Abstract
Frequent extinctions of local populations in metapopulations create opportunities for migrant females to establish new populations. In a metapopulation of the Glanville fritillary butterfly (Melitaea cinxia), more mobile individuals are more likely to establish new populations, especially in habitat patches that are poorly connected to existing populations. Here we show that flight metabolic rate and the frequency of a specific allele of the metabolic enzyme phosphoglucose isomerase (pgi) were both highest in newly established, isolated populations. Furthermore, genotypes with this pgi allele had elevated flight metabolic rates. These results suggest that genetic variation in pgi or a closely linked locus has a direct effect on flight metabolism, dispersal rate, and thereby on metapopulation dynamics in this species. These results also contribute to an emerging understanding of the mechanisms by which population turnover in heterogeneous landscapes may maintain genetic and phenotypic variation across populations.
Keywords: flight physiology, fragmented landscapes, Melitaea cinxia, migration, phosphoglucose isomerase
1. Introduction
Dispersal is one of the key traits determining survival of species in fragmented landscapes. There is an extensive literature examining both theoretically and empirically how changes in landscape structure may influence the evolution of dispersal rate (Clobert et al. 2001; Woiwod et al. 2001; Bullock et al. 2002; Clobert et al. 2004). This work has shown that there is often considerable additive genetic variance for dispersal, and hence dispersal-related traits may respond rapidly to selection (reviewed in Roff & Fairbairn 2001).
The structure of a fragmented landscape may itself contribute to the maintenance of variation in dispersal rate (Travis & Dytham 1999; Hovestadt et al. 2001; Hanski et al. 2004), because dispersive individuals are more likely than sedentary ones to establish new populations and less likely to persist in old populations (Peroni 1994; Olivieri et al. 1995; Taylor & Merriam 1995; Cody & Overton 1996; Piquot et al. 1998; Thomas et al. 1998; Hill et al. 2001; Hanski et al. 2002; Hughes et al. 2003; Hanski et al. 2004). It is clear that variation in dispersal rate has a physiological and genetic basis, but the actual mechanisms remain poorly understood (Roff & Fairbairn 2001). This is especially true for species with continuous variation in dispersal capacity. For wing-dimorphic insects, some of the physiological mechanisms are better known (Zera & Denno 1997; Zhao & Zera 2002) and in some cases wing polymorphism appears to be caused by a single gene with two alleles of unknown molecular function (Roff & Fairbairn 1991; Caillaud et al. 2002; Braendle et al. 2005). However, so far no gene of known molecular function has been found to influence dispersal rate.
Here we report on a candidate gene (Fitzpatrick et al. 2005) and related physiological measurements that are strongly correlated with variation in dispersal rate in the Glanville fritillary butterfly (Melitaea cinxia). Specifically, we relate genetic variation in the glycolytic enzyme phosphoglucose isomerase (pgi) and variation in flight metabolic rate to known spatial variation in dispersal rate in a large and well studied metapopulation of the Glanville fritillary in the Åland Islands in Finland (Hanski et al. 2002; Hanski et al. 2004). This metapopulation occurs in a fragmented landscape consisting of ca 4000 discrete habitat patches, scattered across an area of 50×70 km (Hanski et al. 1995; Hanski 1999; Nieminen et al. 2004). Approximately 500 of the patches are occupied in any given year. The rate of population turnover is high, with ca 100 local extinctions each year and a roughly equivalent number of colonizations of unoccupied patches (Nieminen et al. 2004). A typical female visits at most a few other patches apart from the natal patch during her lifetime (Hanski et al. 1994; Kuussaari et al. 1996; Hanski et al. 2004).
Using a spatially realistic model of the evolution of dispersal rate in this metapopulation, Hanski et al. (2004) predicted that females that establish new local populations have a higher dispersal rate than the average female in the metapopulation. Furthermore, this difference should increase with patch isolation, because higher dispersal capacity makes it more likely that an individual will reach a patch with low connectivity to existing populations. On the other hand, with time, the average mobility of individuals in isolated populations should decrease, because highly mobile individuals are likely to emigrate and, due to patch isolation, emigration is not balanced by the arrival of other mobile individuals (Hanski et al. 2004). The model thus predicted a contrasting relationship between average mobility and spatial connectivity in old versus new populations (figure 1). Results consistent with these predictions were found in a mark–recapture study that examined dispersal rate in the field (Hanski et al. 2002) and in a physiological study that measured the [ATP]/[ADP] ratio in flight muscles of female butterflies after a fixed period of forced flight (Hanski et al. 2004). The latter result suggested that ability to rephosphorylate ADP correlates with variation in dispersal rate, leading to the hypothesis that flight metabolism has an immediate effect on variation in dispersal rate.
Figure 1.
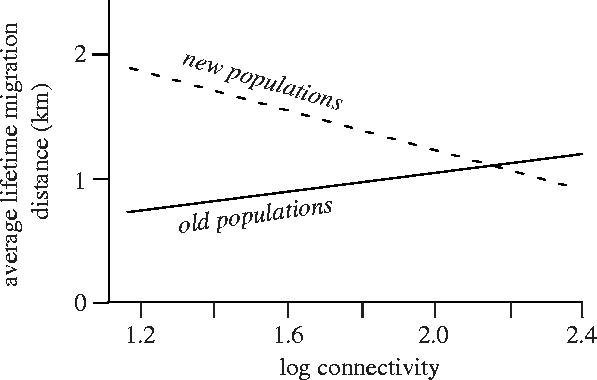
Model-predicted average life-time migration distance of butterflies in relation to population age and connectivity. This schematic figure is based on predictions of an individual-based evolutionary model parameterized with data for the Glanville fritillary (Hanski et al. 2004). The model predicts that females that establish new local populations in isolated habitat patches have a higher dispersal rate than the average female in the metapopulation. On the other hand, highly mobile females have high emigration rates, and hence less mobile individuals accumulate in isolated populations in the course of time (while the population becomes older).
Flying insects have the highest known mass-specific rates of energy consumption, with certain glycolytic enzymes working at rates close to their maximal flux capacity (Suarez 2000). Nonetheless, genetic variation for flight capacity appears to be rampant, as numerous studies have demonstrated heritable variation for flight endurance (Kent & Rankin 2001; Roff & Fairbairn 2001). Maximal flight performance also shows substantial genetic variation that responds to directional selection (Marden et al. 1997) and maps to specific genomic regions (Montooth et al. 2003). It is reasonable to hypothesize that genes responsible for variation in flight metabolic rate may also be responsible for variation in dispersal rate. Thus, in the present study, we link allelic variation at a metabolic enzyme in the Glanville fritillary to variation in flight physiology among individuals that originate from populations of different ages and spatial connectivities.
We identified pgi as a candidate locus for variation in flight metabolic rate and dispersal rate based on previous studies showing that, in Colias butterflies, flight capacity and female fecundity are correlated with genotypic differences in pgi enzyme kinetics and thermal stability (Watt 1977; Watt 1983; Watt 1992; Hughes & Zalucki 1993). The Åland metapopulation of the Glanville fritillary is known to be polymorphic for pgi (Saccheri et al. 1998). We tested whether female butterflies with different pgi genotypes differ in flight metabolic rate, and whether pgi alleles and flight metabolic rate vary among populations of dissimilar ages and spatial connectivities in a manner predicted by the model of Hanski et al. (2004) and consistent with observed phenotypic variation in dispersal rate.
2. Material and methods
(a) Sampling design
A total of 176 adult butterflies were caught in the field during the early flight season in 2004, between 2 and 12 June. Half of the individuals were caught from populations that were newly established by migrants in the previous summer, and half from populations older than 5 years. The spatial connectivity of populations varied widely in both groups of populations. Connectivity of population i was calculated as
where dij is the distance in km between populations i and j and Nj is the number of larval groups in population j in autumn 2002. Connectivity, the inverse of isolation, is expected to be proportional to the number of immigrants arriving in a population (for further details see Hanski et al. 2004). Captured butterflies were individually marked and released into a large outdoor cage (32×26×3 m), where the vast majority of all matings and ovipositions were recorded under practically natural conditions (described in more detail in Hanski et al. in press). It was not possible to determine the age of the individuals transferred to the cage, but they were all caught in the early flight season and thus were at most a few days old. A more detailed description of the long-term field study and methods used to survey the populations is presented by Nieminen et al. (2004).
(b) Flight metabolic rate
From 10 to 16 June, randomly selected females were removed from the cage for one day to measure their flight metabolic rate. Flight metabolic rate was measured using standard respirometry techniques (Lighton 1991) that are described in detail in §S2 in the electronic supplementary material. We stimulated butterflies to fly inside a transparent 1 l jar through which dry CO2-free air was pumped at a regulated flow rate of 0.95 l min−1. Air temperature within the jar was nearly invariant during the testing of an individual butterfly, averaging 31.3 °C across all of the tests (s.d.=0.7). Butterflies flew in a nearly continuous fashion as long as we applied gentle shakes or taps to get them flying again whenever they alighted. Flight was stimulated in this fashion for 15 min, after which the jar was shaded and a steady baseline of resting CO2 emission was reestablished. Many individuals fatigued prior to the end of the 15 min flight test; these were clearly cases of an inability to continue to fly rather than unwillingness. In these cases we continued stimulation so that the butterfly flew again as soon as it was able. At the conclusion of the experiment, the butterfly was removed from the jar, allowed to imbibe 25% honey solution, and returned to the cage. Respirometry experiments were performed blindly with regard to the source population of the butterflies. From the data recorded, we subtracted the mean pre-flight CO2 emission rate (resting metabolism) and used standard equations for open-flow respirometry (Lighton 1991) to determine the rate of CO2 emission attributable to flight metabolism.
In separate experiments we have established that the respiratory exchange quotient (the ratio of CO2 emitted to O2 consumed) does not differ from 1.0 over the 15 min flight test or between strongly and weakly flying individuals. Other experiments showed that flight metabolic rate in this species remains relatively constant with age until it drops precipitously at the oldest ages (K. Niitepõld, unpublished data).
(c) Allozyme data
During 21–24 June, we collected all remaining individuals in the cage and preserved them in liquid nitrogen. Individuals that had been found newly dead or dying before 21 June had been preserved earlier. A total of 75 individuals (32 females, 43 males) were thus preserved and later genotyped for pgi. These 75 individuals originated from 18 new and 21 old populations (one to seven individuals per population), scattered across the Åland Islands (figure S1 in the electronic supplementary material). The material is thus well replicated at the population level.
The sample of 75 individuals was genotyped at the phosphoglucose-isomerase locus (pgi, enzyme commission number EC 5.3.1.9). Half of the thoraces were homogenized in 200 μl water. Allelic variation was assessed using 6 μl homogenate for cellulose acetate electrophoresis (Hebert & Beaton 1993). Gels were run in Tris Glycine buffer at 200 V for 2 h in a refrigerator and scored directly after staining.
Individuals from new populations were the offspring of the original colonizers rather than the colonizers themselves and thus the genotype of the colonizers remained unknown. Hence, for data analysis, we contrasted allele frequencies rather than genotype frequencies between old and new populations. This contrast is slightly conservative, because the allele frequencies represent equally the colonizing females and their mates. Females typically mate soon after eclosion and hence before dispersal (Kuussaari et al. 1998).
(d) Clutch size
All egg clusters laid by individual females in the outdoor cage were collected and the eggs in each cluster were counted. We tested whether average clutch size of individual females was correlated with pgi genotype and flight metabolic rate. Prior to the analysis, average clutch size was corrected for body mass and all statistics were weighted by the number of egg clutches laid by a given female.
3. Results
(a) Polymorphism at pgi in the metapopulation
Among the 75 genotyped individuals, a total of seven alleles could be distinguished at the pgi locus. Allele frequencies were very similar to those recorded in the same metapopulation in 1995 (table 1; Saccheri et al. 1998; Saccheri et al. 2004), suggesting stable allele frequencies over a period of 9 years. Note, however, that our sample is somewhat biased towards new populations, which we specifically sought for the purpose of the present experiment, whereas populations of intermediate age were not sampled.
Table 1.
Relative electromobilities and allele frequencies at the pgi locus in 1995 and 2004.
| allele frequency | |||
|---|---|---|---|
| allele | relative electro-mobility | 1995 (n=1314) | 2004 (n=75) |
| pgi-a | 0 | 0.126 | 0.087 |
| pgi-b | 5 | 0.035 | 0.040 |
| pgi-c | 12 | 0.070 | 0.053 |
| pgi-d | 100 | 0.512 | 0.493 |
| pgi-f | 112 | 0.204 | 0.267 |
| pgi-g | 210 | 0.041 | 0.053 |
| pgi-h | 125 | 0.012 | 0.007 |
Electromobilities (EM) were measured relative to the mobility of the slowest allele (pgi-a, definition: EM=0) and the most common allele (pgi-d, definition: EM=100). Allele frequencies for 1995 are from Saccheri et al. (1998, 2004). N indicates the number of individuals genotyped.
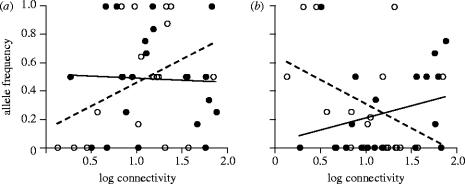
In our sample, the frequencies of the alleles pgi-d and pgi-f varied significantly with population age and connectivity (figure 2, table 2). The frequency of pgi-f showed the same pattern among populations as predicted by a model for phenotypic variation in dispersal propensity (figure 1, Hanski et al. 2004). That is, pgi-f was especially common in new, isolated populations, whereas it was rare in old, isolated populations (figure 2). Hence the interaction between population age and connectivity was significant in determining the frequencies of this allele (table 2). For pgi-d the pattern was inverse (figure 2, table 2). For the rarer alleles sample sizes were too small to test whether their frequencies depended on population age and connectivity.
Figure 2.
The frequencies of the alleles (a) pgi-d and (b) pgi-f as functions of population connectivity. Filled circles represent old populations, open circles new populations. Solid and dotted lines are least-square fits for old and new populations, respectively. The interaction between population age and connectivity was significant (pgi-d: p=0.049; pgi-f: p=0.013; table 2).
Table 2.
Mixed binomial models for the distribution of alleles pgi-d and pgi-f.
| allele pgi-d | allele pgi-f | ||||||
|---|---|---|---|---|---|---|---|
| factor | modelling type | F or z | d.f. | p | F or z | d.f. | p |
| age class | fixed | 2.91 | 1/35 | 0.097 | 5.30 | 1/35 | 0.027 |
| connectivity | fixed | 1.42 | 1/75 | 0.24 | 0.34 | 1/75 | 0.56 |
| age class×connectivity | fixed | 4.02 | 1/75 | 0.049 | 6.45 | 1/75 | 0.013 |
| population (age class) | random | 1.25 | 0.11 | 1.10 | 0.14 | ||
| individual (population) | random | 1.44 | 0.075 | 1.68 | 0.046 | ||
In each genotyped individual an allele could be twice present (1) or absent (0). Population age was modelled as a fixed factor (age class=new, old). F-values for fixed factors were calculated from type III SSQ. For random factors z-values are given.
(b) Flight metabolic rates
We measured the flight metabolic rate of 45 females. A few (four from new populations, three from old populations) were unable to accomplish weight-supported flight and tended to show signs of advanced age or injury. Thus their data were discarded prior to any analyses. The remaining 38 flight-capable butterflies came from 25 distinct populations.
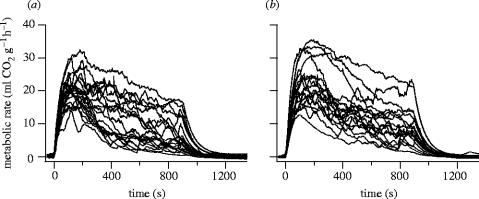
Flight metabolic rate and the ability to maintain flight continuously for 15 min were highly variable (figure 3). All butterflies attained their peak metabolic rate early in the experiment, with varying degrees of decline thereafter. Among females from old populations, 13 of 20 fatigued and were unable to maintain flight for the entire 15 min. Among females from new populations, a smaller proportion fatigued (8 of 18), but the difference was not significant (two-tailed Fisher's exact test, p=0.34).
Figure 3.
Traces of mass-specific metabolic rate of female Glanville fritillary butterflies that were stimulated to fly for 15 min inside a 1 l jar. (a) old populations, (b) new populations.
Both the peak metabolic rate (ml CO2 h−1) and the total volume of CO2 emitted during the 15 min flight test showed a positive relationship with body mass (two-tailed p=0.028 and p=0.090, respectively). To account for this body size effect, we used residuals from these regressions for all further statistical analyses involving metabolic rates.
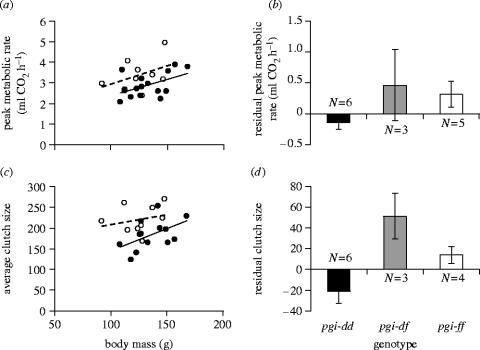
We used a general linear model to test whether flight metabolic rate varied among populations of different age and connectivity in a manner parallel to predicted variation in dispersal rate. This model (r2=0.20) showed that the interaction between population age and connectivity had a significant effect on peak metabolic rate (p=0.040, figure 4). In particular, females from the most isolated (low connectivity) new populations had the highest peak metabolic rates.
Figure 4.
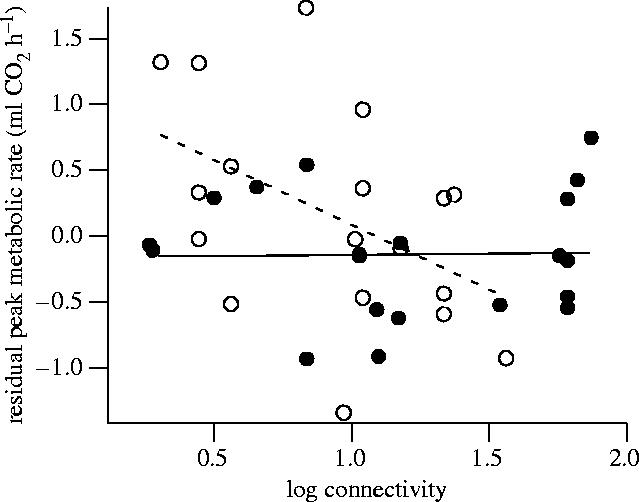
Residual peak metabolic rate as a function of population connectivity in females from new populations (open circles) and old populations (filled circles). Residuals are from a regression accounting for the effect of body mass. Solid and dotted lines are least-square fits for old and new populations, respectively. The interaction between population age and connectivity was significant (p=0.040).
The total volume of CO2 emitted during the 15 min flight test was strongly correlated with peak metabolic rate (r2=0.54), but showed no significant association with population age, connectivity, or the interaction between population age and connectivity. Total metabolism was more variable among individuals than was peak metabolism, which reduced the ability to detect statistically significant differences.
(c) Differences in metabolic rate among pgi genotypes
Twenty-five of the females tested for flight metabolic rate were subsequently genotyped at pgi. Because the distribution of allele frequencies among populations (figure 2) suggested that genotypes with the pgi-f allele were better dispersers, we first tested whether there was a difference in metabolic rate between individuals with the pgi-f allele (either homozygous or heterozygous) as compared to genotypes without this allele. Peak metabolic rate was higher in genotypes with the pgi-f allele than in genotypes without this allele (two-tailed t-test, t=2.37, d.f.=23, p=0.026, figure 5). At average body mass, females with pgi-f had a peak metabolic rate of 3.44 ml CO2 h−1 (s.e.=0.19), compared to 2.85 ml CO2 h−1 (s.e.=0.15) in females without pgi-f (a 17% increase). Overall, 20% of the variance in peak metabolic rate was explained by the presence or absence of pgi-f. Genotypes with pgi-f also differed from genotypes without pgi-f for total CO2 produced during 15 min flight (two-tailed t-test, t=2.40, d.f.=23, p=0.025). For pgi-d as well as for all rarer alleles there were no differences in metabolic rate between individuals with and without the allele.
Figure 5.
(a,b) Peak metabolic rate and (c,d) average clutch size of different pgi genotypes. On the left, (a) peak metabolic rate and (c) average clutch size of genotypes with (open circles) and without (closed circles) the allele pgi-f are shown as a function of body mass. Solid and dotted lines are least-square fits for old and new populations, respectively. Differences between genotypes with and without pgi-f were significant (a: p=0.026, c: p=0.010). The panels on the right show (b) residual peak metabolic rate and (d) residual clutch size in the three most common genotypes. Of the pairwise comparisons, the differences between pgi-dd and pgi-ff and between pgi-dd and pgi-df in (d) were significant (p=0.021 and p=0.006, respectively). All other direct comparisons had p>0.14. Note, however, the low power due to small sample sizes.
Looking at individual genotypes, the sample sizes were very small, but it is noteworthy that the ranking of peak metabolic rate for the three most common genotypes was pgi-df>pgi-ff>pgi-dd (figure 5). This ranking appears to explain why metabolic rate depended on the presence of pgi-f but not of pgi-d, as individuals with the latter allele had a high (heterozygous) or low (homozygous) metabolic rate.
To test whether genotypes with pgi-f had a higher metabolic rate than genotypes without pgi-f independently of the age and connectivity of their population, we used population age, connectivity, and their interaction in a general linear model to examine variation in peak metabolic rate. In this model, the difference between the pgi-f and non-pgi-f was non-significant (p=0.09), though means between the two genotypes were substantially different (mean ±s.e.=0.23±0.18 versus −0.15±0.14). The effects of population age, connectivity, and their interaction were all clearly non-significant (p>0.36). This result indicates that the effect of pgi on peak metabolic rate is unlikely to be due to a spurious correlation of some other factor with the age and spatial location of populations.
(d) Average clutch size in relation to metabolic rate and pgi genotype
Individuals with pgi-f genotypes had significantly higher body mass-adjusted average clutch sizes than individuals without this allele (two-tailed t-test, t=2.85, d.f.=20, p=0.010, figure 5). The means for females with and without pgi-f were 223 (s.e.=12.7) and 184 (9.7) eggs per clutch, with 29% of the total variance in clutch size being explained by the presence of pgi-f. For pgi-d and the rarer alleles, average clutch size did not depend on the presence of the allele. Among the three most common genotypes, the ranking was again pgi-df>pgi-ff>pgi-dd (figure 5), with two of the three contrasts being significant (pgi-dd versus pgi-ff: two-tailed t-test, p=0.021; pgi-dd versus pgi-df: p=0.006; pgi-df versus pgi-ff: p=0.14). The difference in residual clutch size between genotypes with and without pgi-f remained significant (p=0.020) when population age, connectivity, and the interaction between population age and connectivity were included in the model.
Average clutch size was positively related to peak metabolic rate (p=0.045) and total CO2 emitted during the 15 min flight test (p=0.035), but both effects were non-significant (p>0.4) when the presence or absence of the pgi-f allele was included as an independent variable. Thus our data consistently indicate that butterflies with the pgi-f allele have both elevated flight metabolism and larger clutch sizes.
4. Discussion
(a) The links between pgi, flight metabolic rate and dispersal rate
We found a significant association between pgi allele frequencies and flight metabolic rate in the Glanville fritillary butterfly, and that both of these traits varied among local populations with dissimilar ages and spatial connectivities in a manner consistent with field-measured variation in dispersal rate (Hanski et al. 2002; Hanski et al. 2004). A previous study on the Glanville fritillary, which showed parallel variation among populations in the [ATP]/[ADP] ratio in flight muscles following a fixed period of flight (Hanski et al. 2004), suggested that the observed variation in dispersal rate may at least partly be caused by variation in flight physiology. We have now extended this finding by linking variation in dispersal rate and flight physiology to genetic variation in an enzyme with a central role in glycolysis and potentially a direct effect on flight physiology (Watt & Boggs 1987; Watt & Dean 2000).
In Colias butterflies, different pgi-genotypes have different enzyme-kinetic properties (Watt 1977; Watt 1983), which lead to different glycolytic fluxes and flight performance among the pgi genotypes (Watt et al. 1983; Watt & Dean 2000). The pgi polymorphism documented here for the Glanville fritillary appears very similar to that found in Colias butterflies. First, the number and frequency distribution of electrophoretically distinguishable alleles was similar to those in Colias (Watt 1977; Watt 1983; Watt et al. 2003). Second, as in Colias (Watt et al. 2003), the allele frequencies have been stable in samples spanning almost a decade. Third, the ranking of flight capacity among the three most common genotypes is very similar in these two species, with heterozygotes ranking highest (Watt et al. 1983). Nonetheless, it is unlikely that the pgi alleles in the Glanville fritillary are identical to those in Colias, as the two genera diverged ca 40 Myr (N. Wahlberg, personal communication) and structural differences of alleles across species are evident within the genus Colias alone (Watt et al. 1996). Rather, these results suggest that similar forces have shaped the evolution of pgi variation in different butterfly species.
In summary, we have presented correlational evidence suggesting an effect of the pgi locus on dispersal rate in the Glanville fritillary. This correlational evidence is supported by a mechanistic hypothesis, which involves an effect of pgi on flight physiology and an effect of flight physiology on flight capacity and hence on dispersal rate. To prove that pgi has a major effect on variation in dispersal rate one would have to manipulate the pgi genotype of individuals experimentally, which is currently not possible in the Glanville fritillary. But the hypothesis can also be further corroborated by analyzing the mechanistic consequences of allelic variation in pgi in greater detail, as has been done for Colias by analyzing the enzyme kinetic properties of different pgi genotypes. As we have not yet established this mechanistic link in the Glanville fritillary, we cannot exclude the alternative hypothesis that the observed variation is due to an unknown locus in close linkage with pgi. Furthermore, pgi is unlikely to be the only gene involved in determining dispersal rate, as theory and empirical data suggest that variation in most quantitative traits is governed by a few loci of large effects and many loci of smaller effects (e.g. Paterson et al. 1991; Orr 1998). Nonetheless, the present data suffice to strongly indicate that pgi is a candidate for a gene with a major effect on dispersal rate in the Glanville fritillary.
(b) Clutch size differences among pgi genotypes
Allelic variation at pgi and flight metabolic rate were correlated with average clutch size. In Colias, the kinetically superior pgi genotypes have a higher fecundity because they can fly and oviposit at lower temperatures and thus over a longer period of time during the day than the kinetically less competent genotypes (Watt 1992). Similarly, M. Saastamoinen (unpublished data) has observed that Glanville fritillary females with the allele pgi-f lay earlier in the day, when temperatures tend to be higher and when all females typically lay larger clutches. Possibly females with the pgi-f allele become physiologically able to lay earlier in the day than the non-pgi-f females, and can thereby take advantage of the high noon temperatures. Regardless of the actual explanation of why metabolically superior and hence more dispersive females lay larger clutches, this association is consistent with earlier results (Hanski et al. in press) showing no direct trade-off between dispersal rate and fecundity in the Glanville fritillary, in contrast to what is commonly found in wing-dimorphic insects (Roff 1977; Zera & Denno 1997).
(c) Phenotypic and genetic variation in metapopulations
Previous studies on the Glanville fritillary have shown that phenotypic and physiological variation related to dispersal rate and flight capacity are maintained among local populations of different ages and spatial connectivities in a manner predicted by an evolutionary model of dispersal (Hanski et al. 2002; Hanski et al. 2004). Here we have shown that the same is true for genetic variation at a locus that appears to be causally related to variation in dispersal rate and physiology. Recent studies on bird populations illustrate other ways in which non-random dispersal combined with spatial variation in the expression of genetic variation can lead to unexpected population differentiation (Garant et al. 2005; Postma & Noordwijk 2005). Though the mechanisms are different, these studies provide further evidence that dispersal and gene flow among distinct local populations does not necessarily homogenize allele frequencies (Slatkin 1987). In our study system, individuals with a higher dispersal rate are more likely to reach unoccupied patches, especially if isolated from existing populations; this creates a non-random distribution of genotypes among populations. As populations persist, dispersive genotypes appear to be lost due to emigration and populations tend to become more sedentary. Genetic variation for dispersal in this system can be maintained by the spatially and temporally dynamic processes of extinction and recolonization (Hanski et al. 2004). Furthermore, as the ecological metapopulation dynamics depend critically on dispersal rate, variation in the dispersal rate is likely to have a feed-back effect on metapopulation dynamics.
5. Conclusions
This study and the previous work on the Glanville fritillary (Hanski et al. 2002; Hanski et al. 2004) provide the beginnings of a mechanistic explanation of non-random dispersal of butterfly phenotypes in a metapopulation. Frequent extinction of small local populations creates opportunities for migrant females to establish new populations. Our results indicate that females with the pgi-f allele are stronger fliers and hence more likely to establish new populations, especially when the new populations are poorly connected to existing populations and hence the colonizers have to fly for a long distance. The exciting prospect raised by these results is the possibility of developing a truly mechanistic understanding of the spatial and temporal dynamics in genes with major phenotypic effects in metapopulations in real fragmented landscapes. Metapopulation dynamics are a pervasive and general feature of species living in fragmented landscapes (Hanski 1999; Ehrlich & Hanski 2004). Therefore, although this paper examines particular traits of a single species, our approach is likely to be applicable across a wide range of species, including perhaps humans, who show genetic variation in metabolic rates and activity (Kubaszek et al. 2003; Payne & Montgomery 2003; Yang et al. 2003).
Acknowledgments
We thank the staff of Tvärminne Zoological Station and the Åland 2004 team for support and technical assistance. K. Niitepõld helped with the measurements of flight metabolic rate and shared unpublished data. We profited from comments by C. Liautard, C. W. Wheat, and two anonymous reviewers. T. Roslin and T. J. Kawecki provided statistical advice, and E. Meyke helped with figure preparation. The work was supported by the Academy of Finland (grants no. 38604 and 44887 to I.H., Finnish Centre of Excellence Programme, 2000–2005) and grants IBN-0091040 and IBN-0412651 to J.H.M. from the US National Science Foundation.
Footnotes
As this paper exceeds the maximum length normally permitted, the authors have agreed to contribute to production costs.
Supplementary Material
References
- Braendle C, Friebe I, Caillaud M.C, Stern D.L. Genetic variation in an aphid wing polyphenisms is genetically linked to a naturally occurring wing polymorphism. Proc. R. Soc. B. 2005;272:657–664. doi: 10.1098/rspb.2004.2995. 10.1098/rspb.2004.2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock J.M, Kenward R.E, Hails R.S, editors. Dispersal ecology. British Ecological Society, Blackwell; Oxford: 2002. [Google Scholar]
- Caillaud M.C, Boutin M, Braendle C, Simon J.-C. A sexlinked locus controls wing polymorphism in males of the pea aphid, Acyrthosiphon pisum (Harris) Heredity. 2002;89:346–352. doi: 10.1038/sj.hdy.6800146. 10.1038/sj.hdy.6800146 [DOI] [PubMed] [Google Scholar]
- Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford: 2001. [Google Scholar]
- Clobert J, Ims R.A, Rousset F. Causes, mechanisms and consequences of dispersal. In: Hanski I, Gaggiotti O.E, editors. Ecology, genetics, and evolution of metapopulations. Elsevier Academic Press; Amsterdam: 2004. pp. 307–335. [Google Scholar]
- Cody M.L, Overton J.M. Short-term evolution of reduced dispersal in island plant populations. J. Ecol. 1996;84:53–61. [Google Scholar]
- Ehrlich P.R, Hanski I, editors. On the wing of checkerspots: a model system for population biology. Oxford University Press; Oxford: 2004. [Google Scholar]
- Fitzpatrick M.J, Ben-Shahar Y, Smid H.M, Vet L.E.M, Robinson G.E, Sokolowski M.B. Candidate genes for behavioural ecology. Trends Ecol. Evol. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. 10.1016/j.tree.2004.11.017 [DOI] [PubMed] [Google Scholar]
- Garant D, Kruuk L.E.B, Wilkin T.A, McCleery R.H, Sheldon B.C. Evolution driven by differential dispersal within a wild bird population. Nature. 2005;433:60–65. doi: 10.1038/nature03051. 10.1038/nature03051 [DOI] [PubMed] [Google Scholar]
- Hanski I. Oxford University Press; Oxford: 1999. Metapopulation ecology. [Google Scholar]
- Hanski I, Kuussaari M, Nieminen M. Metapopulation structure and migration in the butterfly Melitaea cinxia. Ecology. 1994;75:747–762. [Google Scholar]
- Hanski I, Pakkala T, Kuussaari M, Lei G. Metapopulation persistence of an endangered butterfly in a fragmented landscape. Oikos. 1995;72:21–28. [Google Scholar]
- Hanski I, Breuker C.J, Schops K, Setchfield R, Nieminen M. Population history and life history influence the migration rate of female Glanville fritillary butterflies. Oikos. 2002;98:87–97. 10.1034/j.1600-0706.2002.980109.x [Google Scholar]
- Hanski I, Eralahti C, Kankare M, Ovaskainen O, Siren H. Variation in migration propensity among individuals maintained by landscape structure. Ecol. Lett. 2004;7:958–966. 10.1111/j.1461-0248.2004.00654.x [Google Scholar]
- Hanski, I., Saastamoinen, M. & Ovaskainen, O. In press. Dispersal-related life-history trade-offs in a butterfly metapopulation. J. Anim. Ecol. [DOI] [PubMed]
- Hebert P.D.N, Beaton M.J. 2nd edn. Helena Laboratories; Beaumont, Texas, USA: 1993. Methodologies for allozyme analysis using cellulose acetate electrophoresis. [Google Scholar]
- Hill J.K, Collingham Y.C, Thomas C.D, Blakeley D.S, Fox R, Moss D, Huntley B. Impacts of landscape structure on butterfly range expansion. Ecol. Lett. 2001;4:313–321. 10.1046/j.1461-0248.2001.00222.x [Google Scholar]
- Hovestadt T, Messner S, Poethke H.J. Evolution of reduced dispersal mortality and ’fat-tailed’ dispersal kernels in autocorrelated landscapes. Proc. R. Soc. B. 2001;268:385–391. doi: 10.1098/rspb.2000.1379. 10.1098/rspb.2000.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.M, Zalucki M.P. The relationship petween the pgi locus and the ability to fly at low temperatures in the monarch butterfly Danaus plexippus. Biochem. Genet. 1993;31:521–532. doi: 10.1007/BF02426883. [DOI] [PubMed] [Google Scholar]
- Hughes C.L, Hill J.K, Dytham C. Evolutionary trade-offs between reproduction and dispersal in populations at expanding range boundaries. Proc. R. Soc. B. 2003;270:S147–S150. doi: 10.1098/rsbl.2003.0049. 10.1098/rspb.2002.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent J.W, Rankin M.A. Heritability and physiological correlates of migratory tendency in the grasshopper Melanoplus sanguinipes. Phys. Entomol. 2001;26:371–380. 10.1046/j.0307-6962.2001.00257.x [Google Scholar]
- Kubaszek A, Pihlajamaki J, Punnonen K, Karhapaa P, Vauhkonen I, Laakso M. The C-174G promoter polymorphism of the IL-6 gene affects energy expenditure and insulin sensitivity. Diabetes. 2003;52:558–561. doi: 10.2337/diabetes.52.2.558. [DOI] [PubMed] [Google Scholar]
- Kuussaari M, Nieminen M, Hanski I. An experimental study of migration in the Glanville fritillary butterfly Melitaea cinxia. J. Anim. Ecol. 1996;65:791–801. [Google Scholar]
- Kuussaari M, Saccheri I, Camara M, Hanski I. Allee effect and population dynamics in the Glanville fritillary butterfly. Oikos. 1998;82:384–392. [Google Scholar]
- Lighton J.R.B. Measurements on insects. In: Payne C.A, editor. Concise encyclopedia on biological and biomedical measurement systems. Pergamon Press; Oxford: 1991. pp. 201–208. [Google Scholar]
- Marden J.H, Wolf M.R, Weber K.E. Aerial performance of Drosophila melanogaster from populations selected for upwind flight ability. J. Exp. Biol. 1997;200:2747–2755. doi: 10.1242/jeb.200.21.2747. [DOI] [PubMed] [Google Scholar]
- Montooth K.L, Marden J.H, Clark A.G. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics. 2003;165:623–635. doi: 10.1093/genetics/165.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen M, Siljander M, Hanski I. Structure and dynamics of Melitaea cinxia metapopulations. In: Ehrlich P.R, Hanski I, editors. On the wing of checkerspots: a model system for population biology. Oxford University Press; Oxford: 2004. pp. 63–91. [Google Scholar]
- Olivieri I, Michalakis Y, Gouyon P.H. Metapopulation genetics and the evolution of dispersal. Am. Nat. 1995;146:202–228. 10.1086/285795 [Google Scholar]
- Orr H.A. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Paterson A.H, Damon S, Hewitt J.D, Zamir D, Rabinowitch H.D, Lincoln S.E, Lander E.S, Tanksley S.D. Mendelian factors underlying quantitative traits in the tomato: comparison across species, generations and environments. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne J, Montgomery H. The renin–angiotensin system and physical performance. Biochem. Soc. Trans. 2003;31:1286–1289. doi: 10.1042/bst0311286. [DOI] [PubMed] [Google Scholar]
- Peroni P.A. Seed size and dispersal potential of Acer rubrum (Aceraceae) samaras produced by populations in early and late successional environments. Am. J. Bot. 1994;81:1428–1434. [Google Scholar]
- Piquot Y, Petit D, Valero M, Cuguen J, de Laguerie P, Vernet P. Variation in sexual and asexual reproduction among young and old populations of the perennial macrophyte Sparganium erectum. Oikos. 1998;82:139–148. [Google Scholar]
- Postma E, Noordwijk A.J. Gene flow maintains a large genetic difference in clutch size at a small spatial scale. Nature. 2005;433:65–68. doi: 10.1038/nature03083. 10.1038/nature03083 [DOI] [PubMed] [Google Scholar]
- Roff D.A. Dispersal in dipterans: its costs and consequences. J. Anim. Ecol. 1977;46:443–456. [Google Scholar]
- Roff D.A, Fairbairn D.J. Wing dimorphisms and the evolution of migratory polymorphisms among the Insecta. Am. Zool. 1991;31:243–251. [Google Scholar]
- Roff D.A, Fairbairn D.J. The genetic basis of dispersal and migration, and its consequences for the evolution of correlated traits. In: Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford: 2001. pp. 191–202. [Google Scholar]
- Saccheri I.J, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. 10.1038/33136 [Google Scholar]
- Saccheri I.J, Boggs C.L, Hanski I, Ehrlich P.R. Genetics of checkerspot populations. In: Ehrlich P.R, Hanski I, editors. On the wing of checkerspots: a model system for population biology. Oxford University Press; Oxford, UK: 2004. pp. 199–218. [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Suarez R.K. Energy metabolism during insect flight: biochemical design and physiological performance. Phys. Biochem. Zool. 2000;73:765–771. doi: 10.1086/318112. 10.1086/318112 [DOI] [PubMed] [Google Scholar]
- Taylor P.D, Merriam G. Wing morphology of a forest damselfly is related to landscape structure. Oikos. 1995;73:43–48. [Google Scholar]
- Thomas C.D, Hill J.K, Lewis O.T. Evolutionary consequences of habitat fragmentation in a localized butterfly. J. Anim. Ecol. 1998;67:485–497. 10.1046/j.1365-2656.1998.00213.x [Google Scholar]
- Travis J.M.J, Dytham C. Habitat persistence, habitat availability and the evolution of dispersal. Proc. R. Soc. B. 1999;266:723–728. 10.1098/rspb.1999.0696 [Google Scholar]
- Watt W.B. Adaptation at specific loci. 1. Natural selection on phosphoglucose isomerase of Colias butterflies—biochemical and population aspects. Genetics. 1977;87:177–194. doi: 10.1093/genetics/87.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W.B. Adaptation at specific loci .2. Demographic and biochemical elements in the maintenance of the Colias pgi polymorphism. Genetics. 1983;103:691–724. doi: 10.1093/genetics/103.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W.B. Eggs, enzymes, and evolution—natural genetic variants change insect fecundity. Proc. Natl Acad. Sci. USA. 1992;89:10 608–10 612. doi: 10.1073/pnas.89.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W.B, Boggs C.L. Allelic isozymes as probes of the evolution of metabolic organization. Isozymes: Curr. Topics Biol. Med. Res. 1987;15:27–47. [PubMed] [Google Scholar]
- Watt W.B, Dean A.M. Molecular functional studies of adaptive genetic variation in prokaryotes and eukaryotes. Annu. Rev. Genet. 2000;34:593–622. doi: 10.1146/annurev.genet.34.1.593. 10.1146/annurev.genet.34.1.593 [DOI] [PubMed] [Google Scholar]
- Watt W.B, Cassin R.C, Swan M.S. Adaptation at specific loci 3. Field behavior and survivorship differences among Colias pgi genotypes are predictable from in vitro biochemistry. Genetics. 1983;103:725–739. doi: 10.1093/genetics/103.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W.B, Donohue K, Carter P.A. Adaptation at specific loci 6. Divergence vs parallelism of polymorphic allozymes in molecular function and fitness component effects among Colias species (Lepidoptera, Pieridae) Mol. Biol. Evol. 1996;13:699–709. [Google Scholar]
- Watt W.B, Wheat C.W, Meyer E.H, Martin J.F. Adaptation at specific loci. VII. Natural selection, dispersal and the diversity of molecular–functional variation patterns among butterfly species complexes (Colias: Lepidoptera, Pieridae) Mol. Ecol. 2003;12:1265–1275. doi: 10.1046/j.1365-294x.2003.01804.x. 10.1046/j.1365-294X.2003.01804.x [DOI] [PubMed] [Google Scholar]
- Woiwod I.P, Reynolds D.R, Thomas C.D, editors. Insect movements: mechanisms and consequences. CAB Publications; Wallingford: 2001. [Google Scholar]
- Yang N, MacArthur D.G, Gulbin J.P, Hahn A.G, Beggs A.H, Easteal S, North K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003;73:627–631. doi: 10.1086/377590. 10.1086/377590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zera A.J, Denno R.F. Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 1997;42:207–230. doi: 10.1146/annurev.ento.42.1.207. 10.1146/annurev.ento.42.1.207 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zera A.J. Differential lipid biosynthesis underlies a trade-off between reproduction and flight-capability in a wing-polymorphic cricket. Proc. Natl Acad. Sci. USA. 2002;99:16 829–16 834. doi: 10.1073/pnas.262533999. 10.1073/pnas.262533999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.