Abstract
Human cytomegalovirus blocks cell-cycle progression in the G1 compartment upon infection of primary human fibroblasts. The virus-coded UL69 protein can institute a G1 block when expressed in cells in the absence of virus infection. We have constructed a cytomegalovirus mutant, TNsubUL69, that lacks the UL69 coding region. This virus grows slowly in fibroblasts, but produces a wild-type yield after an extended delay. It grows with normal kinetics in cells coinfected with a recombinant retrovirus, retroUL69, which expresses UL69 protein, demonstrating that its growth defect results from the mutation in the UL69 gene. UL69 protein is packaged within virus particles, and it was possible for us to produce two types of virus stocks. TNsubUL69+pUL69 lacks the UL69 gene but contains UL69 protein in virus particles. It is produced by growth in fibroblasts that are coinfected with retroUL69. TNsubUL69−pUL69 lacks the UL69 gene and protein. It is produced by growth in fibroblasts that do not contain UL69 protein. The mutant virions lacking both the UL69 gene and protein fail to induce a cell-cycle block with normal efficiency, whereas the mutant particles lacking the gene but containing the protein can institute the block. These results are consistent with the view that the UL69 protein contributes to the cytomegalovirus-induced cell-cycle block, and they suggest that UL69 protein delivered to cells within virions can induce the block without the synthesis of additional UL69 protein encoded by the infecting viral genome.
Human cytomegalovirus (HCMV), a β-herpesvirus, is a ubiquitous pathogen. Although HCMV infection generally is asymptomatic in healthy children and adults, it is the leading viral cause of birth defects (1) and a major cause of morbidity and mortality in immunocompromised individuals (2).
HCMV infection blocks cell-cycle progression in primary human fibroblasts (3–6). The arrest occurs at multiple phases of the cell cycle, including a point late in the G1 compartment. We have previously shown that expression of the HCMV UL69 protein in human tumor cells or primary fibroblasts blocks cell-cycle progression (7). Cells expressing UL69 protein after transfection with an expression plasmid or infection with a recombinant retrovirus accumulate in the G1 compartment of the cell cycle. UL69 protein is a constituent of the virion (8) that exhibits transcriptional regulatory activity (9, 10). The mechanism by which UL69 institutes a cell-cycle block is not known.
In this paper, we describe the construction and analysis of a UL69-deficient HCMV mutant, TNsubUL69. The mutant virus lacking the UL69 gene and protein is defective for replication in fibroblasts, producing a normal yield only after a significant delay. The mutant virus lacking the UL69 gene and protein fails to efficiently induce a G1 block after infection of fibroblasts, whereas the mutant virus lacking the UL69 gene but containing UL69 protein can institute the block. These results demonstrate that UL69 protein contributes to the cell-cycle block observed after infection with HCMV, and reveal that UL69 protein delivered to cells within infecting virions is able to induce the block without the synthesis of additional UL69 protein encoded by the infecting genome.
Materials and Methods
Cells and Viruses.
The maintenance of primary human fibroblasts and their infection with HCMV were as described previously (5). HCMV strain Towne (11) was obtained from the American Type Culture Collection. A plaque-purified derivative was prepared and designated TNwt. TNsubUL69 was prepared by mutation of TNwt by homologous recombination within human fibroblasts. Three DNAs simultaneously were introduced into fibroblasts by electroporation: (i) Towne DNA isolated from TNwt virions; (ii) pUL69repEGFP, which contains the enhanced green fluorescent protein (EGFP) coding region flanked by HCMV DNA segments derived from the regions immediately upstream (AD169 sequence 100434–101235, ref. 12; the Towne strain has not been sequenced but is highly related to the AD169 strain) and downstream (AD169 sequence 97216–98289) of the UL69 region that was targeted for deletion; and (iii) pCMV71, which expresses the viral pp71 tegument protein that has been shown to enhance the infectivity of HCMV DNA (13). The resulting fluorescent plaques contained a mixture of mutant and wild-type viruses. To purify the defective mutant away from contaminating wild-type virus, virus from the mixed plaques was used to infect fibroblasts that were infected previously with a recombinant retrovirus, retroUL69 (7). The defective retrovirus was produced in Phoenix A cells (14), and it encodes the UL69 protein. Three sequential rounds of plaque purification in the presence of the complementing retrovirus resulted in the isolation of pure mutant virus. The deletion in TNsubUL69 extends from bp 98290 to bp 100433 (AD169 sequence numbers), removing the ATG and all but the 3′-terminal 91 bp of the UL69 coding region, and the inserted EGFP coding region is expressed under control of the UL69 immediate early promoter. TNsubUL69 propagated in retroUL69-infected human fibroblasts contains UL69 protein, and the virus is designated TNsubUL69+pUL69. TNsubUL69 propagated for one passage in fibroblasts that are not infected with retroUL69 lacks the UL69 protein, and the virus is designated TNsubUL69−pUL69. The infectious titers of these virus stocks were determined by plaque assay on retroUL69-infected fibroblasts.
Analysis of Mutant Virus Phenotype.
To determine the effect of wild-type and mutant viruses on cell-cycle progression, primary human fibroblasts were infected at a multiplicity of 10 plaque-forming units (pfu) per cell. Cultures were treated with nocodozole (100 ng/ml) and phosphonoacetic acid (250 μg/ml) at 12 h after infection to institute a cell-cycle block at G2/M and to prevent HCMV DNA replication. At 30 or 48 h after infection, cells were removed from culture dishes by trypsinization and fixed with 70% ethanol at −20°C for 3 h. They were resuspended in PBS containing propidium iodide (50 μg/ml) and ribonuclease A (100 μg/ml), their DNA content was determined by fluorescence-activated cell sorter (FACS) analysis using a FACScan (Becton Dickinson), and data analysis was performed with cellquest software (Becton Dickinson).
Southern, Northern, and Western blot analyses to monitor viral DNA, mRNA, and protein accumulation, respectively, were performed as described previously (15). For analysis of virion proteins, virus particles were partially purified by centrifugation through a sorbitol cushion (16). The UL69-specific monoclonal antibody used for the Western blots has been described (7).
Results
Construction and Propagation of a Mutant Lacking the UL69 Coding Region.
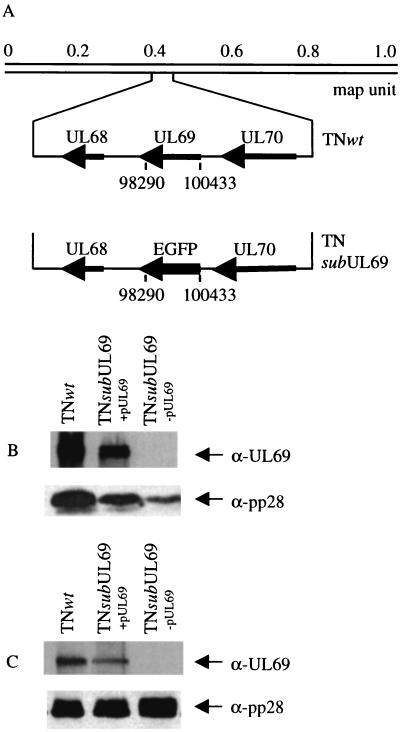
A substitution mutant carrying the EGFP coding region in place of the majority of the UL69 coding region was produced by homologous recombination within infected human fibroblasts (Fig. 1A). The substitution mutant, termed TNsubUL69, was defective, and it was propagated in fibroblasts that previously were infected with a recombinant retrovirus, retroUL69, expressing the UL69 protein. Analysis of the mutant virus DNA by Southern blot confirmed that the variant lacks the UL69 coding region and contains the EGFP marker (data not shown). Extracts of cells produced at 72 h after infection with retroUL69 plus TNsubUL69 contained UL69 protein (Fig. 1B, lane 2), as did mutant virions produced in the coinfected cells (Fig. 1C, lane 2). When mutant virions containing the UL69 protein were used to infect fibroblasts in the absence of the UL69-expressing retrovirus, no UL69 protein was detected in extracts of infected cells (Fig. 1B, lane 3) or in the resulting virions (Fig. 1C, lane 3). Thus, it was possible to generate two types of mutant virus stocks. TNsubUL69+pUL69 lacks the UL69 gene but contains UL69 protein in virus particles, whereas TNsubUL69−pUL69 particles lack both the UL69 gene and protein.
Figure 1.
Construction and analysis of TNsubUL69. (A) A substitution mutant was produced in which a 2,144-bp segment within the UL69 coding region extending from sequence position 98290 to 100433 (AD169 sequence numbers) was replaced with the EGFP coding region. (B) Human fibroblasts were infected with wild-type virus (TNwt), mutant virus grown in the presence of helper retrovirus (TNsubUL69+pUL69), or mutant virus grown for one cycle in the absence of helper retrovirus (TNsubUL69−pUL69), and UL69 protein was assayed by Western blot using a UL69-specific monoclonal antibody (α-UL69) at 72 h after infection. The late protein, pp28, was assayed as a control by using a pp28-specific monoclonal antibody (α-pp28). The expression of late viral gene products is reduced in mutant virus-infected cells. (C) Virus particles were partially purified and virion proteins were assayed by Western blot.
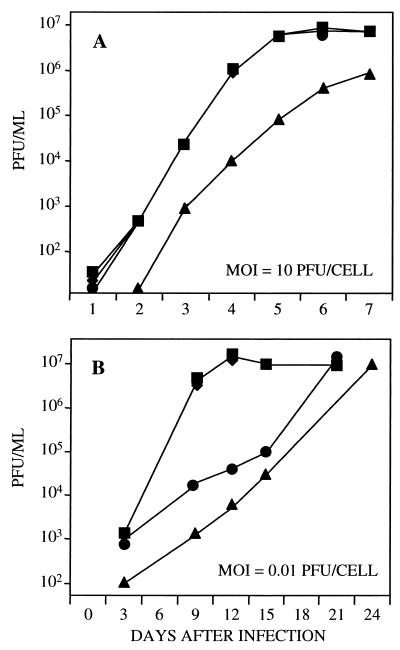
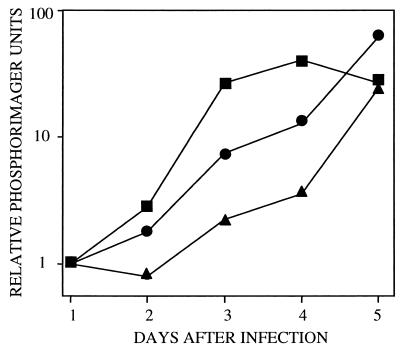
The growth kinetics of the mutant in human fibroblasts was evaluated at two input multiplicities of infection. At a multiplicity of 10 pfu per cell, the yield of TNsubUL69−pUL69 was reduced by a factor of 20–50 in comparison to the wild-type virus, TNwt, through day 5 after infection; by day 7, the mutant yield was reduced by a factor of 10 relative to the wild type (Fig. 2A). The growth defect of TNsubUL69−pUL69 was fully complemented by prior infection of fibroblasts with retroUL69 (Fig. 2A). The efficient complementation demonstrated that the growth defect displayed by TNsubUL69−pUL69 results from the deletion of the UL69 gene and not from other alterations of the mutant virus genome. At this multiplicity, TNsubUL69+pUL69 grew with wild-type kinetics (Fig. 2A), revealing that protein contained within the virion is sufficient to mediate the essential UL69 function throughout the viral growth cycle. At a lower multiplicity of infection, 0.01 pfu per cell, TNsubUL69−pUL69 produced a yield that was reduced by a factor of 2000 on day 9 (Fig. 2B). The mutant ultimately achieved a wild-type yield, but long after the wild-type parent. Thus, the lack of UL69 dramatically slows viral replication, but does not prevent the mutant from eventually producing a wild-type yield. Prior infection of fibroblasts with retroUL69 accelerated the replication of TNsubUL69−pUL69 to wild-type kinetics (Fig. 2B), confirming that the phenotype of the mutant results from the lack of UL69 protein. Like TNsubUL69−pUL69, TNsubUL69+pUL69 exhibited delayed growth kinetics when used to infect cells at a multiplicity of 0.01 pfu per cell (Fig. 2B). This result is in contrast to infection at a multiplicity of 10 pfu per cell, where TNsubUL69+pUL69 replicated with wild-type kinetics (Fig. 2A). We interpret this result to indicate that insufficient UL69 protein is delivered to cells within virions at a low multiplicity of infection, which results in delayed growth.
Figure 2.
Growth kinetics of wild-type and mutant viruses. Human fibroblasts were infected at a multiplicity of 10 pfu per cell (A) or 0.01 pfu per cell (B) with TNwt (■), TNsubUL69+pUL69 (●), TNsubUL69−pUL69 (▴), or TNsubUL69−pUL69 plus retroUL69 (♦). Cultures were harvested at the indicated times after infection, and infectious virus was assayed by plaque assay on retroUL69-infected fibroblasts. Similar results were obtained in two independent experiments.
UL69 Contributes to the G1 Block in HCMV-Infected Cells.
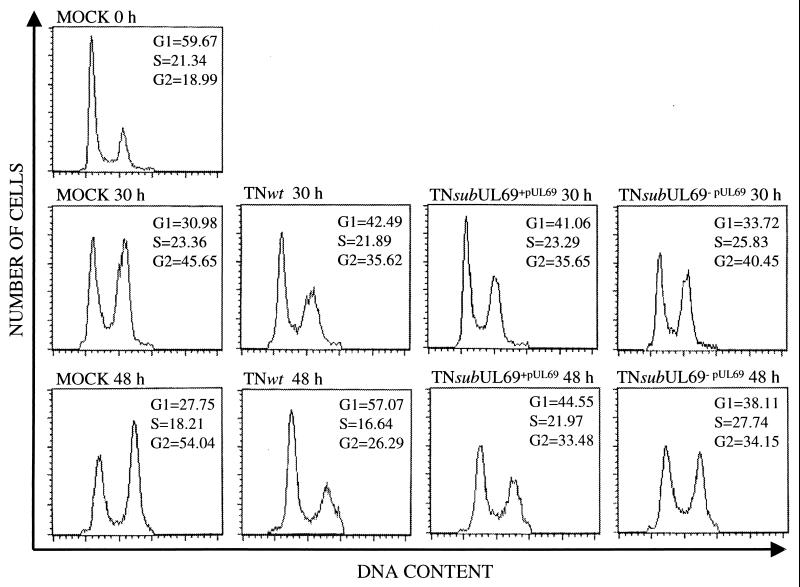
As noted above, we previously have reported that UL69 protein blocks cell-cycle progression in the G1 compartment when it is expressed in cells that are not infected with HCMV (7). To test whether UL69 protein is required for the G1 block observed within infected cells (4–6), we evaluated the ability of TNsubUL69+pUL69 and TNsubUL69−pUL69 to inhibit cell-cycle progression (Fig. 3). At 12 h after mock infection or infection at a multiplicity of 10 pfu per cell, cultures of fibroblasts were treated with nocodozole to block cell-cycle progression at the G2/M boundary and with phosphonoacetic acid to prevent viral DNA replication. This procedure enabled us to measure the portion of cells that remained in G1 by FACS analysis without the confounding effects of additional cells cycling into the compartment or the accumulation of viral DNA. At the time of drug addition, 59.7% of mock-infected human fibroblast cells resided in the G1 compartment (Fig. 3, Mock 0 h). At 30 h after infection, when the culture had been maintained in nocodozole for 18 h, 31% of mock-infected cells were in G1 (Fig. 3, Mock 30 h), reflecting the movement of cells out of G1 and through S and G2 to the drug-induced block. When TNwt-infected cells were subjected to the same 18-h drug treatment, 42.5% of the culture remained in G1 (Fig. 3, TNwt 30 h). The increase in the number of cells in the G1 compartment in TNwt-infected as compared with mock-infected cells is caused by the ability of HCMV to institute a G1 block (4–6). In contrast to the result with TNwt, only 33% of TNsubUL69−pUL69-infected cells remained in G1 (Fig. 3, TNsubUL69−pUL69 30 h), indicating that the G1 block was not instituted efficiently in the absence of UL69 protein. Whereas mutant virions lacking UL69 protein were less effective than wild-type virus in holding cells in G1, mutant virus particles containing the UL69 protein behaved like the wild-type parent (Fig. 3, TNsubUL69+pUL69 30 h).
Figure 3.
Effect of wild-type and mutant viruses on cell-cycle progression. Human fibroblasts were mock-infected or infected at a multiplicity of 10 pfu per cell with TNwt, TNsubUL69+pUL69, or TNsubUL69−pUL69. Cultures were treated with nocodozole and phosphonoacetic acid 12 h later. Cells were harvested, fixed, and stained at either 30 or 48 h after infection, and then they were subjected to FACS analysis. The proportions of cells in G1, S, and G2 were determined by using cellquest software. As a control, mock-infected cells that were not treated with drugs (MOCK 0 h) were also analyzed.
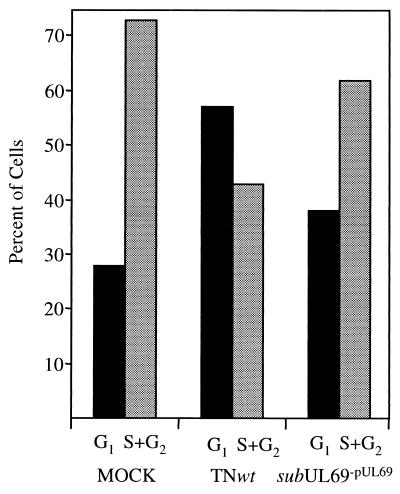
TNsubUL69−pUL69 also held cells in G1 less efficiently than did TNwt in a second experiment, where nocodozole was again added at 12 h after infection but cells were maintained in the drug for 36 h before FACS analysis (Fig. 3, TNsubUL69−pUL69 48 h). The TNsubUL69−pUL69 cell-cycle phenotype readily is apparent when the number of cells in G1 is compared with the portion of the culture in S + G2 (Fig. 4). Mock-infected cultures contain fewer cells in G1 than in the S + G2 compartments, whereas the ratio is reversed in TNwt-infected cultures. The TNsubUL69−pUL69 ratio is more similar to the mock-infected ratio than the TNwt-infected G1/(S + G2) ratio. In the experiment analyzed at 48 h after infection, mutant virus containing UL69 protein, TNsubUL69+pUL69, was intermediate between wild type and mutant virions lacking the protein in its ability to hold cells in G1 (Fig. 3, TNsubUL69+pUL69 48 h).
Figure 4.
Effects of wild-type and mutant viruses on cell-cycle progression. The bar graph compares the number of fibroblasts in G1 versus S + G2 for cultures that were either mock-infected or infected, treated with nocodozole and phosphonoacetic acid 12 h later, and then harvested and analyzed after an additional 36 h (at 48 h after infection). The primary data used to generate this graph are displayed in Fig. 3.
We conclude that UL69 protein contributes to the G1 block that is induced by infection with HCMV. Further, when cells are infected at a multiplicity of 10 pfu per cell, the UL69 protein in virions is sufficient to induce a block in a substantial portion of infected cells. Wiebusch and Hagemeier (17) recently have reported that UV-irradiated virus is not able to block cell-cycle progression. It is unlikely that UL69 protein contained within virus particles was inactivated by this treatment. Conceivably, UL69 protein must reach an intracellular threshold level to block cell-cycle progression, and more of the virion-associated UL69 protein was delivered to the infected cells in our experiments.
UL69 Is Required for Efficient HCMV Replication and Late Gene Expression.
Viral DNA accumulation was monitored by Southern blot analysis after infection of fibroblasts with the wild-type or mutant virus at a multiplicity of 10 pfu per cell (Fig. 5). A substantial delay was evident in TNsubUL69−pUL69-infected cells, but, given sufficient time, viral DNA accumulated to a wild-type level in mutant-infected cells. TNsubUL69+pUL69 exhibited a more modest defect in the accumulation of viral DNA than did the mutant particles lacking UL69 protein.
Figure 5.
Accumulation of wild-type and mutant HCMV DNA. Human fibroblasts were infected at a multiplicity of 10 pfu per cell with TNwt (■), TNsubUL69+pUL69 (●), or TNsubUL69−pUL69 (▴). Cells were harvested at the indicated times after infection, viral DNA accumulation was assayed by Southern blot using 32P-labeled HCMV DNA as a probe, and radioactivity was quantified with a phosphorimager. Similar results were obtained in two independent experiments.
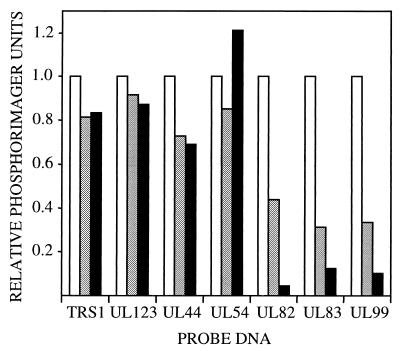
Viral mRNA accumulation was monitored by Northern blot assay after infection at a multiplicity of 10 pfu per cell (Fig. 6). Similar amounts of immediate-early, UL123 (IE1) and TRS1, and early, UL44 and UL54, mRNAs accumulated after infection with mutant as compared with wild-type viruses. However, late mRNAs accumulated to substantially reduced levels in TNsubUL69−pUL69-infected cells. UL82 (pp71), UL83 (pp65), and UL99 (pp28) mRNAs were reduced by factors of 24, 9, and 11, respectively. It is possible that late viral mRNAs accumulate to normal levels after a delay, but this possibility has not been tested. Late mRNA accumulation was also deficient in TNsubUL69+pUL69-infected cells, but to a lesser extent than that observed for TNsubUL69−pUL69.
Figure 6.
Accumulation of wild-type and mutant HCMV mRNA. Human fibroblasts were infected at a multiplicity of 10 pfu per cell with TNwt (white bars), TNsubUL69+pUL69 (gray bars), or TNsubUL69−pUL69 (black bars), and polyadenylated RNA was prepared at 6 h (UL123, TRS1), 24 h (UL44, UL54), or 72 h (UL82, UL83, UL99) to monitor immediate-early, early, and late mRNAs, respectively. Viral mRNA was assayed by Northern blot using 32P-labeled gene-specific probes, and radioactive bands were quantified with a phosphorimager. Similar results were obtained in two independent experiments.
Our results demonstrate that UL69 protein is required for efficient DNA replication and late mRNA accumulation. UL69 protein delivered to infected cells in mutant virions can partially alleviate both of these defects.
Discussion
Our results lead to three principal conclusions. First, it is possible to complement fully the growth of mutant HCMV in human fibroblasts by using a recombinant retrovirus. Second, UL69 is required for HCMV efficiently to induce a G1 block within infected cells. Third, UL69 protein packaged in virus particles is sufficient to induce a G1 block and to generate a normal yield of virus when cells are infected at a relatively high multiplicity. It is not necessary to express UL69 protein from the infecting viral genome.
TNsubUL69 is profoundly defective when used to infect cells at a low multiplicity of infection in the absence of UL69 protein (Fig. 2B). The defect is not absolute. Given time, the mutant virus produces an infectious yield similar to that of the wild-type virus. It was not practical to generate a derivative of human fibroblasts containing a constitutively expressed UL69 gene, because expression of the gene is toxic and because the primary cells have a limited lifespan. However, TNsubUL69 produced plaques and grew with normal kinetics (Fig. 2) on fibroblasts that first were infected with retroUL69. The full complementation demonstrates that the TNsubUL69 phenotype results entirely from the lack of UL69, obviating the need to produce and analyze a revertant virus.
Complementation of TNsubUL69 led to an interesting and useful complexity. Because UL69 protein is a constituent of the virion (8), mutant virions that are produced in a complementing infection contain UL69 protein (Fig. 1C, lane 2). TNsubUL69+pUL69 particles contain UL69 protein but lack the UL69 gene. Because TNsubUL69+pUL69 can be propagated in the absence of complementation, it is possible to produce TNsubUL69−pUL69, which is lacking detectable amounts of the protein (Fig. 1C, lane 3) and lacking the gene. TNsubUL69+pUL69 grows as well as TNwt when fibroblasts are infected at a multiplicity of 10 pfu per cell, but it grows slowly when fibroblasts are infected at a multiplicity of 0.01 pfu per cell. Thus, the mutant replicates normally if sufficient UL69 protein is delivered to the cell as a constituent of the virion. It is not necessary to produce UL69 protein from the genome during the late phase of infection to generate a normal virus yield.
TNsubUL69−pUL69 exhibited a slowed replication cycle (Fig. 2B) and failed to block efficiently cell-cycle progression in the G1 compartment (Fig. 3). The alteration in cell-cycle regulation was predicted by our earlier work (7) in which we demonstrated that UL69 protein blocks cell-cycle progression when it is expressed in the absence of HCMV infection. However, our present results do not rule out the possibility that a subset of cells infected with TNsubUL69−pUL69 remain blocked in G1. It has been reported recently that the HCMV IE2 protein can block cell-cycle progression (17). TNsubUL69−pUL69 produces normal levels of IE2 mRNA (Fig. 5), and it is possible that IE2 protein is responsible for a residual but incomplete cell-cycle block in cells infected with TNsubUL69−pUL69. As yet, the mechanism by which UL69 institutes a G1 block is unknown.
TNsubUL69−pUL69-infected cells contained normal levels of the immediate-early and early viral mRNAs that were tested (Fig. 5). However, viral DNA replication was delayed (Fig. 4), and the expression of the late mRNAs that were tested was reduced (Fig. 5). We do not know yet the mechanistic basis for these defects. UL69 might act directly to facilitate viral DNA replication and late gene expression. Although UL69 is not known to have a direct role in DNA replication (18, 19), it does exhibit transcriptional regulatory activity (9, 10). Further, UL69 is related in its sequence to the herpes simplex virus ICP27 protein (12), and ICP27 is known to mediate the cytoplasmic accumulation of intronless mRNAs (20). It is possible that UL69 directly influences late HCMV mRNA accumulation at a posttranscriptional level. However, UL69 cannot substitute for ICP27 in a herpes simplex virus infection (9), so a direct role for the HCMV protein in mRNA biogenesis remains uncertain.
Conceivably, the defect in DNA replication and late gene expression results from the inability of TNsubUL69−pUL69 to induce a block in the G1 compartment of the cell cycle. Future studies will be needed to test this hypothesis.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (CA82396). C.B. received fellowship support from Bristol-Myers Squibb, M.L.H. was supported by a fellowship from the American Heart Association, and T.S. is an American Cancer Society Professor.
Abbreviations
- HCMV
human cytomegalovirus
- EGFP
enhanced green fluorescent protein
- pfu
plaque-forming units
- FACS
fluorescence-activated cell sorter
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050587597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050587597
References
- 1.Alfred C A, Stagno S, Pass R F, Britt W. Rev Infect Dis. 1990;12:S745–S753. doi: 10.1093/clinids/12.supplement_7.s745. [DOI] [PubMed] [Google Scholar]
- 2.Britt W, Alfred C A. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2493–2523. [Google Scholar]
- 3.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richmann D D, Spector D H. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 5.Lu M, Shenk T. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittmer D, Mocarski E S. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu M, Shenk T. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. .A. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler M, Stamminger T. J Virol. 1996;70:8984–8987. doi: 10.1128/jvi.70.12.8984-8987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler M, Rice S A, Stamminger T. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romanowski M, Garrido-Guerrero E, Shenk T. J Virol. 1997;71:5703–5705. doi: 10.1128/jvi.71.7.5703-5705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plotkin S A, Furukawa T, Zygraich N, Huygelen C. Infect Immun. 1975;12:521–527. doi: 10.1128/iai.12.3.521-527.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, III, Kouzarides T, Martignetti J A, et al. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Baldick C J, Marchini A, Patterson C E, Shenk T. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 15.Patterson C E, Shenk T. J Virol. 1999;73:7126–7131. doi: 10.1128/jvi.73.9.7126-7131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stinski M F. J Virol. 1976;19:594–609. doi: 10.1128/jvi.19.2.594-609.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiebusch L, Hagemeier C. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pari G S, Anders D G. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandri-Golden R M. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]