Abstract
The Drosophila protein Chip potentiates activation by several enhancers and is required for embryonic segmentation. Chip and its mammalian homologs interact with and promote dimerization of nuclear LIM proteins. No known Drosophila LIM proteins, however, are required for segmentation, nor for expression of most genes known to be regulated by Chip. Here we show that Chip also interacts with diverse homeodomain proteins using residues distinct from those that interact with LIM proteins, and that Chip potentiates activity of one of these homeodomain proteins in Drosophila embryos and in yeast. These and other observations help explain the roles of Chip in segmentation and suggest a model to explain how Chip potentiates activation by diverse enhancers.
The Drosophila Chip protein is provided maternally and is also expressed zygotically, and both supplies are required for normal development (1). Embryos lacking maternal Chip die without forming segments, and those lacking zygotic Chip form segments and hatch but die as larvae (2). It has been proposed that Chip facilitates enhancer–promoter communication at many genes (1–3). That idea was suggested by genetic studies indicating that the Su(Hw) insulator protein counteracts Chip activity at the cut locus (2). Thus, when the gypsy transposon is inserted at virtually any place between a wing-specific enhancer and the promoter of cut, Su(Hw) binds to gypsy and blocks communication between the enhancer and promoter, resulting in a cut wing phenotype (4, 5, and reviewed in refs. 3 and 6). Chip was identified as a mutation that further reduced expression of a cut gene bearing a gypsy insertion (2). These studies also indicated that the Su(Hw) bound to gypsy DNA in a cut gene on one chromosome works, to some extent, to block activation by an enhancer in a wild-type cut gene on the homologous chromosome. Under this situation of inter-chromosomal enhancer-blocking, expression of the wild-type cut gene is much more sensitive to reduction of Chip activity than to reductions in the activities of all other known cut regulators, including the enhancer-binding protein Scalloped (2).
Several recent studies have described interactions of Chip and homologous proteins with other proteins in vitro. Vertebrate Chip homologs were isolated as proteins that bind zinc-chelating structures called LIM domains that mediate diverse protein interactions (7–9). Nuclear LIM proteins include those that consist solely of a few LIM domains, and certain DNA-binding proteins that also contain homeodomains (LIM-HD proteins, reviewed in ref. 10). The Chip mammalian homologs promote formation of homotypic and heterotypic dimers of LIM-HD proteins in vitro (11). Drosophila Chip also interacts with LIM domains and supports activity of the Apterous LIM-HD protein in wings (1, 12–16). It also has been reported that a mammalian Chip homolog interacts with a mammalian HD protein (9).
Interactions between Chip and LIM proteins do not explain the full range of Chip's in vivo functions. None of the known Drosophila LIM proteins is needed for segmentation, and none of the known LIM proteins is required for activation of most genes, e.g., cut and even-skipped (eve), known to be regulated by Chip. In contrast, many segmentation genes, such as eve, are activated by HD proteins. Here we show that Drosophila Chip interacts with diverse HD proteins and with the Su(Hw) insulator protein through a domain distinct from the region that interacts with LIM proteins. We also present two experiments showing that Chip potentiates the activity of the Bicoid (Bcd) HD protein in both Drosophila embryos and in yeast.
Materials and Methods
Affinity Chromatography.
Glutathione-S-transferase (Gst)-Chip and mutant Gst-Chip fusion proteins (see below) were expressed in Escherichia coli (1). Glutathione-agarose beads (Sigma) were incubated with bacterial extracts overnight at 4°C and washed with 4 vol of extraction buffer (20 mM Hepes, pH 7.5/0.1 M NaCl/10% glycerol/0.1% Nonidet P-40/1 mM 2-mercaptoethanol), 2 vol of binding buffer (10 mM Pipes, pH 6.8/0.45 M NaCl/5% glycerol/0.1% Nonidet P-40/1 mg/ml BSA/1 mM DTT), and 8 vol of phosphate-buffered saline (1 mM KH2PO4/10 mM Na2HPO4/137 mM NaCl/2.7 mM KCl, pH 7.0) containing 1 mM DTT. Washes contained protease inhibitors (1 mM phenylmethylsulfonyl fluoride/1 μg/ml each of leupeptin, aprotinin, and pepstatin). The beads were equilibrated in PBS containing 1% Triton X-100, 1 mM DTT, and 25 mg/ml BSA and protease inhibitors and stored at 4°C. Before binding reactions, beads were incubated overnight at 4°C with 1 ml of blocking solution (PBS with 2% nonfat milk/1% Triton X-100/1 mM DTT/protease inhibitors) per 100 μl of beads.
[35S]proteins for binding to Gst-Chip beads were produced by in vitro translation using the TNT kit (Promega), [35S]methionine (NEN, 1,000 Ci/mmol), and transcription templates described below. ZnCl2 (1 mM) was added to Apterous and Su(Hw) translations.
Binding reactions were conducted for 1 hr at 4°C in blocking solution with a total volume of 200 μl, ≈10–20 μg of Gst fusion protein, and 10−13 mmol of [35S]protein. The beads were washed three times with blocking solution for 10 min, twice with PBS containing 1% Triton X-100 for 10 min, and twice with PBS for 5 min. The beads were boiled in 40 μl of 2× SDS-PAGE sample buffer, and the retained [35S]protein quantified with an phosphorimager after SDS-PAGE.
Gst-Chip Fusion Constructs.
Gst-Chip was made from pGEX2T-Chip (5). Gst-Chip:Δ472–577 was made by PCR amplification (17) of the Chip cDNA sequence encoding residues 1–471 using a 5′-primer with a BglII site and a 3′-primer with a stop codon and an EcoRI site and cloning into the BamHI and EcoRI sites of pGEX2T (Pharmacia). Gst-Chip:Δ441–454 was made by cloning a BglI to SmaI linker into the blunted BamHI and AvaI sites of pGEX2T-Chip. Gst-Chip:Δ404–465 was made by substituting the MaeI (blunted) to EcoRI Chip cDNA fragment for the AlwNI (blunted) and EcoRI fragment of pGEX2T-Chip. Gst-Chip:Δ404–519 was made by deleting the AlwNI to BstXI fragment of pGEX2T-Chip and blunt religation. Gst-Chip:Δ294–381 was made by deleting the SphI to BssHII fragment of pGEX2T-Chip. Gst-Chip:Δ384–438 was made by substituting a SmaI linker for the BssHII to BamHI fragment of pGEX2T-Chip. To make Gst-Chip:Δ457–471, the sequence encoding residues 471–577 was amplified by PCR with primers containing AvaI and AflII sites and substituted for the 507-bp AvaI to AflII fragment in the Chip cDNA. The resulting BamHI to EcoRI Chip cDNA fragment was cloned into pGEX2T. Gst-Chip:Δ1–381 was made by cloning the blunted 856-bp BssHII to EcoRI Chip cDNA fragment into the blunted XhoI site in pGEX1ZT (Pharmacia).
In Vitro Translation Vectors.
The apterous cDNA was amplified by PCR with a 5′-primer containing the Kozak initiation sequence (18) and an NcoI site, and a 3′-primer containing an EcoRV site, and cloned into the NcoI and EcoRV sites of the pING14.1 expression vector (S. Ingles and I. Brierly, cited in ref. 19). pTN3-Bcd (20), pAR-Ftz (20), and pAR-Eve (20) were used to produce Bcd, Ftz, and Eve. The Orthodenticle (Otd) expression vector contains the otd cDNA in a pBluescript vector (Frieder Schöck, Max Planck Institute for Biophysical Chemistry, personal communication).
Bcd:1–255 was made by deleting the AccI to EcoRI fragment of pTN3-Bcd. Bcd:1–190 was made by deleting the 1.6-kb BseRI to EcoRI fragment of pTN3-Bcd. Bcd:1–166, Bcd:57–166, and Bcd:157–255 were made by PCR amplification of the appropriate coding sequences with a 5′-primer containing an NcoI site and initiation codon and a 3′-primer with an EcoRV site, and cloning the resulting DNA fragments into the NcoI and EcoRV sites of pING14.1.
The Chip cDNA-coding sequence was amplified by PCR using a 5′-primer with an NcoI site and a Kozak initiation site and a 3′-primer with an EcoRV site, and cloned into the NcoI and EcoRV sites of pING14.1. Chip:Δ404–519 was made by replacing the BssHII to HindIII 668-bp fragment of pING14.1-Chip with the BssHII to HindIII 322-bp fragment of pGEX2T-ChipΔ:404–519. Chip:Δ2–381 was made by cloning the blunted BssHII to EcoRI Chip cDNA fragment into the blunted NcoI site in pING14.1.
The su(Hw) cDNA was cloned as an XhoI to SmaI fragment (21) into the SalI and SmaI sites of pGEM4Z. Circular pGEM4Z-Su(Hw) was used to produce full-length Su(Hw), and Su(Hw):ΔCTD (truncated at residue 780) was produced after linearization with BamHI. Su(Hw):1–190 was made after cutting pGEM4Z-Su(Hw) with BsaBI or XhoII. The Su(Hw) zinc finger domain (residues 204–672) was made by cloning the SalI to NheI fragment from pSJ-su(Hw)Zn (22) into the SalI and NheI sites of pGEM4Z. Su(Hw):706–944 was made by cloning the PvuII to HindIII 713-bp fragment of the su(Hw) cDNA into the EcoRV and HindIII sites of pING14.1. Mutant su(Hw) zinc finger domains described previously (21) were amplified by PCR with a 5′-primer with an NcoI site and an initiation codon, and a 3′-primer with an EcoRV site, and the resulting DNA fragments were cloned upstream of the HindIII (blunted) to SspI su(Hw) cDNA fragment with the termination codon into the NcoI and BamHI (blunted) sites of a pGEM3Z (Promega) vector modified to lack the polylinker HindIII site.
Sequencing of the Chipg96.1 Mutation.
The Chipg96.1 ORF was amplified by PCR (using primers 5′-CGGAATTCAGTGCATACACATACGCATG-3′ and 5′-ATTAAGATCTGTGTGTAGAGTAGACGAC-3′) from genomic DNA isolated from y w; P[w+]57B Chipg96.1/Chipk04405 larvae according to Levis et al. (23), cloned into the EcoRI site of pBluescript (SK) (Stratagene), and sequenced using Sequenase II (U.S. Biochemicals).
Genetic interaction between Chipg96.1 and bcdE3. y w; P[w+] 57B Chipg96.1/+; ru st bcdE3 ca e females and sibling y w; ru st bcdE3 ca e females were crossed to y w males and allowed to lay embryos on apple juice agar plates at 25°C. After 48 hr, embryonic cuticles were prepared (1) and the segments counted under dark field illumination. Partially fused segments were counted as loss of one-half a segment. Calculation of standard errors and statistical significance was aided by the statview computer program (SAS Institute, Cary, NC).
Effects of Chip on Activation by Bcd in Yeast.
The lacZ yeast reporter plasmids and the Bcd and Gal4-ER-VP16 expression vectors are described by Burz et al. (24). The Chip expression vector was made by cloning the Chip cDNA as an MslI to EcoRI fragment downstream of a KpnI to NcoI (blunted) fragment with the yeast ACT1 promoter (25) into the YCplac22 ARS-CEN vector (26). Chip mutants were made by substituting appropriate fragments from the pGEX2T-Chip mutant vectors described above into Ycplac22-Chip.
Yeast strain NLY2 (MATα ura3 his3Δ200 leu2–1 lys- trp1Δ63 gal4- gal80-) was sequentially transformed using LiCl (27) with the reporters and expression vectors and cultured in appropriate drop-out media with 2% glucose at 30°C. Mid-log cultures were diluted to an OD600 of 0.2, and β-estradiol was added to induce Bcd expression. Four hours postinduction, extracts were assayed for β-galactosidase activity (27). All experiments were performed at least twice with four independent clones for each Bcd transformant and two independent clones for wild-type and mutant Chip.
Results
Chip Interacts with Diverse HD Proteins and with the Su(Hw) Insulator Protein.
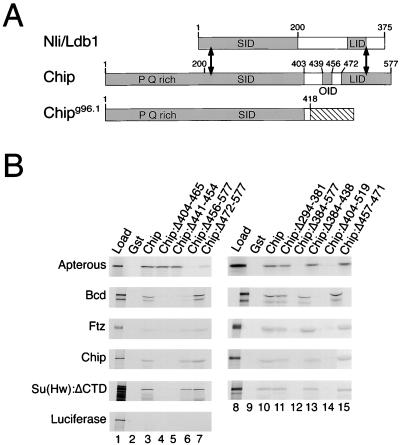
The experiments of Fig. 1 demonstrate that Chip interacts in vitro with various HD proteins and with Su(Hw). To perform these experiments we prepared [35S]-labeled HD and Su(Hw) proteins using in vitro translation, and tested their abilities to bind to Gst-Chip fusion protein, expressed in and purified from bacteria and attached to glutathione-bearing beads. Fig. 1B shows that full-length Chip interacted with the HD proteins Bicoid (Bcd) and Ftz, and with a fragment of the Su(Hw) insulator protein (lanes 3 and 10). Fig. 1B also shows, consistent with previous results, that Chip interacted with the Apterous LIM-HD protein and with itself (lanes 3 and 10; refs. 1, 12, 13, 15, and 16). In experiments not shown, we found that the HD protein Otd bound almost as efficiently as did Bcd and Ftz to Chip, but that the Eve HD protein bound poorly, a result possibly attributable to improper folding of the in vitro-translated protein. The depicted experiment with Su(Hw) was performed with a fragment lacking approximately one-third of its carboxyl residues [Su(Hw):ΔCTD]; full-length Su(Hw) also bound but less efficiently (not shown). As expected, Gst-Chip beads did not bind luciferase (Fig. 1B, lane 3), and none of the [35S]proteins bound Gst control beads (Fig. 1B, lanes 2 and 9).
Figure 1.
Chip interacts with HD proteins and the Su(Hw) insulator protein. (A) The homologous region (≈60% identity) of Chip and mouse Nli (Ldb1) is between the double-headed arrows. The protein interaction domains of Chip and Nli are shaded. These are the SID, LID, and OID (this work and refs. 7, 8, 11, 15, 16, 28, and 29). The OID interacts with HD proteins, Chip and Su(Hw). The Chipg96.1 mutant contains the SID but lacks the OID and LID. (B) Autoradiograms of SDS-PAGE gels from representative affinity chromatography experiments with the Gst-Chip fusion protein beads indicated at the tops of the lanes and the in vitro translated [35S]proteins indicated on the left. The left lanes (Load) contain the amount of input protein, and the second lanes (Gst) contain the amounts retained by beads with Gst alone. The other lanes contain the protein retained by the indicated fusion proteins. Residues deleted from the Chip mutants are indicated after the colons. Apterous is a LIM-HD protein; Bcd and Ftz are HD proteins. Su(Hw):ΔCTD is a Su(Hw) mutant truncated after residue 780, leaving the region containing 12 zinc fingers. Smaller products seen with some [35S]proteins are assumed to result from incomplete translation or proteolysis. All experiments were reproduced at least three times. With all proteins except luciferase, a minimum of 10-fold more protein bound to Gst-Chip beads than to Gst control beads.
The domains of Chip involved in homotypic and heterotypic interactions as deduced from our experiments, and from previous experiments with Chip and its homologs, are diagrammed in Fig. 1A. These include the LIM interaction domain (LID) and the self-interaction domain (SID) (this work and refs. 7, 8, 11, 15, 16, 28, 29). Consistent with the work of others, deletion of the LID reduced interaction with Apterous (Chip:Δ472–577, Fig. 1B, lane 7). That deletion, however, had no effect on interaction with Bcd, Ftz, Su(Hw):ΔCTD, or Chip (Fig. 1B, lane 7). In contrast, two other deletion mutants, Chip:Δ404–465 and Chip:Δ441–454, reduced binding to Bcd, Ftz, Su(Hw):ΔCTD, and Chip but had little effect on binding to Apterous (Fig. 1B, lanes 4 and 5). On the basis of this and additional deletion mutants (Fig. 1B, lanes 11, 13, and 15), we identified Chip residues 439–456 as the region that interacts with the HD proteins, Su(Hw), and with Chip itself. This region is labeled the other interaction domain (OID) in Fig. 1A.
Previous studies suggested that the SID is sufficient for self-interaction of Chip, but as noted above, Fig. 1B shows that Chip self-interaction was reduced by deletions affecting the OID (Chip:Δ404–465 and Chip:Δ441–454, lanes 4 and 5) but unaffected by a deletion that removes much of the SID (Chip:Δ294–381, lane 11). Further affinity chromatography experiments show that an isolated SID fragment (Chip:Δ404–519) interacted with itself but did not interact well with intact Chip, whereas a Chip fragment lacking the SID (Chip:Δ2–381) interacted both with itself and with intact Chip (not shown). The previous experiments showing interactions between the SID and intact Chip were performed by translating the two interaction partners together in vitro (16). Evidently, cotranslation permits an interaction not seen by affinity chromatography. We conclude that Chip interacts with itself through both the SID and the OID.
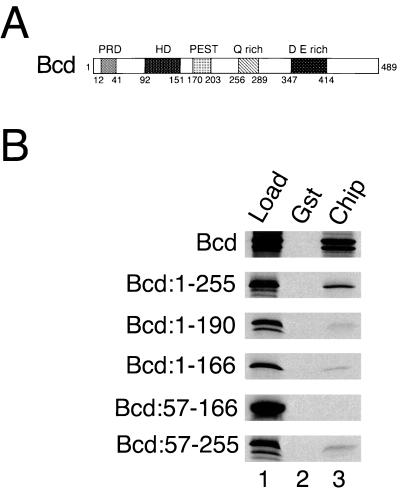
We mapped the portions of Bcd and Su(Hw) that interact with Chip to determine if the OID recognizes a common motif in its diverse interaction partners. The N-terminal half of Bcd (residues 1–255) contains the HD (Fig. 2A) and everything needed to rescue bcd mutants in vivo (30, 31). The N-terminal half of Bcd interacted with Gst-Chip (Fig. 2B, lane 3), whereas the C-terminal half (residues 246–489) did not (not shown). Smaller Bcd fragments containing the HD (residues 1–190, 1–166 or 57–255) bound more weakly than did the 1–255 fragment, and a fragment (residues 57–166) consisting mostly of the HD (residues 92–151) did not bind (Fig. 2B, lane 3). Thus, residues on both sides of the HD are required for strong binding. Similar results were obtained with the Otd HD protein (C.R., D.D., Frieder Schöck, and Herbert Jäckle, unpublished data). The region of Su(Hw) that contains 12 zinc fingers (residues 204–672) interacted with Gst-Chip, whereas the N-terminal region (residues 1–190) and the C-terminal region (residues 706–944) did not (not shown). Mutation of any one of the 12 zinc fingers (21) did not significantly affect binding to Chip (not shown). The regions of Bcd, Su(Hw), and Chip that interact with the Chip OID are not homologous at the primary sequence level.
Figure 2.
Regions flanking the Bcd HD interact with Chip. (A) Bcd contains paired repeats (PRD), a HD, a PEST protein degradation motif, glutamine-rich (Q rich), and acidic (D E rich) regions. (B) Autoradiograms of SDS-PAGE gels from representative affinity chromatography experiments with Gst and Gst-Chip beads, and the [35S]Bcd proteins containing the indicated residues on the left.
Chip Supports Bcd HD Protein Activity in Drosophila Embryos.
The interactions between Chip and HD proteins in vitro, as well as other considerations discussed above raised the question of whether Chip affects the activities of HD proteins in vivo. We chose to test the effect of Chip on Bcd activity in embryos because both Chip and Bcd are provided maternally and do not regulate each other's expression. Thus, any effect of Chip on Bcd is likely to be direct. The design of the experiment in Fig. 3 showing that, in embryos, reducing Chip activity decreases the activity of a partially defective Bcd protein, was guided by the following considerations.
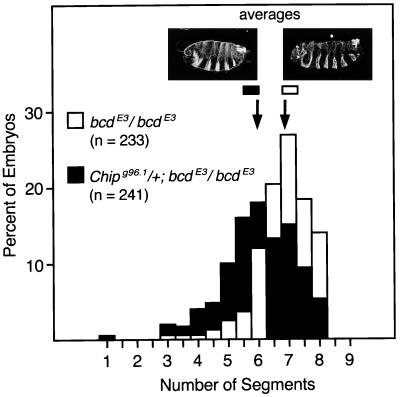
Figure 3.
Chip potentiates Bcd activity in embryos. The histograms show the percentage of embryos with the indicated number of segments produced by each maternal genotype. Embryos from +/+; bcdE3/bcdE3 mothers (white bars) crossed to +/+; +/+ males produced an average of 6.84 ± 0.06 segments (standard error, n = 233), whereas embryos from their Chipg96.1/+; bcdE3/bcdE3 sisters (black bars) crossed to +/+; +/+ males produced an average of 6.02 ± 0.08 segments (n = 241). This difference is significant (P < 0.0001, unpaired Student's t test), and virtually identical results were obtained in an independent experiment. Cuticles approximating the average phenotypes are shown for each maternal genotype.
To demonstrate a helping effect of Chip on Bcd activity, we could not simply eliminate maternal Chip because that manipulation results in a more severe segmentation defect than does elimination of Bcd itself (1, 32). Nor could we merely halve the dosage of maternal Chip because that change has no effect, even if the maternal Bcd level is also reduced by one-half (not shown). Moreover, zygotic Bcd makes no contribution to segmentation (32), and heretofore no effect has been seen on segmentation by changing the level or nature of zygotically expressed Chip (1). To detect an effect of Chip on Bcd activity, therefore, we reduced the activities of both Bcd and Chip to less than that provided by a single maternal dose of each. This was accomplished by producing doubly mutant mothers: these mothers were homozygous for the bcdE3 allele, which encodes a mutant with reduced DNA-binding activity (33), and were also heterozygous for the Chipg96.1 allele. This latter mutant allele encodes the SID fragment (Fig. 1A), which acts as a dominant negative, inhibiting, but not eliminating, maternal Chip activity. We deduced that the SID fragment inhibits maternal Chip activity from the observations that Chipg96.1/Chipg96.1 embryos produced by Chipg96.1/+ mothers die before reaching the larval stage (some display a mild segmentation defect), whereas all Chip−/Chip− embryos produced by Chip−/+ mothers segment normally and die as larvae. We further deduced that at least one maternal and two zygotic doses of the SID fragment are required to cause embryonic lethality from the fact that Chipg96.1/+ embryos from Chipg96.1/+ mothers segment normally and survive to adulthood. Presumably the SID fragment, produced in our experiment both maternally and zygotically, forms nonfunctional multimers with maternal wild-type Chip.
The key result is shown in Fig. 3: on average, embryos from Chipg96.1/+; bcdE3/bcdE3 mothers (and wild-type fathers) produced nearly one segment less than did embryos from bcdE3/bcdE3 mothers (6.02 ± 0.08 vs. 6.84 ± 0.06, respectively). This effect is almost certainly an underestimate of the effect of Chip on Bcd activity because the embryos are zygotically of two different Chip genotypes, Chipg96.1/+ and +/+, which cannot be distinguished by inspection. Presumably, the Chipg96.1/+ embryos have more SID fragment and display a larger effect than do the +/+ embryos. Moreover, embryos adapt to large changes in Bcd activity (34), and this adaptation may partly counteract the effects of reducing Chip activity.
We deduce that the Chipg96.1 mutation is solely responsible for the effect on segmentation observed in Fig. 3 because a wild-type Chip gene introduced on a P element (1) completely rescues Chipg96.1 homozygous embryos (not shown). We cannot formally rule out the possibility that part of the effect of the Chipg96.1 allele shown in Fig. 3 is due to reduced activities of other HD proteins, even though Bcd was the only HD protein that was mutant in this experiment. If Chipg96.1 were to substantially reduce the activities of other HD proteins, however, we would expect that posterior segments would be affected. This expectation arises because in contrast to Bcd, many other HD proteins have effects on posterior segments, as does the complete loss of maternal Chip activity. Fig. 3 shows, however, that posterior segments were not affected. We conclude, therefore, that most, if not all of the effect of partially reducing maternal Chip activity observed in Fig. 3 reflects a decrease in BcdE3 protein activity.
Chip Aids Transcriptional Activation by Bcd from Multiple Bcd-Binding Sites in Yeast.
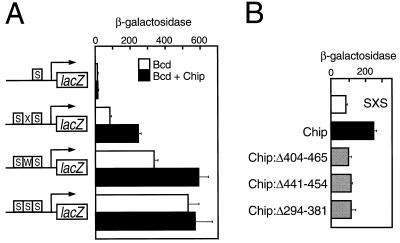
The experiments of Fig. 4 provide additional evidence that Chip directly affects Bcd activity by showing that, in yeast, Chip can potentiate the transcriptional activating function of Bcd (30, 33) at reporter genes that have multiple Bcd-binding sites. Yeast do not contain endogenous Chip or Bcd homologs, and exogenously added Bcd activates a reporter bearing Bcd-binding sites without contributions from other activators and repressors (unlike the situation with the typical Drosophila gene). This experiment thus provides a direct assay of the effect of Chip on Bcd activity. We used various combinations of Bcd-binding sites driving a yeast GAL1-lacZ reporter (Fig. 4A), and a vector producing Bcd under control of a β-estradiol sensitive activator (24). Increasing the β-estradiol concentration increases the amount of Bcd, which in turn increases activation of the reporter. These constructs were previously used to demonstrate that activation of a gene with multiple Bcd-binding sites is cooperative, involving pairwise interactions between Bcd molecules (24).
Figure 4.
Chip potentiates transcriptional activation by Bcd in yeast. (A) Effects of Chip on expression of GAL1-lacZ reporter genes with Bcd-binding sites. The reporters are: S, single strong Bcd-binding site; SXS, two strong binding sites flanking a nonbinding sequence; SWS, two strong binding sites flanking a weak binding site; SSS, three tandem strong binding sites (24). The bars indicate the β-galactosidase activity (Miller units per A595 Bradford protein unit) produced by yeast containing the reporter genes in the presence of nonsaturating levels of Bcd (induced with 0.5 nM β-estradiol) and in the absence (white bars) or presence (black bars) of Chip. The β-galactosidase levels obtained with the S reporter are >10-fold higher than the background activity. Error bars are standard errors. (B) The Chip SID and OID are required to increase activation of the SXS reporter. Bars indicate the β-galactosidase activity produced by the SXS GAL1-lacZ reporter gene in the presence of Bcd and the indicated Chip proteins.
Activation of a reporter containing three tandem, 11-bp strong Bcd-binding sites (SSS) was approximately half maximal at 0.5 nM β-estradiol, as measured by β-galactosidase activity. Fig. 4A shows that under these conditions, activation of a reporter bearing two strong Bcd-binding sites flanking a weak site (SWS) was ≈65% of that obtained with the SSS reporter. The figure also shows that two strong sites flanking a nonbinding spacer (SXS) mediated ≈15%, and a single strong site (S) ≈2% the activity observed with the SSS reporter. In the absence of either β-estradiol or the Bcd expression vector, there was <0.2% of the β-galactosidase activity obtained with the SSS reporter. These results confirm previous observations that activation increases with the strength and number of Bcd-binding sites (24).
Chip had little effect on activation of the SSS reporter by Bcd, but it increased activation of the SWS reporter to the level obtained with the SSS reporter (i.e., nearly 2-fold; Fig. 4A). Chip also increased activation of the SXS reporter 2.5- to 3-fold (Fig. 4A). Chip did not increase activation with a single binding site (S) (Fig. 4A). Chip mutants lacking the OID (Chip:Δ404–465 and Chip:Δ441–454) or the SID (Chip:Δ294–381) did not increase activation by Bcd (Fig. 4B), although Western blots confirmed that they were present at levels similar to wild-type Chip (not shown).
Chip did not increase activation above the level that can be obtained with higher concentrations of Bcd alone. For instance, when higher levels of Bcd were induced (using 2.5 nM β-estradiol), expression of the SXS reporter was increased to maximal levels and Chip did not significantly further increase activation (not shown). These results, taken together, are consistent with the idea that the effect of Chip on Bcd activity in yeast is to promote binding of Bcd to multiple sites (Fig. 5).
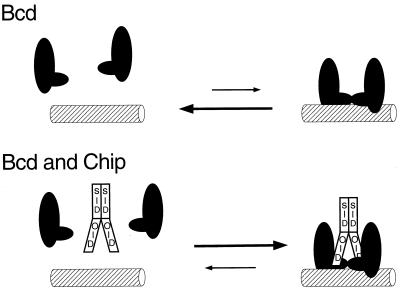
Figure 5.
Model for how Chip potentiates cooperative interactions between Bcd molecules. Bcd interacts cooperatively with itself through residues flanking the HD (Upper, ref. 36). We posit that a Chip dimer interacts with two Bcd molecules to fortify the Bcd interactions and increase DNA binding (Lower).
Discussion
We have shown that Chip has a region, the OID, that interacts in vitro with diverse HD proteins (Bcd, Ftz, and Otd) and with the Su(Hw) insulator protein. We have also shown that Chip potentiates the activity of Bcd in embryos and in yeast. These results help explain the broad segmentation and gene expression defects displayed by Chip mutants. The interaction between Chip and Su(Hw) supports previous genetic data indicating that Chip and the Su(Hw) insulator protein are antagonists in vivo, and therefore the notion that Chip facilitates enhancer–promoter communication.
Effects of Chip on HD Protein Activity.
Our results suggest that Chip increases interactions between Bcd molecules (Fig. 5). Thus, in yeast with nonsaturating levels of Bcd, Chip increased activation by Bcd from two strong binding sites separated by a weak site or by a nonbinding spacer, but not from one or three contiguous strong sites. Moreover, Chip did not increase activation above levels that were achieved with high concentrations of Bcd itself. Bcd binds DNA cooperatively, mediated by interactions of regions overlapping those that interact with Chip (Fig. 5; refs. 24 and 35–37), and we suggest that Chip interacts with Bcd to amplify that cooperativity (Fig. 5). It is unlikely that Chip itself is a transcriptional activator. Previous experiments have shown that Chip does not activate when tethered upstream of yeast promoters but that it can induce activation by recruiting an activation domain fused to LIM domains (1).
The idea that Chip increases interactions between certain other proteins agrees with all previous observations on Chip and its homologs. In transient transfection experiments with mammalian cells, Chip homologs increased transcriptional activation by the combination of the P-Otx HD and the Lhx3 LIM-HD proteins from a promoter containing a single binding site for each molecule (9). The Chip homologs had little effect with P-Otx or Lhx3 alone, indicating that they aid P-Otx-Lhx3 interactions. Furthermore, the nuclear LIM interactor (Nli) homolog of Chip aids formation of different LIM-HD protein dimers in vitro, an effect requiring the Nli SID (11). Finally, an Apterous-Chip fusion protein, in which the LIM domains of Apterous are replaced by the Chip SID, can replace wild-type Apterous in Drosophila wings, suggesting that Chip aids formation of Apterous dimers in vivo (15, 16).
Effects of Chip on Segmentation Gene Expression.
Our experiments demonstrate that Chip potentiates Bcd activity in the Drosophila embryo when the Bcd activity is low. This effect is consistent with previous studies on the expression of segmentation genes in embryos lacking maternal Chip activity. Embryos contain a gradient of Bcd protein, with a high concentration at the anterior end and a low concentration at the posterior end (38). Loss of maternal Chip strongly reduces all seven blastoderm stripes of Eve protein produced by the eve pair-rule gene (1). Many, if not all of these stripes are also regulated by Bcd, even though most occur in regions with low to intermediate Bcd concentrations (39). The eve stripes are activated by several remote enhancers located ≈1.5–9 kb from the promoter (40, 41) and Bcd-binding sites are critical for activation by at least the stripe 2 enhancer (42, 43). It is likely, therefore, that Chip increases eve expression at least in part by increasing binding of Bcd to the enhancers.
Accumulation of the Hb protein encoded by the hunchback (hb) gap gene is not substantially affected by loss of maternal Chip (1) even though hb expression is dependent on Bcd and several Bcd-binding sites just upstream of the promoter (33, 44, 45). This lack of an effect of Chip is not unexpected, however, because hb is expressed in the anterior end where the Bcd concentration is the highest (38).
The effects of Chip on Bcd activity that we have demonstrated cannot fully explain the roles of Chip in segmentation because Chip affects all segments (1) whereas Bcd does not (32). Proper formation of every segment does, however, depend on multiple HD proteins, and we show here that Chip also interacts with Otd and Ftz, and it is likely that it interacts with other HD proteins as well. Potentiation of the activities of other HD proteins besides Bcd would explain many of the additional effects of Chip on segmentation.
Effects of Chip on Enhancer–Promoter Communication.
Based on observations presented here and elsewhere, we suggest that Chip plays two roles in the regulation of gene expression. First, as discussed above, Chip is likely to aid binding of proteins to enhancers, and second, as previously proposed, Chip is also likely to function between enhancers and promoters to support enhancer–promoter communication (1–3). The in vitro interaction between Chip and the Su(Hw) insulator protein shown here is consistent with the notion that Su(Hw) is directly antagonistic to Chip activity as previously demonstrated genetically at the cut locus (2).
It remains to be seen how, if our speculations are correct, Chip facilitates enhancer–promoter communication and how that communication is disrupted by Su(Hw). It is believed that Su(Hw) blocks activation not by reducing the binding of proteins to enhancers, but rather by hindering enhancer-promoter communication (see reviews in refs. 3 and 6). For instance, an enhancer blocked in its interaction with one promoter by Su(Hw) can nevertheless activate a second promoter located on the opposite side of the enhancer from Su(Hw) (46, 47). Thus, although Su(Hw) is antagonistic to Chip, it is unlikely to affect binding of proteins to enhancers. It is also unlikely that Chip functions merely by preventing binding of Su(Hw) to gypsy because Chip is also important for the expression of several genes, e.g., cut and eve, in the absence of gypsy and Su(Hw) (1, 2)
It has been suggested that, in vivo, different HD proteins, including Bcd and Ftz, bind both to sites in the eve stripe enhancers and to many sites between the enhancers and the promoter (48, 49). It is conceivable that Chip aids binding of Bcd and Ftz to the sites between the enhancers and the promoter as well as to the sites in the enhancers. This could help form a series of loops that brings the enhancers and the promoter closer together, or help HD proteins bind near the promoter to serve as surrogate activators (3). The latter possibility is similar to the “linking” model put forth to explain long range activation of human β-globin genes by their locus control region (50). According to either scenario, Su(Hw) would block enhancer–promoter communication by interacting with the Chip molecules that function between the enhancer and the promoter and somehow interfering with their activities.
Acknowledgments
We thank Claude Desplan, Frieder Schöck, Steven Hanes, Mary K. Baylies, and Daniel Gietz for plasmids, Kathryn Anderson for flies, Linda Jurata, Gordon Gill, Don van Meyel and Mark Biggin for discussions, and Pamela Meluh, Mary K. Baylies, Kathryn Anderson, and Richard S. Mann for comments on the manuscript. This work was supported by grants from the National Institutes of Health to D.D., M.P., and the Memorial Sloan-Kettering Cancer Center, and a grant from the U.S.–Israel Binational Science Foundation to D.D.
Abbreviations
- Gst
glutathione-S-transferase
- HD
homeodomain
- Su(Hw)
suppressor of hairy-wing
- LID
LIM interaction domain
- SID
self-interaction domain
- Otd
Orthodenticle
- OID
other interaction domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050586397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050586397
References
- 1.Morcillo P, Rosen C, Baylies M K, Dorsett D. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morcillo P, Rosen C, Dorsett D. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsett D. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 4.Jack J, Dorsett D, DeLotto Y, Liu S. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- 5.Dorsett D. Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell A C, Felsenfeld G. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 7.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 8.Jurata L W, Kenney D A, Gill G N. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bach I, Carriere C, Ostendorff H P, Anderson B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 10.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 11.Jurata L W, Pfaff S L, Gill G N. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Funez P, Lu C H, Rincon-Limas D E, Garcia-Bellido A, Botas J. EMBO J. 1998;17:6846–6853. doi: 10.1093/emboj/17.23.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milan M, Diaz-Benjumea F J, Cohen S M. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoresh M, Orgad S, Shmueli O, Werczberger R, Gelbaum D, Abiri S, Segal D. Genetics. 1998;150:283–299. doi: 10.1093/genetics/150.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milan M, Cohen S M. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- 16.van Meyel D J, O'Keefe D D, Jurata L W, Thor S, Gill G N, Thomas J B. Mol Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- 17.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannister A J, Cook A, Kouzarides T. Oncogene. 1991;6:1243–1250. [PubMed] [Google Scholar]
- 20.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Shen B, Rosen C, Dorsett D. Mol Cell Biol. 1996;16:3381–3392. doi: 10.1128/mcb.16.7.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Shen B, Dorsett D. Genetics. 1993;135:343–355. doi: 10.1093/genetics/135.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levis R, Bingham P M, Rubin G M. Proc Natl Acad Sci USA. 1982;79:564–568. doi: 10.1073/pnas.79.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burz D S, Rivera-Pomar R, Jäckle H, Hanes S D. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 27.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 28.Jurata L W, Gill G N. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breen J J, Agulnick A D, Westphal H, Dawid I B. J Biol Chem. 1998;273:4712–4717. doi: 10.1074/jbc.273.8.4712. [DOI] [PubMed] [Google Scholar]
- 30.Driever W, Ma J, Nüsslein-Volhard C, Ptashne M. Nature (London) 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- 31.Schaeffer V, Janody F, Loss C, Desplan C, Wimmer E A. Proc Natl Acad Sci USA. 1999;96:4461–4466. doi: 10.1073/pnas.96.8.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fröhnhoefer H G, Nüsslein-Volhard C. Nature (London) 1986;324:120–125. [Google Scholar]
- 33.Struhl G, Struhl K, Macdonald P M. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 34.Namba R, Pazdera T M, Cerrone R L, Minden J S. Development. 1997;124:1393–1403. doi: 10.1242/dev.124.7.1393. [DOI] [PubMed] [Google Scholar]
- 35.Ma X, Yuan D, Diepold K, Scarborough T, Ma J. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- 36.Yuan D, Ma X, Ma J. J Biol Chem. 1996;271:21660–21665. [PubMed] [Google Scholar]
- 37.Ma X, Yuan D, Scarborough T, Ma J. Biochem J. 1999;338:447–455. [PMC free article] [PubMed] [Google Scholar]
- 38.Driever W, Nüsslein-Volhard C. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 39.Reinitz J, Sharp D H. Mech Dev. 1995;49:133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- 40.Fujioka M, Emi-Sarker Y, Yusibova G L, Goto T, Jaynes J B. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goto T, Macdonald P, Maniatis T. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 42.Small S, Kraut R, Hoey T, Warrior R, Levine M. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 43.Stanojevic D, Small S, Levine M. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 44.Driever W, Nüsslein-Volhard C. Nature (London) 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 45.Schröder C, Tautz D, Seifert E, Jäckle H. EMBO J. 1988;7:2881–2887. doi: 10.1002/j.1460-2075.1988.tb03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai H, Levine M. Nature (London) 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 47.Scott K S, Geyer P K. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter J, Dever C A, Biggin M D. Genes Dev. 1994;8:1678–1692. doi: 10.1101/gad.8.14.1678. [DOI] [PubMed] [Google Scholar]
- 49.Carr A, Biggin M D. EMBO J. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulger M, Groudine M. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]