XRCC4, and Lig4 but not DNA-PKcs homologs have been identified in the yeast NHEJ pathway (1).
Despite the substantial overlap, there are notable differences in the phenotypes of mice with targeted inactivation of the individual NHEJ proteins. DNA-PKcs-deficient mice have a severe combined immunodeficient phenotype because of defective coding joining but no other major observed phenotypes. In contrast, Ku70- and Ku80-deficient mice, which are viable and fertile, have a severe combined immunodeficient phenotype with severely impaired joining of both coding and RS ends, as well as a profound growth defect (3, 4, 6, 7). XRCC4- and Lig4-deficient embryos also have growth defects and severely impaired lymphocyte development in association with defective coding and RS end joining (9, 12). However, in contrast to the Ku70-, Ku80-, and DNA-PKcs-deficient mice, XRCC4- and Lig4-deficient mice experience late embryonic lethality accompanied by massively increased cell death within the developing nervous system (8, 9, 12).
Embryonic neurogenesis in the mammalian central nervous system (CNS) begins with progenitor cell proliferation in anatomically defined zones surrounding the ventricles of the neural tube (ventricular zone, VZ; ref. 18). As progenitor neuroblasts differentiate, they exit the cell cycle and migrate through the embryonic intermediate zone/mantle layer (IZ/ML) to the regions that will become the adult gray matter (15). In XRCC4- or Lig4-deficient embryos, the aberrant apoptotic neurons are predominantly postmitotic cells, which are localized to the IZ/ML (8, 12). The known NHEJ functions of XRCC4 and Lig4 raised the possibility that increased neuronal cell death in the absence of either protein results from a NHEJ defect. However, the lack of embryonic lethality and apparently normal nervous systems of adult Ku70-, Ku80-, and DNA-PKcs-deficient mice seemed to contradict this interpretation (12, 15). In the current report, we resolve this potential contradiction and firmly implicate a requirement for the four evolutionarily conserved NHEJ proteins in neuronal development.
Materials and Methods
Mice.
All mutant mice were housed in a pathogen-free facility. References for each of the mutant mouse studies in this report are as follows: Ku70−/− (6) Ku80−/− (4), Lig4−/− (9), DNA-PKcs−/− (11), and XRCC4−/− (12).
Histological Analyses.
Whole embryos were fixed in either Bouin's solution or 4% (wt/vol) paraformaldehyde in PBS, paraffin embedded, serially sectioned (5 μm per section), and stained with hematoxylin and eosin following a standard protocol. The terminal deoxynucleotidyl transferase-mediated dUTP biotin nick end-labeling (TUNEL) assay was performed on paraformaldehyde-fixed sections with the DeadEnd colorimetric apoptosis detection kit (Promega).
Quantitation.
Four pairs of age-matched Ku70- and Lig4-deficient embryos were used to quantitate the levels of pyknosis at embryonic day (E)12.5 in coronal sections, at E13.5 in coronal sections, at E13.5 in transverse sections, and at E14.5 in sagittal sections. Sections through a designated CNS subdivision in each pair of mutant embryos were grouped as a set for comparison. The numbers of pyknotic cells from at least four randomly selected sections (separated by at least two consecutive sections to avoid double counting) in each set were averaged, and the standard error was determined. Individual pyknotic cells within a cluster of dying cells were also counted. The average numbers of pyknotic cells per section in the developing cerebral cortex, diencephalon, and midbrain (but not the spinal cord) of Lig4−/− embryos were significantly higher than those of Ku70−/− embryos at all developmental stages examined. The counting results were confirmed by an independent observer who performed the analysis blind.
Transient V(D)J Recombination Assay.
The RAG1 and RAG2 expression constructs and the signal-joining substrate pJH200 have been described (6). Murine embryonic fibroblasts (MEFs) are derived from E12.5 embryos of a heterozygous breeding following a standard protocol. The constructs were cotransfected into passage 2–3 MEFs with the Superfect reagent (Qiagen, Chatsworth, CA). Recombinant pJH200 products were recovered from the Ampr/Camr double-selection plates. The fidelity of the RS joins was determined by PCR amplification of the recombination products followed by digestion of the PCR products with the restriction enzyme ApaLI. For analyses of the RS join sequences, recombinant clones were picked from the double-selection plates, purified, and sequenced.
Results
Increased Death of Developing Ku-Deficient but Not DNA-PKcs-Deficient Neuronal Cells.
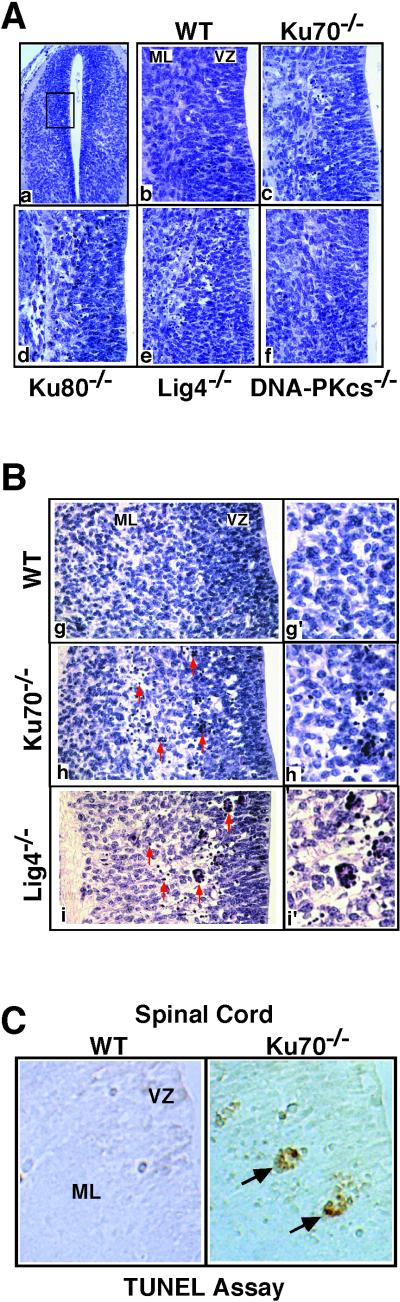
We performed histological analyses on E11–E15 Ku70−/−, Ku80−/−, and DNA-PKcs−/− embryos and compared these analyses to those of littermate controls and of age-matched Lig4−/− embryos. Based on findings with XRCC4- and Lig4-deficient embryos, initial studies focused on portions of the neural tube that actively undergo neurogenesis over this developmental period. We found substantially increased numbers of pyknotic cells in developing spinal cord of E12.5 Ku70−/− and Ku80−/− embryos compared to wild-type controls (Fig. 1A). To confirm that pyknotic cells were apoptotic, we assayed for DNA fragmentation by TUNEL. This assay identified significantly increased numbers of TUNEL-positive nuclei in E12.5 Ku70−/− sections as compared to controls (Fig. 1C), indicating that the pyknotic cells resulted from programmed cell death. We also found substantially increased numbers of pyknotic cells in other developing CNS subdivisions of Ku70-deficient mice, such as cerebral cortex, diencephalon, midbrain, and hindbrain (Fig. 2A and data not shown). However, there was no obvious, aberrant pyknosis in any region of age-matched DNA-PKcs-deficient embryos (n = 7; between E12.5 and E14.5; Figs. 1A and 2A) as measured by the hematoxylin and eosin staining.
Figure 1.
Increased cell death in spinal cord of Ku70−/−, Ku80−/−, and Lig4−/− but not DNA-PKcs−/− embryos at E12.5. (A) Hematoxylin- and eosin-stained transverse sections. Sections of spinal cord from different mutant embryos were compared with the sections of spinal cord from their littermate controls [one representative wild-type (WT) control is shown] at ×250 (a) and ×400 (b–f, boxed area of spinal cord from noted embryos). (B) Hematoxylin- and eosin-stained coronal sections. Cervical area of the spinal cord from control (g), Ku70−/− (h), and Lig4−/− (i) embryos. (g′–i′) Enlarged images from (g–i). (C) TUNEL staining of coronal sections (×400) of the spinal cord. Arrows indicate clusters of TUNEL-positive pyknotic cells in the Ku70−/− spinal cord.
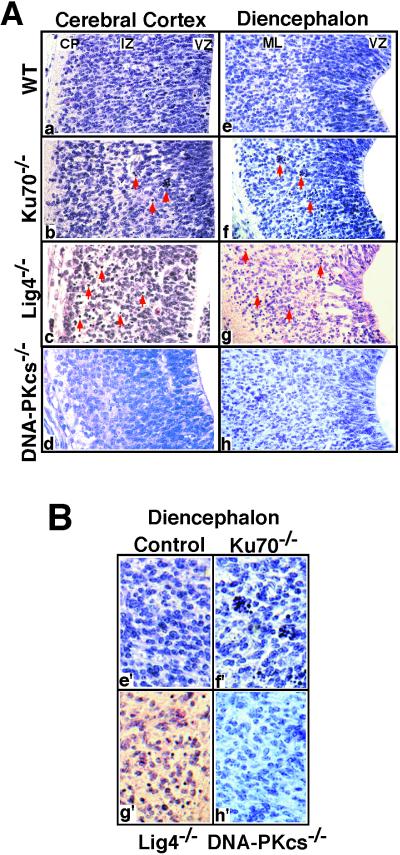
Figure 2.
Increased cell death in other subdivisions of developing CNS of Ku70−/− and Lig4−/− but not DNA-PKcs−/− embryos. (A) Hematoxylin- and eosin-stained sections (×400). Arrows point to the typical pyknotic cells, either isolated or clustered. (a–d) Cerebral cortex of E13.5 embryos (coronal sections). WT, wild type; CP, cortical plate. (e–h) Hypothalamic sulcus of the diencephalon of E13.5 embryos (transverse sections). (B) Enlarged images of Ae–Ah. Note pyknosis and hypocellularity in the ML of the diencephalon in the Ku70−/− (f′) and Lig4−/− (g′) embryos compared to DNA-PKcs−/− (h′) and control (e′) embryos. The darkly staining mitotic figures can be identified within the VZ of all control and mutant sections.
Differences in the Extent of Neuronal Death in Ku70-Deficient Versus XRCC4- or Lig4-Deficient Mice.
Pyknotic cells in Ku70−/− embryos were present either as clusters or as isolated cells (Figs. 1B and 2B). Individual pyknotic cells were mostly scattered within the IZ/ML, superficial to the VZ (Figs. 1B and 2B). Comparison of the average numbers of pyknotic cells indicated significant differences between the Ku70−/− versus Lig4−/− embryos within certain subdivisions of the CNS (see Materials and Methods for details). We found that the average number of pyknotic cells within the developing cerebral cortex in Ku70−/− embryos was 10–45% of that observed in the Lig4−/− embryos. In addition, we found that the average number of pyknotic cells in the diencephalon and midbrain in Ku70−/− embryos was 15–40% of that observed in the Lig4−/− embryos. However, we did not observe significant differences in the number of pyknotic cells in the spinal cord between the Ku70- and Lig4-deficient embryos. Finally, we did not observe a significant number of pyknotic cells in either control or DNA-PKcs-deficient embryos in any region of the developing brain examined (one pyknotic cell per approximately 6–10 sections examined). However, we cannot rule out the possibility that DNA-PKcs deficiency might result in a subtle increase in neuronal cell death that is below the sensitivity of our assay.
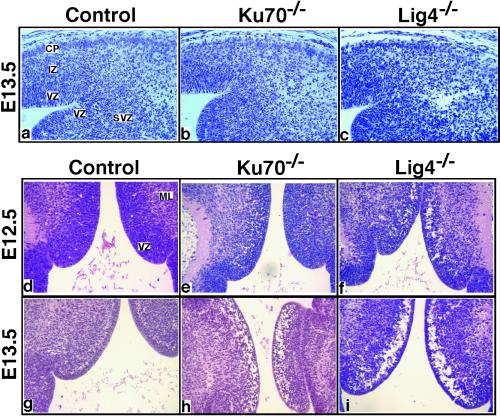
We observed that most clusters of dying or dead cells in the Ku70−/− embryonic CNS were localized either within or superficial to the VZ (Figs. 1B and 2B), the region of the developing XRCC4−/− and Lig4−/− CNS where we observed pronounced hypocellularity (12). We speculate that apoptotic cells may be aggregated as clusters and engulfed by microglial cells, resulting in the cavity formation. Analyses of sections from multiple different embryos identified a substantial number of large cavities in Lig4−/− embryonic diencephalon (Fig. 2A), striatum (Fig. 3 a–c), and brainstem (midbrain, pons, and medulla oblongata; Fig. 3 d–i); however, the occurrence of such cavities in Ku70−/− embryos appeared more variable in frequency and less severe in extent (Fig. 3 g–i; representative data shown).
Figure 3.
Differential CNS lesions in Ku70−/− and Lig4−/− embryos. (a–c) Coronal sections of the cerebral cortex and striatum at E13.5 (×200). CP, cortical plate; SVZ, sub-VZ. (d–i) Coronal sections of ventral midbrain and posterior pons at E12.5 (d–f) and E13.5 (g–i).
Increased Death of Ku70-Deficient Neuronal Cells Coincides with Embryonic Neurogenesis.
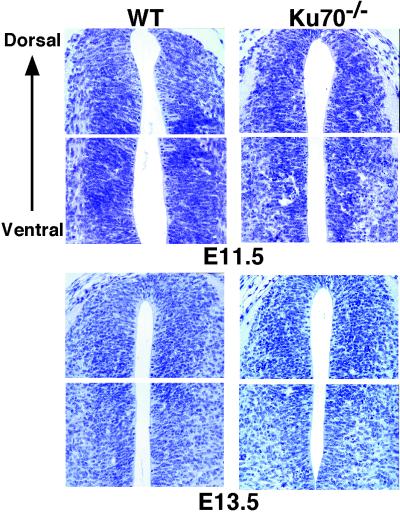
The onset and peak of cell death in various CNS subdivisions in the Ku70−/− embryos correlate with the known gradient of neuronal differentiation and normal apoptosis (19), as similarly observed for XRCC4−/− embryos (12). For example, the peak of cell death in Ku70−/− embryos first appears in the ventral portions of spinal cord at around E11.5 and then shifts to the dorsal portions at E13.5 (Fig. 4). In addition, when the dying cells appear in the ventrolateral portion of the forebrain at E12.5, pyknosis has markedly increased in the spinal cord and brainstem (Figs. 1 and 3). As noted previously (12), surviving neurons are observed in XRCC4- or Lig4-deficient embryos subsequent to the differentiation stage when increased apoptosis occurs. For example, a thin cortical plate is still formed in the cerebral cortex of Lig4−/− and XRCC4−/− embryos (Figs. 2 Ab and Ac and 3 b and c).
Figure 4.
Temporal patterns of increased cell death in developing spinal cord of Ku70−/− embryos. Hematoxylin- and eosin-stained transverse sections show the peak of dying cells at E11.5 (ventral section; Upper) and at E13.5 (dorsal section; Lower). WT, wild type.
The fact that adult Ku-deficient mice have a nervous system that is, at least in large part, functional strongly indicates that some fraction of most or all types of neurons can survive. However, our analyses of Ku70-deficient mice also suggest that certain neuronal populations are relatively refractory to effects of Ku deficiency. Thus, with respect to cerebellar neurons (which develop and differentiate over several weeks of embryonic and postnatal development; ref. 20), there was no obviously increased pyknosis in Ku70−/− mice—during either embryonic (e.g., at E13.5 or E17.5) or postnatal (e.g., between P4 and P14; data not shown) development. In addition, there was no overtly increased pyknosis in migrating and differentiating olfactory neurons or in neurons of the hippocampus in postnatal Ku70−/− mice (data not shown).
Leaky RS End Joining in Ku-Deficient but Not XRCC4/Lig4-Deficient Mice.
Mice deficient in different components of the V(D)J recombination reaction have varying degrees of impaired lymphocyte development. RAG deficiency results in a complete block because of the absolute requirement for these proteins to initiate the reaction (21, 22). In contrast, DNA-PKcs or Ku deficiency is associated with leakiness (6, 23, 24), probably because developing lymphocytes can, at low levels, use other pathways to repair RAG-liberated ends. In this context, we considered the possibility that the differential effects of DNA-PKcs, Ku70, Ku80, XRCC4, and Lig4 deficiency on neuronal development could be related to their relative effects on NHEJ pathways. To clarify this issue, we characterized the relative ability of Ku70−/−, Lig4−/−, and XRCC4−/− MEFs to join RS ends after transient transfection of RAG expression constructs plus a V(D)J recombination substrate. We focused on RS ends, because we had observed that the DNA-PKcs deficiency has no apparent effect on neurogenesis and, at most, modest effects on RS end joining, as opposed to major negative effects on coding end joining (11).
In Ku70−/− MEFs, the level of RS joining, as described (6), was significantly reduced; however, a low level of imprecise RS joints was consistently recovered (Table 1). All recombination products analyzed were RAG-initiated; however, all junctions contained deletions, and the majority contained short homology repeats (data not shown; ref. 6). In contrast, in multiple assays, we were unable to recover any RS joins from the Lig4−/− MEFs and only a single, aberrant join from XRCC4−/− MEFs (Table 1). Therefore, the impact of Ku70 deficiency on RS end joining, although severe, is substantially leakier than that observed in the context of XRCC4 and Lig4 deficiency. The ability to recover imprecise RS joins from Ku70-deficient cells may reflect a low-level ability of XRCC4 and Lig4 to join the RS ends in the absence of Ku. Alternatively, another DSBR pathway, such as the homology-mediated single-strand annealing pathway, may more effectively join the ends in the absence of Ku than in the absence of XRCC4 or Lig4 (25).
Table 1.
Analysis of signal join formation in MEFs by transient V(D)J recombination assay
| Experiment no./cell line | pJH200 (signal joining)
|
|||
|---|---|---|---|---|
| (Ampr + Camr)/Ampr | Percentage | Relative level | Fidelity, % | |
| I | ||||
| Wild type | 286 /13,447 | 2.1 | 1.0 | 100 (12/12) |
| Lig4−/− | 0 /543,020 | <0.0002 | <9.5 × 10−5 | — |
| XRCC4−/− | 0 /127,940 | <0.0008 | <0.0004 | — |
| Ku70−/− | 12 /119,200 | 0.1 | 0.005 | 0 (0/12) |
| II | ||||
| Wild type | 1,022 /72,600 | 1.4 | 1.0 | 100 (12/12) |
| Lig4−/− | 5 /30,853* | <0.016 | <0.01 | 0 (0/5) |
| XRCC4−/− | 1 /30,200 | 0.003 | 0.002 | 0 (0/1) |
| Ku70−/− | 82 /64,500 | 0.13 | 0.09 | 0 (0/12) |
| III | ||||
| Wild type | 3,037 /55,800 | 5.4 | 1.0 | 100 (12/12) |
| Lig4−/− | 0 /48,400 | <0.002 | <0.0004 | — |
| XRCC4−/− | 0 /106,600 | <0.001 | <0.0002 | — |
| Ku70−/− | 192 /94,867 | 0.2 | 0.04 | 0 (0/12) |
Relative levels are normalized to 1.0 for wild type. Fidelity is expressed as the percentage of recovered RS joins that were susceptible to ApaLI digestion.
*Spontaneous deletions, not RAG-mediated events.
Discussion
Normal Neuronal Development Requires Essential Components of a Conserved NHEJ Pathway.
We demonstrate that Ku-deficient but not DNA-PKcs-deficient embryos experience increased death of newly generated neurons. These findings were unanticipated, because no neuronal defects were originally ascribed to either Ku70 or Ku80 deficiency in adult animals (3, 6, 7). We conclude that essential components of a conserved NHEJ pathway, including Ku70, Ku80, XRCC4, and Lig4, also are essential for normal embryonic neurogenesis. The lack of an obvious neuronal phenotype in DNA-PKcs-deficient mice may well reflect the dispensability of DNA-PKcs in certain end-joining reactions, such as RS joining or DSBR in ES cells (11). The same NHEJ components necessary for normal neurogenesis also are necessary for normal growth. However, deficiencies in these components do not lead to any other obvious, specific histologic defects in tissues or organs outside of the nervous and immune systems (refs. 6 and 12; D.F., Y. Gu, and F.W.A., unpublished data). Therefore, normal development of the nervous system seems to have a particular reliance on NHEJ factors and, by extension, the ability to perform NHEJ.
The Level of NHEJ May Be Responsible for the Severity of Neuronal Defects.
Despite the overall similarity of CNS lesions, there were clear differences in the extent of aberrant neuronal cell death in several examined regions of the developing CNS in Ku70- versus Lig4-deficient embryos (Figs. 1–3). Although the direct cause of embryonic death in either XRCC4- or Lig4-deficient embryos has yet to be determined, the massive neuronal cell death in these embryos may cause irreparable CNS damage, such as the large cavities within the sub-VZ of the brainstem (Fig. 3 d–i). Such severe damage could prevent appropriate neural connections from being established and lead to an overall CNS failure. Perhaps the less severe aberrant neuronal cell death in the regions of the developing nervous system of Ku-deficient embryos allows enough neurons to survive to form a functioning nervous system in adults.
How do we explain the differential impact of deficiencies of DNA-PKcs versus Ku70/80 versus XRCC4/Lig4 on neurogenesis? One conceivable possibility might involve differential requirements for these factors in downstream cell-cycle/survival pathways (26). Another, not mutually exclusive possibility is that neuronal development and/or survival depends on DNA repair via NHEJ, with relative neuronal survival in the different mutant backgrounds resulting from the relative leakiness in the NHEJ defects. This notion, which is raised by analogy to the effects of these different mutations on lymphocyte development (e.g. 6, 9, 11), gains substantial support from our finding that the presence and/or relative severity of the CNS lesions in DNA-PKcs-, Ku70-, and Lig4-deficient embryos is reflected by the relative ability of corresponding mutant cells to join RAG-liberated RS ends (Table 1). If this interpretation is correct, the generation of a functioning nervous system in Ku-deficient adults implies that its development can tolerate a substantial loss of developing neurons. In addition, this interpretation further suggests that, although NHEJ per se is critical, there is not a strict requirement for absolutely precise end joining to allow neuronal differentiation.
Why Do Developing Neuronal Cells Depend on NHEJ?
An intriguing question is how deficiencies in NHEJ proteins result in the widespread apoptosis of developing neurons, while having no obvious in vivo effects on cell types other than lymphocytes? We previously considered a number of different mechanisms by which XRCC4 or Lig4 deficiency might result in increased neuronal apoptosis (12). Our current findings of similar defects in association with Ku70 and Ku80 deficiency lead us now to focus on the involvement of DSBR, because that is the only process currently known to employ these four evolutionarily conserved NHEJ proteins. In this context, there are several possible scenarios—none of which are mutually exclusive—whereby DSBR defects could lead to prominent apoptosis of developing neurons with little apparent effect on most other cell types.
One possibility is that neuronal cells might be especially prone to apoptosis in response to DSBs, perhaps to eliminate aberrant neurons from further development. There are several observations that support this notion. First, both proliferating neuroblasts and newly generated neurons in rat embryos are highly sensitive to ionizing radiation (27). In addition, Ku70-, XRCC4-, and Lig4-deficient fibroblasts seem to respond to DSBs by cell-cycle arrest but not apoptosis (data not shown; refs. 6 and 12). Conceivably, cell-cycle arrest of other cell types in response to DSBs might also contribute to the decreased size of Ku-deficient mice. A related scenario is suggested from studies of Drosophila embryonic development that indicated that rapidly proliferating neuroprogenitor cells suspend normal cell-cycle checkpoints (28, 29). If such suspension also occurs in the analogous populations of developing murine neurons, it might facilitate accumulation of DSBs and lead to massive death if checkpoints were reestablished after differentiation. Another possible scenario for increased reliance of neuronal cells on NHEJ would be that these cells lack the ability to employ repair pathways, such as homologous recombination, that are overlapping with NHEJ with respect to DSBR. Finally, with respect to potential mechanisms for increased neuronal cell death that involve increased DNA damage sensitivity, it is also conceivable that similar phenotypes might be observed in the context of defects in other types of DNA repair processes.
There are two general etiologies of DSBs, nonspecific and specific. Increased occurrence of DSBs from either source, potentially in concert with increased sensitivity to such breaks, also could explain our findings. Sources of nonspecific DSBs theoretically could be increased levels of metabolic products such as free radicals or breaks linked to DNA replication or transcription of unstable genomic sequences (2, 30, 31). The notion of specific DSBs as a part of neuronal development has been widely discussed, particularly in the context of RAG1 expression in the brain (15). However, in other studies, we have shown that the neuronal defects in XRCC4-deficient mice do not depend on RAG proteins (26). Thus, if a recombinase specific for developing neurons did exist, it would likely have a unique set of targets. Attractive candidates for rearrangement in the nervous system include multigene families such as the odorant/pheromone receptors (32, 33) and the protocadherin gene family, which has an organization reminiscent of antigen receptor variable region loci (34–36). However, Ku70-, Lig4-, and XRCC4-deficient embryos do not have overtly, as determined in preliminary studies, impaired olfactory receptor expression (K.F., Y. Gao, J.S., and F.W.A., unpublished observations) or increased neuronal cell death in developing olfactory organs. In addition, we have, in preliminary attempts, failed to detect rearrangements within protocadherin gene loci (D.F., G. Rathbun, J.S., A. Eggleston, and F.W.A., unpublished observations). However, more detailed studies of these and other potential candidate families are warranted.
In summary, our studies show that the Ku70-, Ku80-, XRCC4-, and Lig4-deficient phenotypes are, in fact, highly similar, with apparent differences potentially attributable only to the degree to which NHEJ is impaired. In particular, Ku70, Ku80, XRCC4, and Lig4 deficiencies result in a striking neuronal death phenotype, with a number of intriguing parallels to effects on lymphocyte development. It remains to be determined whether the implicated DSBs are specific or nonspecific. In either case, characterization of these animal models should be telling with respect to aspects of neurogenesis and, potentially, neurodegenerative diseases.
Acknowledgments
We thank Drs. David Schatz and David Weaver for critical reviews of the manuscript. This work was supported in part by National Institutes of Health Grants A.I.35714 (to F.W.A.), A.I.01428 (to K.F.), and R01NS36949 (to J.C.). J.S. is the Richard D. Frisbee, III, Foundation Fellow of the Leukemia Society of America. Y. Gu and Y. Gao are associates and F.A. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- DSB
double-strand break
- NHEJ
nonhomologous end joining
- DSBR
DSB repair
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- RS
recombination signal sequences
- CNS
central nervous system
- VZ
ventricular zone
- IZ
intermediate zone
- ML
mantle layer
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP biotin nick end labeling
- En
embryonic day n
- MEF
murine embryonic fibroblast
- Pn
postnatal day n
References
- 1.Jeggo P A. Radiat Res. 1998;150:S80–S91. [PubMed] [Google Scholar]
- 2.Lieber M R. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 3.Nussenzweig A, Chen G, da Costa Soares M, Sanchez M, Sokol K, Nussenzweig M D, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 5.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H-L, Sekiguchi J, Frank K, et al. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang H, Nussenzweig A, Kurimasa A, Soares V C, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Illakis G, et al. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes D E, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 9.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H-L, Davidson L, Kangaloo L, Alt F W. Nature (London) 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 10.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 13.Taccioli G, Amatucci A, Beamish H, Gell D, Xiang X, Arzayus M, Priestley A, Jackson S, Rothstein A, Jeggo P, et al. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 14.Kurimasa A, Ouyang H, Dong L J, Wang S, Li X, Cordon-Cardo C, Chen D J, Li G C. Proc Natl Acad Sci USA. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun J, Schatz D G. Neuron. 1999;22:7–10. doi: 10.1016/s0896-6273(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 16.Willerford D M, Swat W, Alt F W. Curr Opin Genet Dev. 1996;6:603–609. doi: 10.1016/s0959-437x(96)80090-6. [DOI] [PubMed] [Google Scholar]
- 17.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 18.Angevine J B., Jr J Comp Neurol. 1970;139:129–187. doi: 10.1002/cne.901390202. [DOI] [PubMed] [Google Scholar]
- 19.Blaschke A J, Weiner J A, Chun J. J Comp Neurol. 1998;396:39–50. doi: 10.1002/(sici)1096-9861(19980622)396:1<39::aid-cne4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 20.Hatten M E, Heintz N. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 21.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 22.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 23.Bosma G C, Fried M, Custer R P, Carroll A, Gibson D M, Bosma M J. J Exp Med. 1988;167:1016–1033. doi: 10.1084/jem.167.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussenzweig A, Sokol K, Burgman P, Li L, Li G C. Proc Natl Acad Sci USA. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thacker J. C R Acad Sci Ser III. 1999;322:103–108. doi: 10.1016/s0764-4469(99)80030-4. [DOI] [PubMed] [Google Scholar]
- 26.Sekiguchi, J. M., Gao, Y., Gu, Y., Frank, K., Chaudhuri, J., Zhu, C., Cheng, H.-L., Manis, J., Ferguson, D., Davidson, L., et al. (2000) Cold Spring Harbor Symp. Quant. Biol.64, in press. [DOI] [PubMed]
- 27.Bayer S A, Altman J. Neocortical Development. New York: Raven; 1991. pp. 128–149. [Google Scholar]
- 28.Edgar B. Curr Opin Cell Biol. 1995;7:815–824. doi: 10.1016/0955-0674(95)80065-4. [DOI] [PubMed] [Google Scholar]
- 29.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 30.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krasilnikova M M, Samadashwily G M, Krasilnikov A S, Mirkin S M. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 33.Mombaerts P. Annu Rev Neurosci. 1999;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- 34.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 35.Obata S, Sago H, Mori N, Davidson M, St. John T, Suzuki S T. Cell Adhes Commun. 1998;6:323–333. doi: 10.3109/15419069809010791. [DOI] [PubMed] [Google Scholar]
- 36.Wu Q, Maniatis T. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]