Abstract
A unique cohort of HIV-1-infected long term nonprogressors (LTNP) with normal CD4+ T cell counts and <50 copies/ml of plasma were prospectively recruited for study. HLA typing revealed a dramatic association between the HLA B*5701 class I allele and nonprogressive infection [85% (11 of 13) vs. 9.5% (19 of 200) in progressors; P < 0.001]. Antigen-specific CD8+ T cells were enumerated by flow cytometric detection of intracellular IFN-γ in response to HIV antigens and HLA B*57-gag tetramer staining. No quantitative differences in the total HIV-specific CD8+ T cell responses were observed between B*57+ LTNP and five B*57+ progressors (P = 0.4). Although similar frequencies of peptide specific CD8+ T cells were also found, the gag-specific CD8+ T cell response in the LTNP group was highly focused on peptides previously shown to be B*57-restricted. These findings indicate that, within this phenotypically and genotypically distinct cohort, a host immune factor is highly associated with restriction of virus replication and nonprogressive disease. They also strongly suggest a mechanism of virus specific immunity that directly operates through the B*5701 molecule. Further characterization of qualitative differences in the virus-specific responses that distinguish HLA B*57+ LTNP from progressors may ultimately define mechanisms of effective immune mediated restriction of virus replication.
After infection with HIV, progression to AIDS typically occurs within 10 years. An unusually benign clinical course extending beyond the median time to AIDS is observed in a subset of HIV-1-infected patients variably referred to as long term asymptomatics or nonprogressors (LTNP). Considerable efforts have been focused on determining the host or virus factors that might cause or be associated with a nonprogressive clinical course. Extensive studies have demonstrated strong cellular and humoral HIV-directed responses in patients with slowly progressive or nonprogressive infection (1–7). However, it has been difficult to conclude whether such vigorous HIV-directed responses are the cause of nonprogression, given that comparisons are made with responses of patients with advanced disease and global decline in immunity. To date, decreases in virus replication caused by deletions within a virus co-receptor (human cc-chemokine receptor-5) of the host (8, 9) or within the virus nef (10–12) gene have accounted for many patients that meet commonly used definitions of nonprogressors. Although these and other factors not believed to operate directly through virus-specific immunity account for many patients with nonprogressive disease, there remain patients in whom these virus or host factors have not been found (13).
It is now apparent that a large fraction of patients previously considered LTNP ultimately show a decline in CD4+ T cell numbers (14, 15). A small subpopulation of patients (<0.8% of HIV-infected individuals) show no signs of progression over a 10-year period (14, 16). These patients are characterized by stable CD4+ T cell counts and <50 copies of viral RNA/ml plasma in the absence of antiretroviral therapy. Because these patients are rare, previous studies even in large cohorts have not included more than two such patients. We have recently described a small cohort of four such patients with <50 copies of viral RNA/ml plasma and strong proliferative responses to HIV antigens (17). These patients were also phenotypically distinguished by the ability of their peripheral blood mononuclear cells (PBMC), when engrafted into severe combined immunodeficient animals, to restrict autologous or challenge virus replication. In the present study, we have prospectively recruited a larger cohort of LTNP with these clinical characteristics. We demonstrate a striking association with the HLA B*57 allele and investigate its possible functional significance.
Materials and Methods
Patients.
Patients that were HIV-infected for more than 2 years with <50 copies of viral RNA/ml plasma in the absence of antiretroviral therapy were prospectively recruited. Patient 3 previously received IFN-α/AZT (1/90–12/95) or IFN-α/AZT/DDI (1/96–12/96) as part of a National Institute of Allergy and Infectious Diseases protocol. This patient has remained off of antiretrovirals since that time. All other patients have not received antiretrovirals during or before the study period. HIV infection in study participants was documented by HIV-1/2 enzyme immunoassay. All subjects signed informed consent approved by the National Institute of Allergy and Infectious Diseases investigational review board. Patients 3–6(17), 7(18), and 10(C135) (11) have previously been reported in other studies. HLA class I and II typing was performed by hybridization with sequence specific oligonucleotide probes after amplification of the corresponding genes by using PCR as described elsewhere (19). To detect polymorphisms within HLA B*57, cDNA was amplified by PCR using primers that span the 5′ to 3′ untranslated regions, was cloned, and was sequenced as described elsewhere (20). CCR5 deletion mutations were detected as described (21).
Intracellular Cytokine Detection Assay and Flow Cytometry.
Peripheral blood lymphocytes were obtained by sodium diatrizoate density centrifugation (Organon Teknika, Durham, NC) of apheresis donor packs. Either fresh or cryopreserved PBMC were used as effectors. PBMC were cryopreserved in RPMI media with 10% FBS and 7.5% DMSO at −140°C. Cryopreserved PBMC were cultured overnight at 37°C in RPMI 10% FBS before use as effectors. Preliminary experiments yielded similar results with fresh or cryopreserved PBMC. Patients 12, 13, and 30 were not included in this analysis because sufficient numbers of cells of these patients were not available at the time of this assay.
Autologous EBV-transformed B cells were infected for 16 h at 37°C with the recombinant vaccinia viruses vVK1 (containing the HIV-1HXB2 gag-pol gene), vP1287 (HIV-1IIIB gag), vP1289 (HIV-1IIIB p24), vP1290 (HIVIIIB p17), vP1288 (HIVIIIB pol), vPE16 (HIV-1BH10 env), vTFnef (HIV-1pNL432 nef), vP1490 (HIVIIIB rev), HIVIIIBtat, or the negative control virus vSC8 (Escherichia coli β-galactosidase) as previously described (22). The vP1287 (HIV-1IIIB gag), vP1289 (HIV-1IIIB p24), vP1290 (HIVIIIB p17), vP1288 (HIVIIIB pol), and vP1490 (HIVIIIB rev) viruses were contributed to the National Institutes of Health AIDS Research and Reference Reagent Program by Virogenetics (Troy, NY). The vTFnef virus was contributed by MedImmune (Gaithersburg, MD), and vPE16, vVK1, and VSC8 viruses were contributed by Bernard Moss (National Institute of Allergy and Infectious Diseases). HIVIIIBtat (23) was supplied by Bernard Moss.
Intracellular cytokine detection was performed as described (24). In brief, 4 million PBMC were incubated with 4 × 105 uninfected, vac-β-galactosidase, or vac-HIV-recombinant infected autologous EBV transformed B cells in a final volume of 2 ml of media. This effector-to-target ratio was previously demonstrated to give optimal responses with low background or bystander activation (J. Gea-Banacloche, personal communication). At 2 h of incubation, brefeldin-A (Sigma) was added to the medium at a final concentration of 10 μg/ml to inhibit cytokine secretion. At 6 h of incubation, the cells were washed twice, were fixed in 4% paraformaldehyde (Sigma), and were permeabilized and blocked or frozen for future use.
Four color flow cytometry was performed according to standard protocols (25). Surface or intracellular staining was performed by using the following antibodies: FITC-conjugated anti-IFN-γ (PharMingen) and anti-CD3 (Becton Dickinson); PE-conjugated anti-CD3, anti-CD8, and anti-CD69 (Becton Dickinson); APC-conjugated anti-CD3 (Becton Dickinson); PerCP-conjugated anti-CD3 and anti-CD8 (Becton Dickinson). Gating on CD3+CD8+ lymphocytes, 15,000–200,000 events (100,000–700,000 total cells) were collected. Data were analyzed by using either cellquest (Becton Dickinson) or flowjo software (TreeStar, Cupertino, CA). Color compensation settings were made with each round of data acquisition by using patient cells labeled with specific fluorochrome-conjugated anti-CD3 antibody. In experiments using HLA tetramers, 0.5 μl of APC-conjugated B*5701(KAFSPEVIPMF) or A*0201(SLYNTVATL) tetrameric complex was used to stain 2 × 106 PBMC in a 50-μl volume at 4°C for 30 min. The tetrameric complexes used in this study were provided by the National Institutes of Health AIDS Reagent Program Tetramer Facility.
To map the gag-specific CD8+ T cell responses, autologous EBV-transformed B cells were pulsed for 1 h in 50 μl of RPMI containing 10% FBS with a single 20-aa peptide. Overlapping peptides spanning the HIVHXB2 gag sequence were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. A total of 5 × 104 targets were mixed with 2 × 106 PBMC in 5-ml round bottom polystyrene tubes (Becton Dickinson). In preliminary experiments the final effector:target ratio of 40:1 and peptide concentration of 10 μM used in these experiments gave optimal responses under most conditions.
In experiments that used HIV-infected primary CD4+ T cells, autologous CD4+ T cells were isolated by using magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) and were stimulated with media containing anti-CD3 (OKT3; Coulter), anti-CD28 (PharMingen), and 40 million units of IL-2/ml as described (26). On day 3, 5 × 106 cells were infected with 5,000 TCID50 of HIVSF162 in 200 μl of media for 1 h at 37°C and then were maintained in culture at 106 cells/ml in 24-well plates for 6 days further. On day 6, CD8+ cells were depleted by using magnetic beads. CD8+ depletion (>99%) was confirmed by flow cytometry. The percent of infected cells (20–40%) was documented by intracellular staining for p24 with Kc57-FITC or Kc57-RD1 (Coulter). The effector-to-target ratio used (1:1) was found in preliminary experiments to give the maximal response with low (<1%) background.
Statistics.
The comparison of frequencies of the HLA B*57 allele between LTNP and progressors was done by the Fisher exact test. The exact method of Gart (27) was used to determine the statistical significance of the odds ratio and its 95% confidence interval. The mean total specific CD8+ T cell responses and tetramer staining cells between LTNP and progressors were compared by Student's t test.
Results
Study Population. Thirteen patients that have maintained plasma virus below 50 copies/ml of plasma in the absence of antiretroviral therapy were recruited for study (Table 1). The majority of patients were infected through unprotected homosexual activity with the exception of patient 7 (unprotected heterosexual contact), patient 10 (blood transfusion), and patient 12 (hemophiliac). Patients 3–8, 12, 13, 17, and 25 have been infected for a minimum of 13 years with stable peripheral CD4+ T cell counts ranging from 277 to 1,105 cells/mm3. Although initially found to be seropositive in 1997, by history, patient 9 was likely infected before 1994. Patient 10 is part of a cohort of patients that received blood products contaminated with nef-defective HIV in 1985 (11). Ten patients have had levels of plasma viral RNA consistently below 50 copies/ml of plasma. In the remaining three individuals, increases in plasma virus have occurred during the study period in association with febrile illnesses but returned to levels below 50 copies/ml of plasma thereafter. All patients have maintained strong proliferative responses to the HIV p24 antigen (Δcpm 4,000–40,000) in conventional lymphoproliferation assays. Of the 13 nonprogressors, only patients 10, 12, and 25 were found to be heterozygous for the Δ32 deletion within the CCR5.
Table 1.
Clinical characteristics and MHC class I haplotypes of long term nonprogressors
| Patient | HLA
class I
|
Year of diagnosis | CD4+T cell count; cells/μl* | CD8+T cell count; cells/μl† | Plasma HIV RNA; copies/ml‡ | ||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| 4 | 1, 31 | 57, 8 | 6, 7 | 1985 | 1,063 | 1,088 | <50 |
| 5 | 2, 24 | 57 | 6, 7 | 1985 | 1,105 | 835 | <50 |
| 6 | 11, 30 | 57, 52 | 7, 12 | 1986 | 760 | 803 | <50–62 |
| 7 | 1, 2 | 57 | 6 | 1985 | 277 | 385 | <50 |
| 8 | 11, 23 | 57, 44 | 4, 6 | 1985 | 664 | 1,120 | <50–325 |
| 9 | 23, 26 | 57, 44 | 1, 7 | 1997 | 1,079 | 985 | <50 |
| 10 | 1, 33 | 57, 50 | 6 | 1996 | 602 | 584 | <50 |
| 12 | 3, 11 | 57, 7 | 6, 7 | 1986 | 500 | 218 | <50 |
| 13 | 1, 11 | 57, 35 | 4, 6 | 1986 | 1,016 | 767 | <50 |
| 25 | 3, 24 | 57, 27 | 2, 6 | 1986 | 1,028 | 1,082 | <50–1,089 |
| 30 | 31, 74 | 57, 51 | 7, 16 | 1990 | 422 | 592 | <50 |
| 3 | 2, 3 | 13, 39 | 6, 7 | 1985 | 915 | 1,079 | <50 |
| 17 | 2, 26 | 27, 38 | 1, 12 | 1985 | 1,073 | 1,051 | <50 |
*Mean CD4+ T cell counts of uninfected individuals are 912 ± 24.08 cells/mm3 in this laboratory.
†Mean CD8+ T cell counts of uninfected individuals are 528 ± 20.8 cells/mm3 in this laboratory.
‡Plasma virus was quantified by the bDNA assay (Chiron) with a 50 copies/ml plasma sensitivity.
HLA class I and class II typing of these patients revealed a dramatic overrepresentation of the B*57 class I allele (Table 2). HLA B*57 was present in 11 of the 13 patients (85%) compared with 11% in the Caucasian U.S. population (28). Subtyping revealed the allele to be B*5701 in each case. We then determined the frequency of the B*57 allele in patients with progressive disease. Of 200 patients that were screened for HLA B*57, 19 (9.5%) carried the B*57 allele, which is close to the expected frequency in this population. The frequency of the B*57 allele was significantly greater in LTNP than progressors (P < 0.001). The odds of the presence of B*57 was 52 times greater (95% CI: 10, 501) than the odds of the presence of this allele in progressors. In the majority of cases the allele was B*5701 and in one case was B*5703. Among all nonprogressors and all B57+ progressors, patients 30, 105, and 108 were Hispanic, and the remaining were Caucasian. Although the linked A1 and Cw6 alleles, which are part of an ancestral haplotype (29), were found in high frequency with B*5701, this was not universally the case. No association with a class II allele was found. Taken together, these results suggest that, although the B*57 allele is very highly associated with the LTNP phenotype, the allele is not sufficient by itself to confer long term restriction of virus replication.
Table 2.
MHC class I haplotypes of HLA B*57+ progressors
| Patient | HLA class I
|
||
|---|---|---|---|
| A | B | C | |
| 101 | 1, 31 | 57, 51 | 3, 6 |
| 102 | 24, 68 | 57, 15 | 6, 7 |
| 103 | 2, 11 | 57, 55 | 3, 6 |
| 104 | 2 | 57, 58 | 3, 6 |
| 105 | 2, 80 | 57, 8 | 2, 7 |
| 106 | 1, 2 | 57, 40 | 3, 6 |
| 107 | 1, 2 | 57, 7 | 6, 7 |
| 108 | 32, 68 | 57 | 7, 3 |
| 109 | 1, 3 | 57, 7 | 6, 7 |
| 110 | 36, 68 | 57, 45 | 7, 16 |
| 111 | 1, 3 | 57, 14 | 6, 8 |
| 112 | 1, 24 | 57, 8 | 6, 7 |
| 113 | 30, 32 | 57, 18 | 5, 6 |
| 114 | 2 | 57, 51 | 6, 14 |
| 115 | 2, 25 | 57, 44 | 4, 6 |
| 116 | 1 | 57, 35 | 4, 6 |
| 117 | 1, 11 | 57, 55 | 3, 6 |
| 118 | 1, 2 | 57, 51 | 6, 14 |
| 119 | 1, 11 | 57, 55 | 3, 6 |
HIV-Specific CD8+ T Cell Responses.
Previous studies in severe combined immunodeficient animals and in vitro showed that no residual restriction of R4 or R5 challenge virus replication in the PBMC of patients 3–6 was observed after complete depletion of CD8+ T cells (17). This result indicated that restriction of virus replication in these patients was not caused by passive restriction at the level of the CD4+ T cell. The prevalence of the B*5701 allele in the extended group of LTNP in the present study further suggests that the difference between these patients and B*57 positive progressors may lie in the number or phenotype of HIV-specific class I restricted CD8+ T cells. To determine whether LTNP patients have higher numbers of HIV-specific CD8+ T cells, quantitative single cell assays were used to enumerate HIV specific cells. We have recently modified a method of measuring the frequency of HIV specific CD8+ T cells by using flow cytometric detection of intracellular IFN-γ accumulation in response to autologous HIV-vaccinia recombinant infected autologous EBV transformed B cells (J. Gea-Banacloche, personal communication). It has been previously shown that the frequency of CD8+ T cells that respond to a given peptide by secretion of IFN-γ correlates well with the numbers obtained by tetramer staining (30). This method provides a highly reproducible and quantitative measure of the HIV specific CD8+ T cell response. In addition, it permits the study of the total numbers of CD8+ T cells specific for entire products of HIV genes in the context of each of the patient's MHC molecules.
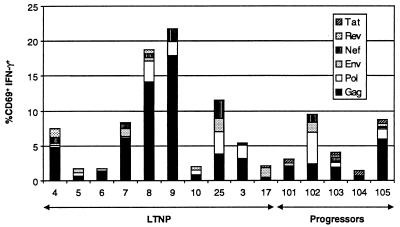
Summary data of the fraction of CD3+CD8+ 69+ lymphocytes producing IFN-γ in response to various HIV gene products are shown in Fig. 1. Antiretroviral therapy has been previously shown to rapidly reduce the numbers of HIV-specific CD8+ T cells (31). Although all 19 progressors were receiving antiretroviral therapy, 5 patients (101–105) were found with greater than 1,000 copies of viral RNA/ml plasma, and the CD8+ T cell responses are shown for comparison. By this technique, the percentage of HIV-specific CD8+ T cells in progressors and LTNP ranged from 1.7 to 22%. There were no significant differences in the total frequency of HIV-specific CD8+ T cells between the LTNP and patients with progressive disease (mean 8.1 vs. 5.3%, respectively; P = 0.4). Because antiretroviral therapy lowers the numbers of HIV-specific CD8+ T cells, the frequency found in progressors likely underestimates the frequency that would be found if therapy were discontinued. These data then suggest that that the LTNP within this cohort do not restrict virus replication because of higher numbers of HIV specific CD8+ T cells.
Figure 1.
The percent of CD3+CD8+ that are CD69+IFN-γ+ in response to autologous B cells infected with HIV-vaccinia recombinants encoding the indicated gene product. Background activity against β-galactosidase has been subtracted from the percents shown.
Similar frequencies of CD8+ T cells specific for individual HIV gene products were found in LTNP and progressors. In all but one case (patient 17), the predominant activity was directed against the gag gene product. It has been shown previously that some patients with nonprogressive HIV infection have HLA B*57-restricted responses specific for tat or rev (32). It has been suggested that these responses may mediate immunologic control over HIV by targeting these genes that are expressed early in the virus replication cycle. However, in most cases, the responses to tat or rev were low or absent and did not differ between progressors or LTNP.
Mapping of Gag-Specific CD8+ T Cell Responses.
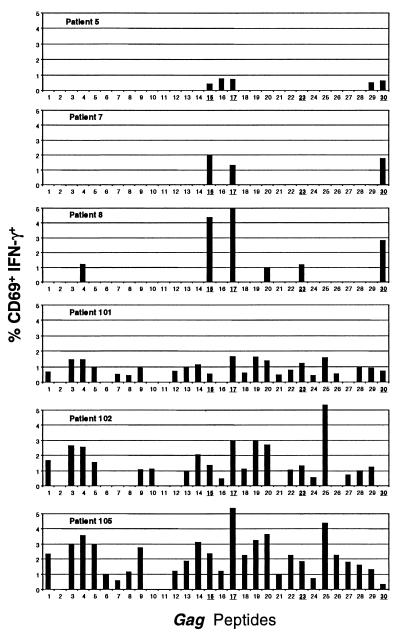
Although comparable frequencies of HIV-specific CD8+ T cells were found in LTNP and progressors, it remained possible that differences might lie in the peptides targeted by CD8+ T cells in the two groups. Given the majority of the HIV specific CD8+ T cells were gag specific, this response was mapped by using overlapping peptides. The percent of CD8+ T cells that produced IFN-γ in response to peptide pulsed autologous EBV transformed B cells is shown in Fig. 2. Consistent with one previous report in HLA B*57+ patients (33), the responses of five LTNP studied were highly focused on peptides that contain previously described B*57 epitopes. These responses were largely confined to four peptides. In each case, similar responses were observed when the optimized peptides were used (see Fig. 2 legend). Only the QASQEVKNW peptide was recognized by all of the LTNP tested. Responses to peptides within p17 and N-terminal regions of gag were low or absent in the LTNP studied.
Figure 2.
The percent of CD3+CD8+ cells that are CD69+IFN-γ+ in response to autologous B cells pulsed with overlapping peptides spanning the gag protein are shown for three HLA B*57-positive LTNPs (patients 5, 7, and 8) and three HLA B*57-positive progressors (patients 101, 102, and 105). Peptides 15, 17, 23, and 30 contain previously described highly conserved HLA B*57-restricted epitopes shown here in bold (15-QMVHQAISPRTLNAWVKVVE, 17-EKAFSPEVIPMFSALSEGAT, 23-PRGSDIAGTTSTLQEQIGWM and 30-YKTRAEQASQEVKNWMTET). Peptides 1–13 include p17 sequences. The N-terminal sequences of gag beyond peptide 30 (amino acid 320) elicited weak responses and are not shown.
Responses to the B*57-restricted peptides were also determined in five HLA B*57-positive patients with progressive HIV infection. However, the breadth of the gag-specific CD8+ T cell responses in the progressors exceeded those seen in the LTNP. Responses restricted to other MHC alleles of the patient were commonly observed. Overall, responses to the B*57-restricted peptides were not lower in patients with progressive disease. Thus, patients with progressive disease maintained a considerably broader response to HIV peptides. It was possible that the greater focus on B*57-restricted peptides in LTNP was caused by sequence polymorphisms within peptide binding regions. Alternatively, polymorphisms within the cytoplasmic tail might diminish MHC down-regulation by nef protein (34). However, sequence analysis of the cDNA from cells of five LTNP and five progressors confirmed the sequence to be B*5701 without polymorphisms throughout, including the signal peptide and cytoplasmic tail.
HIV peptide-specific cells were further quantified by using MHC-tetramers. Attempts were made to construct HLA B*5701 tetramers with each of the four peptides to which the LTNP responded. However, the expressed B*57 molecule did not successfully fold with each of the peptides and only made complexes with the KAFSPEVIPMF peptide. A high correlation was found between the percentage of CD8+ T cells staining with this HLA B*57 gag tetramer and the fraction of CD69+IFN-γ+ cells responding to autologous B cells pulsed with KAFSPEVIPMF (r = 0.84; P = 0.005). The percent of CD8+ T cells that stain with the A*2 gag SLYNTVATL tetramer was low (0–0.31%) in A*2+B*57+ LTNP when compared with progressors, confirming the high degree of focus of the response on B*57-restricted peptides in these patients. No significant difference was found in the frequency of HLA B*57 gag tetramer+ CD8+ T cells between progressors and nonprogressors (P = 0.20). To the contrary, there was a trend toward greater tetramer staining in the CD8+ T cells of the patients with progressing HIV infection and higher plasma viral RNA levels.
Responses to Autologous and Heterologous HIV-Infected Primary CD4+ T Cells.
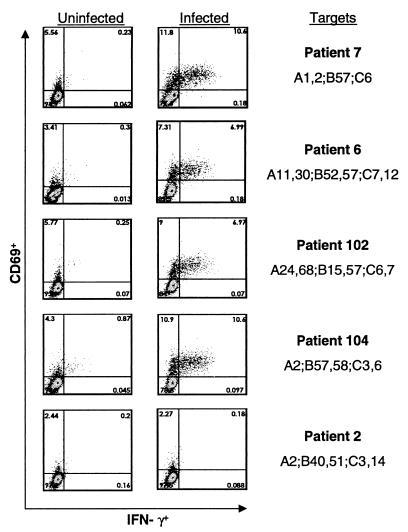
Although the frequencies of HIV-specific CD8+ T cells did not distinguish LTNP from patients with progressing infection, it remained possible that other differences might be found in the avidity of the effector cells or differences in antigen processing or presentation at the level of the CD4+ T cell. Each of these might not be detected by using EBV-transformed B cells that present high levels of MHC on the cell surface, express co-stimulatory molecules and, in the case of gene products other than nef, do not undergo MHC down-regulation. We adapted a recently described technique (26) to use HIV-infected autologous primary CD4+ T cells to stimulate CD8+ T cells to produce IFN-γ. To determine whether differences in the responses of LTNP might lie in effectors or antigen presentation by the target cell, effectors from LTNP were combined with HIV-infected CD4+ T cells from other B*57+ LTNP or progressors.
The responses of CD8+ T cells of patient 7 to autologous or heterologous HIV infected CD4+ T cells is shown in Fig. 3. Similar frequencies of HIV specific CD8+ T cells were detected by this method when compared with the total response in the vaccinia/EBV transformed B cell system. The CD8+ T cells of patient 7 produced IFN-γ in response to autologous or heterologous B*57+ cells from a LTNP or progressor. No response was observed to infected cells of another patient that expressed A*2, showing very low alloreactivity in this 6-h assay and confirming the low or absent HIV-specific A*2-restricted responses in B*57+ LTNP. Higher frequencies of responding cells were intermittently detected when the CD4+ T cells were homozygous for B*57. This phenomenon was also observed if the target cells also expressed B*5801, to which several of the B*57-restricted peptides are cross-restricted (33). Responses of similar magnitude were obtained when the cells of B*57+ progressors were stimulated with B57-matched target cells from progressors or nonprogressors. Thus, we did not detect differences between LTNP and progressors in the responses of effectors, or the ability of their CD4+ T cells to activate CD8+ T cells under these experimental conditions.
Figure 3.
The percent of CD3+CD8+ cells of patient 7 that are CD69+IFN-γ+ in response to autologous and heterologous HIVSF162-infected CD4+ T cell targets. Background activity against uninfected CD4+ T cells from each patient is included (left column).
Discussion
The patients described in this report represent a phenotypically and genotypically distinct subgroup of LTNP that likely make up <0.8% of the HIV infected population. These patients are characterized by no decline in CD4+ T cell numbers despite prolonged infection, viral RNA below 50 copies/ml plasma, and strong proliferative responses to HIV antigens. The PBMC of several of these patients have also shown the ability to restrict autologous or challenge virus replication when engrafted into severe combined immunodeficient animals. Although each of these phenotypes are infrequent in HIV-infected patients (14, 16), all are associated in this cohort. The present study provides evidence that such patients are also genotypically distinguished by a very high level association with the HLA B*57 allele. The level of this association is similar to the highest described for other diseases, such as that between HLAB*27 and ankylosing spondylitis (35, 36). Several previous studies have not reliably found phenotypic or genotypic characteristics that distinguish patients with nonprogressive disease. The majority of patients previously described as LTNP, using clinical definitions that comprise up to 15% of the HIV infected population, ultimately develop progressive disease, suggesting that they represent one extreme of a normal distribution of disease progression. Although a very large number of publications on associations between HLA alleles and HIV disease exist, consistent associations between HLA alleles and slow disease progression have not been made. In one study, B*57 was second to B*27 as the allele most commonly associated with nonprogression (37). The highest prevalence of the B*57 allele previously reported in a nonprogressor cohort was 30% compared with 11% in an ethnically similar uninfected population (33). In two recent large studies, only B14 and C8 were significantly associated with nonprogression (38), or no protective effect of individual class I alleles was found (39). However, because the prevalence of patients with <50 copies of viral RNA/ml of plasma in the absence of antiretroviral therapy after 2 years of infection is very low, the associations described in the present study would not be detected in phenotypic and genotypic studies of large cohorts. Detection of these patients is also complicated by the need for corrections for the large number of comparisons made when examining many HLA alleles. Only when a very strict definition of LTNP is applied to detect extreme outliers do these phenotypic and HLA associations become apparent.
The results of the present study also indicate that a host immune factor is responsible for restriction of virus replication in this cohort. However, the precise mechanism by which the HLA B*57 molecule mediates a protective effect in HIV infection remains unclear. The dramatic overrepresentation of this class I allele in the LTNP, peptide mapping results, and observations from the severe combined immunodeficient/hu animals suggest an important role for class I restricted CD8+ T cells. However, quantitative differences in total HIV-specific or peptide-specific CD8+ T cell responses between the B*57+ LTNP and progressors were not observed in the present study. Thus far, we have also not detected differences in cytotoxicity, suppressive activity, and chemokine secretion in previous studies (17) or the ability to respond to infected primary CD4+ T cells in the present study. It remains likely that phenotypic differences may reside in the ability of CD8+ T cells to mediate cytolysis, secrete suppressive factors, or proliferate in vivo, yet these properties are not adequately modeled by current in vitro assays for HIV-specific CD8+ T cell-mediated immunity. It should be noted that both HLA B*57 and B*27 are associated with autoimmune diseases. Although highly speculative, it is possible that a common mechanism exists between these diseases and restriction of virus replication in LTNP. The finding of similar numbers of cells specific for B*57-restricted peptides by intracellular cytokine and MHC tetramer staining in LTNP and progressors suggests that differences do not lie in the peptides targeted or in the number of peptide-specific cells but possibly in a qualitatively greater ability of the cells of these LTNP to restrict virus replication.
One phenotypic difference between CD8+ T cell responses of B*57+ LTNP and progressors was that the gag-specific response of LTNP was highly focused on peptides that are B*57 restricted. This result strongly suggests that the B*57 molecule is directly involved in the restriction of virus replication and that this association is not simply the result of the function of another linked gene. A highly focused response in LTNP may appear somewhat unexpected given previous descriptions of greater breadth of CD8+ T cell expansions during acute infection in patients that go on to nonprogressive disease (40), the protective effect of heterozygosity at MHC loci (39), or associations of broader HIV-specific CD8+ T cell responses with lower viral loads (41). The observation of a high degree of focus on B*57-restricted peptides in nonprogressors corroborates one previous report (33) and provides quantitative evidence that the response is less focused in progressors. However, it is possible that a highly focused response is not the cause of greater restriction of virus replication but may be the result. It is possible that the provision of CD4+ T cell help during acute infection permits expansion of these immunodominant clones above a critical threshold for immunologic control. Effective restriction of virus replication then does not permit the expansion of clones with specificities for peptides restricted by other MHC alleles (42). The finding of B*57 at the expected frequency in progressors may be interpreted to suggest that, although the allele is highly associated with the LTNP phenotype, it is not sufficient by itself to confer nonprogression. The observed focused response in LTNP may also then be the result of a second factor or response modifier that alters the CD8+ T cell response to favor expansion of B*57-restricted clones in these individuals.
Regardless of the events early in HIV infection that may have formed the HIV-specific cellular immune response of these patients, they likely hold further important clues to immune-mediated restriction of HIV replication. Further study may provide important insights into the critical mechanisms by which HIV circumvents the cellular immune response of patients with progressive disease or the parameters that should be measured to predict restriction of virus replication. Perhaps most importantly, they may provide mechanisms that could be exploited in vaccines or therapies that might result in effective control of virus replication.
Acknowledgments
The authors thank Julie Metcalf, Betsey Herpin, Shuying Liu, Linda Ehler, and Stephanie Mizell for arranging patient apheresis and handling of clinical samples. We also thank Dr. Juan Gea-Banacloche and Dr. Kent Weinhold for discussions. We especially thank the patients involved in this study for their time and dedication to its completion.
Abbreviations
- LTNP
long term nonprogressor
- PBMC
peripheral blood mononuclear cell
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050567397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050567397
References
- 1.Barker E, Mackewicz C E, Reyes-Teran G, Sato A, Stranford S A, Fujimura S H, Christopherson C, Chang S Y, Levy J A. Blood. 1998;92:3105–3114. [PubMed] [Google Scholar]
- 2.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L J, et al. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 3.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 4.Pilgrim A K, Pantaleo G, Cohen O J, Fink L M, Zhou J Y, Zhou J T, Bolognesi D P, Fauci A S, Montefiori D C. J Infect Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 5.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinaldo C, Huang X L, Fan Z F, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, et al. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrer T, Harrer E, Kalams S A, Elbeik T, Staprans S I, Feinberg M B, Cao Y, Ho D D, Yilma T, Caliendo A M, et al. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 8.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, et al. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 9.de Roda Husman A M, Koot M, Cornelissen M, Keet I P, Brouwer M, Broersen S M, Bakker M, Roos M T, Prins M, de Wolf F, et al. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 11.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Zhang L, Ho D D. J Virol. 1995;69:93–100. doi: 10.1128/jvi.69.1.93-100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen O J, Vaccarezza M, Lam G K, Baird B F, Wildt K, Murphy P M, Zimmerman P A, Nutman T B, Fox C H, Hoover S, et al. J Clin Invest. 1997;100:1581–1589. doi: 10.1172/JCI119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefrere J J, Morand-Joubert L, Mariotti M, Bludau H, Burghoffer B, Petit J C, Roudot-Thoraval F. Blood. 1997;90:1133–1140. [PubMed] [Google Scholar]
- 15.Vesanen M, Stevens C E, Taylor P E, Rubinstein P, Saksela K. J Virol. 1996;70:9035–9040. doi: 10.1128/jvi.70.12.9035-9040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefrere J J, Mariotti M, Morand-Joubert L, Thauvin M, Roudot-Thoraval F. J Infect Dis. 1999;180:526–529. doi: 10.1086/314906. [DOI] [PubMed] [Google Scholar]
- 17.Lopez J C, Shupert W L, McNeil A C, Gea-Banacloche J C, Flanigan M, Savage A, Martino L, Weiskopf E E, Immamichi H, Zhang Y M, et al. J Virol. 2000;74:2023–2028. doi: 10.1128/jvi.74.4.2023-2028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz D, Sharma U, Busch M, Weinhold K, Matthews T, Lieberman J, Birx D, Farzedagen H, Margolick J, Quinn T, et al. AIDS Res Hum Retroviruses. 1994;10:1703–1711. doi: 10.1089/aid.1994.10.1703. [DOI] [PubMed] [Google Scholar]
- 19.Bunce M, O'Neill C M, Barnardo M C, Krausa P, Browning M J, Morris P J, Welsh K I. Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 20.Bettinotti, M. P., Norris, R. D., Hackett, J. A., Thompson, C. O., Simonis, T. B., Stroncek, D. & Marincola, F. M. (2000) J. Immunother. in press. [DOI] [PubMed]
- 21.Cohen O J, Paolucci S, Bende S M, Daucher M, Moriuchi H, Moriuchi M, Cicala C, Davey R T, Jr, Baird B, Fauci A S. J Virol. 1998;72:6215–6217. doi: 10.1128/jvi.72.7.6215-6217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker B D. Techniques in HIV Research. New York: Stocton; 1990. [Google Scholar]
- 23.Falkner F G, Fuerst T R, Moss B. Virology. 1988;164:450–457. doi: 10.1016/0042-6822(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 24.Prussin C, Metcalfe D D. J Immunol Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 25.Holmes K, Fowlkes B, Schmid I, Giorgi J. In: Current Protocols in Immunology. Coligan J, Kruisbeek A, Margulies D, Sheevac E, Strober W, editors. Vol. 1. New York: Green; 1995. pp. 5.3.1–5.3.23. [Google Scholar]
- 26.Ferrari G, Humphrey W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gart J. Biometrika. 1970;57:471–475. [Google Scholar]
- 28.Marincola F, Stroncek D, Simonis T. In: HLA 1997. Terasaki P, Gjerston D, editors. Los Angeles: UCLA Tissue Typing Labs; 1997. pp. 348–349. [Google Scholar]
- 29.Dawkins R L, Degli-Esposti M P, Zhang W. In: Molecular Evolution of the Major Histocompatibility Complex. Klein J K A D, editor. Berlin: Springer; 1991. pp. 391–402. [Google Scholar]
- 30.Larsson M, Jin X, Ramratnam B, Ogg G S, Engelmayer J, Demoitie M A, McMichael A J, Cox W I, Steinman R M, Nixon D, Bhardwaj N. AIDS. 1999;13:767–777. doi: 10.1097/00002030-199905070-00005. [DOI] [PubMed] [Google Scholar]
- 31.Ogg G S, Jin X, Bonhoeffer S, Moss P, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Hurley A, et al. J Virol. 1999;73:797–800. doi: 10.1128/jvi.73.1.797-800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Baalen C A, Pontesilli O, Huisman R C, Geretti A M, Klein M R, de Wolf F, Miedema F, Gruters R A, Osterhaus A D. J Gen Virol. 1997;78:1913–1918. doi: 10.1099/0022-1317-78-8-1913. [DOI] [PubMed] [Google Scholar]
- 33.Goulder P J, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips R E, McMichael A J. AIDS Res Hum Retroviruses. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 34.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 35.Schlosstein L, Terasaki P I, Bluestone R, Pearson C M. N Engl J Med. 1973;288:704–706. doi: 10.1056/NEJM197304052881403. [DOI] [PubMed] [Google Scholar]
- 36.Brewerton D A, Hart F D, Nicholls A, Caffrey M, James D C, Sturrock R D. Lancet. 1973;1:904–907. doi: 10.1016/s0140-6736(73)91360-3. [DOI] [PubMed] [Google Scholar]
- 37.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O'Brien S J, Rinaldo C, et al. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 38.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O'Brien S, Andrieu J M, Schachter F, Zagury D, Rappaport J, Winkler C, et al. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 39.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 40.Pantaleo G, Soudeyns H, Demarest J F, Vaccarezza M, Graziosi C, Paolucci S, Daucher M, Cohen O J, Denis F, Biddison W E, et al. Proc Natl Acad Sci USA. 1997;94:9848–9853. doi: 10.1073/pnas.94.18.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalod M, Dupuis M, Deschemin J C, Sicard D, Salmon D, Delfraissy J F, Venet A, Sinet M, Guillet J G. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak M A, May R M, Phillips R E, Rowland-Jones S, Lalloo D G, McAdam S, Klenerman P, Koppe B, Sigmund K, Bangham C R, et al. Nature (London) 1995;375:606–611. doi: 10.1038/375606a0. [DOI] [PubMed] [Google Scholar]