Abstract
Studies of natural killer (NK) cell function in vivo have been challenging primarily due to the lack of animal models in which NK cells are genetically and selectively deficient. Here, we describe a transgenic mouse with defective natural killing and selective deficiency in NK1.1+ CD3− cells. Despite functionally normal B, T, and NK/T cells, transgenic mice displayed impaired acute in vivo rejection of tumor cells. Adoptive transfer experiments confirmed that NK1.1+ CD3− cells were responsible for acute tumor rejection, establishing the relationship of NK1.1+ CD3− cells to NK cells. Additional studies provided evidence that (i) NK cells play an important role in suppressing tumor metastasis and outgrowth; (ii) NK cells are major producers of IFNγ in response to bacterial endotoxin but not to interleukin-12, and; (iii) NK cells are not essential for humoral responses to T cell-independent type 2 antigen or the generalized Shwartzman reaction, both of which were previously proposed to involve NK cells.
NK cells were discovered because of their in vitro ability to kill tumor cells without prior sensitization, a process termed natural killing (1, 2). Over the years, this process has become the functional definition and hallmark of NK cells, but it has also led to confusion over their identity with other cells having similar activity. As other cell lineages became molecularly characterized, natural killing against certain tumor cells, YAC-1 in mouse and K562 in human, has become attributable to NK cells. However, there is no consensus on the molecular definition of NK cells; they are still distinguished from other major lymphocyte populations in a negative sense, by the absence of antigen receptors such as the CD3/T cell antigen receptor complex. Although this distinction is useful, CD3− NK cells share many functions with T cells, such as cytolytic effector mechanisms and cytokine production. Without reliable means to positively identify NK cells, there still are difficulties in discriminating the activities of NK cells, particularly in vivo, from those of other cells.
Unlike B and T cells, NK cells develop normally in mutant mice with defective antigen receptor rearrangement. Splenocytes from scid mice, e.g., are capable of natural killing even though mature T cell populations are essentially absent (3). By contrast, beige (bg) mice display defective natural killing (4). Yet, the effect of the bg mutation is neither selective nor complete because it affects the granules of many other cell types and it does not ablate other NK cell activities such as cytokine production (5, 6). Similarly, natural killing capacity is defective in perforin or granzyme B-deficient mice, but cytotoxic T cells are also defective whereas other NK cell activities are presumably normal (7, 8). NK cell development is variably impaired in mice with targeted mutations in Ikaros, IL-2Rγ, IL-2Rβ, IRF-1, Ets-1, Id2, or Ltα genes, but all of these mice have other profound abnormalities due to the pleiotropic roles of these molecules (9–15). Although tgɛ26 mice (transgenic for human CD3ɛ) have been used for in vivo studies of certain NK cell functions, they have profound defects in T cell as well as NK cell development (16). Thus, there is as yet no animal model in which NK cell activities are genetically and selectively deficient.
In various experimental models, NK cells have been proposed to play an important role in tumor rejection in vivo. In addition, NK cells are thought to be key players in innate immunity because of their potent capacity to produce cytokines, particularly IFNγ (17). Relevant to IFNγ production, NK cells may contribute to T cell-independent (TI) immune responses and the generalized Shwartzman reaction, an experimental septic shock model whereby mice die from repeated lipopolysaccharide (LPS) exposure (18, 19). Importantly, however, most previous in vivo studies of NK cell functions have relied heavily on antibody (Ab) administration to eliminate NK cells, a strategy that may be revealing but with significant limitations (1, 17). In particular, Abs used in these studies are not specific for NK cells. Even administration of the currently favored reagent, mAb PK136 (anti-NK1.1), which recognizes the most specific serological marker on NK cells in certain strains of mice (2), may affect a CD3+ T cell subpopulation, termed NK/T cells. These cells are potent producers of IL-4 and express the NK1.1 molecule as well as members of the Ly49 family, most of which are MHC class I-specific inhibitory receptors expressed on NK cell subpopulations (20). Although potential Ab cross-reactivity with non-NK cells was primarily a theoretical concern, recent work suggesting that NK/T cells play major roles in tumor rejection and IFNγ production under certain conditions (21–23) raised an important issue that these NK/T functions may have been mistaken for NK cell-mediated functions. Moreover, Ab administration may induce undesirable side effects. For example, anti-NK1.1 crosslinking can induce both NK/T and NK cells to produce cytokines (24, 25). It is also conceivable that administrated Abs may cause nonspecific effects through interactions with Fc receptors expressed on many cell types. Finally, the Ab depletion strategy is limited in reconstitution studies to formally demonstrate the function of NK cells. Thus, animal models with selective deficiency in NK cells are necessary to allow reliable progress beyond the existing correlative data on in vivo NK cell function.
Herein, we describe a transgenic (Tg) mouse with a profound and selective deficiency in NK1.1+ CD3− cells and defective natural killing. Studies with the Tg mice provided opportunities to establish and enhance understanding of the relationship of NK1.1+ CD3− cells to NK cell functions and ascertain the significance of NK cells in vivo.
Materials and Methods
Mice.
C57BL/6 and C57BL/6-Prkdcscid/Sz mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained in a specific pathogen-free barrier facility at Washington University.
Generation of Transgenic Mice.
A murine granzyme A genomic construct (carrying 11-kb putative promoter and 7-kb structural gene with poly-A addition signal) (26) was used for expression of a Ly49A cDNA. The granzyme A-Ly49A transgene was constructed by standard methods. The granzyme A translation start codon was deleted by replacing an 1,032-bp NaeI–SmaI fragment (carrying the putative promoter and start codon) with an 1,008-bp NaeI–SmaI fragment (carrying only the putative promoter) generated by PCR using the primers CCTGAAGCATGCTATCTCACGA and GTTCCCGGGCTCTCCCACCCCAATCA. The Ly49A cDNA was excised as a 1.2-kb XhoI fragment from pA1.3 (27), filled by Klenow fragment, and inserted into the SmaI site of the modified granzyme A gene. The granzyme A-Ly49A transgene was excised as a 19-kb SalI–KpnI fragment and micro-injected into the pronucleus of fertilized C57BL/6 eggs by standard methods. Initial screening for Tg mice was performed by Southern analysis of tail DNA with the Ly49A cDNA probe. After Tg founders were established, Tg mice were screened by PCR analysis of tail DNA using transgene-specific primers. Hemizygous Tg mice were used in all experiments. They were healthy, fertile, and had no apparent abnormalities in lymphoid organs.
Cell Lines and Reagents.
RMA-S, a Tap-2-deficient mutant originated from RBL-5, was provided by K. Kärre (Karolinska Institute, Stockholm, Sweden). B16 was provided by W. Seaman (VA Medical Center, San Francisco, CA). YAC-1 and hybridomas for the following mAbs were obtained from American Type Culture Collection (Manassas, VA): anti-NK1.1 (PK136), anti-CD3ɛ (145–2C11), and anti-FcγRII/III (2.4G2). Anti-Ly49A (JR9–318) (28) was obtained from J. Roland (Pasteur Institute, Paris, France) and conjugated to fluorescein isothiocyanate. The following fluorochrome-conjugated mAbs were purchased from PharMingen (La Jolla, CA): fluorescein isothiocyanate or Cy-Chrome-conjugated anti-CD3ɛ (145–2C11), phycoerythrin-conjugated anti-NK1.1 (PK136) and anti-CD4 (RM4–5), and fluorescein isothiocyanate-conjugated anti-CD8 (53–6.7). Recombinant murine IL-12 was a gift from M. Gately (Hoffmann-La Roche, Nutley, NJ). LPS from Escherichia coli serotype 0127:B8 or Serratia marcescens was purchased from Sigma. The 2,4,6,-trinitrophenyl (TNP)-Ficoll and TNP-BSA were purchased from Biosearch Technologies (Novato, CA).
Cell Preparation and Flow Cytometry.
Under anesthesia, mice were exsanguinated by cardiac puncture, and then organs were collected and cell suspensions were prepared as described (29–31). Peripheral blood was directly treated with red blood cell lysis solution (0.14 M NH4Cl/0.017 M Tris, pH 7.2). For flow cytometry, cells were incubated with anti-FcγRII/III Ab to block nonspecific binding then stained with combinations of indicated fluorochrome-conjugated mAbs and analyzed with a FACScalibur (Becton Dickinson). Dead cells were removed by prior centrifugation on lympholyte-M (Cedarlane Laboratories) gradient or excluded from analysis by propidium iodide staining. Each organ was analyzed from at least three mice per group.
In Vitro Cytotoxicity and In Vivo Tumor Rejection Assays.
Mice received i.p. injection of poly-I:C (150 μg). Freshly prepared splenocytes were tested 24 hr later in a standard 51Cr -release assay as described (29). Lung clearance assay was performed as described (32). In brief, tumor cells were preincubated with 5-fluoro-2-deoxyuridine (FUdr; Sigma) then radiolabeled with 125I-5-iodo-2-deoxyuridine (125I-Udr; Amersham). Mice were injected i.v. (tail vein) with 3 × 104 to 3 × 105 cells/200 μl PBS. Mice were killed 4–6 hr later and the lungs were counted with a γ-counter. The percentage of residual radioactivity was calculated as being equal to (residual radioactivity in the lungs/total injected radioactivity) × 100. For tumor metastasis assay, mice were injected i.v. (tail vein) with 3 × 104 B16 melanoma cells/200 μl of PBS. Visible, black metastatic foci were counted on lung surfaces 14 d later. For long-term tumor outgrowth assay, mice were injected s.c. in the left flank with RMA-S cells/100 μl. Mice were monitored for palpable tumors twice weekly.
Ig Isotypes and Cytokines.
Serum levels of Ig isotypes were determined by using the Clonotyping system-horseradish peroxidase kit (Southern Biotechnology Associates) according to the manufacturer's instructions. For the detection of TNP-specific Igs, Immulon2 microtiter plates (Dynex Technologies) were coated with TNP-BSA in PBS overnight at 4°C and washed with PBS + 0.3% Tween 20. The plates were then incubated with PBS containing 2% BSA for 1 hr at room temperature. Serum samples serially diluted in PBS + 2% BSA were added, incubated for 2 hr at room temperature, and washed with PBS containing 0.3% Tween 20, and plate-bound Igs were determined with the Clonotyping system-horseradish peroxidase kit. Optical density was measured with a ELISA plate reader. Cytokines were measured by using IL-4 and IFNγ ELISA kits (Endogen, Woburn, MA).
Shwartzman Reaction.
The generalized reaction was induced as described (19). In brief, mice were first primed with a footpad injection of 5 μg of Serratia marcescens-derived LPS (Sigma) in 40 μl PBS. Mice were injected i.v. 24 hr later with LPS as indicated. Susceptibility was scored by mortality.
Results and Discussion
Production of Transgenic Mice with Defective Natural Killing.
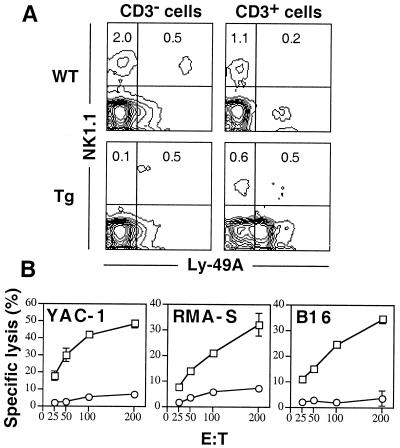
In an attempt to direct expression of Ly49A, an inhibitory MHC class I-specific receptor, on all NK cells, we produced Tg mice expressing the Ly49A cDNA under control of the mouse granzyme A genomic sequence. Among eight potential founders, all derived by pronuclear injection of C57BL/6 fertilized ova, Tg mice derived from founder no. 7 expressed Ly49A on nearly all splenic NK1.1+ CD3− cells (Fig. 1A), a cell population previously correlated with in vitro NK cell activity (33). By contrast, wild-type (WT) littermates expressed Ly49A on only 20% of these cells as previously reported (29). The Tg mice also expressed Ly49A on approximately one-half of T cells but not on other cell types. Surprisingly, the number of NK1.1+ CD3− splenic cells was markedly reduced in Tg mice derived from founder no. 7, which is different from results published on two other Ly49A Tg mice (34, 35). Tg mice derived from other founders expressed Tg Ly49A mainly on T cells and had normal numbers of NK1.1+ CD3− splenic cells (data not shown). Because the Tg mice from founder no. 7 were informative, this line was extensively studied to determine the relevance of the markedly diminished number of NK1.1+ CD3− cells.
Figure 1.
In vitro natural killing is defective in a Ly49A Tg mouse strain. (A) Spleen cells from WT and Ly49A Tg mice were stained with anti-Ly49A, anti-NK1.1, and anti-CD3. Profiles for Ly49A expression on cells gated on CD3− or CD3+ populations are shown. The numbers represent the percentage of cells within the quadrant among all viable cells. (B) Splenocytes from WT (□) and Ly49A Tg (○) mice were used in 4-hr 51Cr-release cytotoxicity assays against YAC-1, RMA-S, and B16 targets at varying effector/target (E:T) ratios as indicated. Results are expressed as the mean percentage specific lysis ± SD of triplicate wells.
Functional studies demonstrated that unlike splenocytes isolated from WT mice, splenocytes from Tg mice had undetectable in vitro natural killing activity against YAC-1 (Fig. 1B), a target widely accepted as being sensitive to mouse NK cells. Although YAC-1 is susceptible to lysis by Ly49A+ IL-2-activated NK cells from WT mice, YAC-1 cells express an MHC class I ligand for Ly49A (H2Dd) at low levels that can inhibit killing by Ly49A+ NK cells when expressed on other targets (29). However, Tg splenocytes also failed to kill B16 (H2b) and MHC class I-deficient RMA-S targets (Fig. 1B), indicating that the impaired killing capacity is not due to an inhibitory effect of Ly49A-MHC class I interaction. Thus, the data demonstrate that Tg mice have no in vitro natural killing activity, an activity defining NK cells, and this absence is correlated with reduction of splenic NK1.1+ CD3− cells.
Selective Deficiency in the NK Cells.
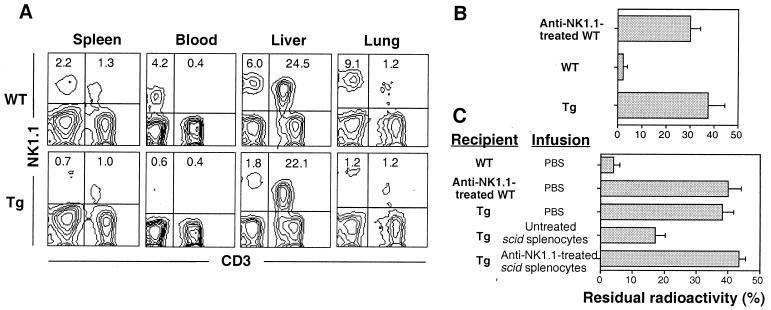
The number of NK1.1+ CD3− cells also was markedly reduced in other peripheral tissues of Tg mice (Fig. 2A). For example, there were virtually no NK1.1+ CD3− cells in peripheral blood. By contrast, albeit showing a slight reduction in the spleen, Tg mice had essentially normal numbers of NK/T cells. In particular, NK/T cells constituted >20% of leukocytes in the livers of Tg mice like WT mice as previously reported (30). The functional activities of NK/T cells were examined by measuring ex vivo IL-4 and IFNγ production by splenocytes from mice injected with anti-CD3 mAb (36). The levels of both IL-4 and IFNγ produced by Tg splenocytes were ≈70% of normal when compared to WT splenocytes (data not shown). The production was proportional to the number of splenic NK/T cells, suggesting that NK/T cells in Tg mice are functionally normal.
Figure 2.
Impaired acute in vivo rejection of tumor cells due to selective reduction of peripheral NK1.1+ CD3− cells. (A) Selective reduction of peripheral NK1.1+ CD3− cells in Tg mice. Cells prepared from indicated tissues were stained with anti-NK1.1 and anti-CD3. The numbers represent the percentage of cells within the quadrant among all viable cells. (B) Mice were inoculated i.v. with 125I-Udr-labeled 3 × 105 YAC-1 tumor cells. After 4 hr, the residual lung radioactivity was measured. Results are expressed as the mean % residual radioactivity ± SD from three to five mice per group. (C) Reconstituted tumor clearance by scid splenocytes. Scid mice that were pretreated with anti-NK1.1 or untreated were injected i.p. with poly-I:C on day −1. Two hours after i.v. infusion of PBS or 8 × 106 splenocytes isolated from anti-NK1.1-treated or untreated scid mice, the recipient mice were inoculated i.v. with 125I-Udr-labeled 3 × 104 RMA-S tumor cells. After 6 hr, the residual lung radioactivity was measured. Results are expressed as the mean percentage residual radioactivity ± SD from three mice per group. Where indicated for B and C, anti-NK1.1-treated mice received an i.p. injection of 200 μg of anti-NK1.1 mAb 2 days before tumor inoculation.
The other lymphocytes in Tg mice also appeared to be normal. There were normal numbers of total cells, T cells, T cell subsets, and B cells in lymphoid organs, including spleen, peripheral blood, and thymus, as determined by cell counting and expression of CD3, CD4, CD8, and sIg in flow cytometric analysis (data not shown). The functional activities of T and B cells in Tg mice appeared to be normal. Between Tg and WT mice, there were no apparent differences in proliferative responses to anti-CD3 mAb in vitro, allo-MHC-specific and cytotoxic T cell activities except for the known inhibitory effect of Ly49A in response to H2Dd (34), rejection of skin allografts, and serum levels of IgM and IgG subclasses (data not shown). Finally, Tg mice appeared to have normal numbers of myeloid cells as determined by analysis of Gr-1, Mac-1, and CD11c expression (data not shown). Taken together, Tg mice appeared to have a selective deficiency in NK1.1+ CD3− cells.
To evaluate the capacity of Tg mice to eliminate tumor cells in vivo, radiolabeled YAC-1 cells were injected; WT mice completely eliminated the tumor cells within 4 hr as evidenced by only low levels of lung radioactivity (Fig. 2B). By contrast, >70-fold higher levels of radioactivity were present in Tg lungs. Similar results were obtained with RMA-S cells (Fig. 2C). Treatment of WT mice with anti-NK1.1 mAb resulted in impaired tumor clearance at levels comparable to those of unmanipulated Tg mice (Fig. 2 B and C). The results demonstrate that Tg mice have a profound functional defect, apparently involving NK1.1+ CD3− cells.
To establish that the defect was in the NK1.1+ CD3− population, we performed adoptive transfer studies with splenocytes from scid mice that have NK1.1+ CD3− cells but no mature B, T, and NK/T cells. After transfer of scid splenocytes prior to tumor inoculation, Tg mice demonstrated markedly improved capacity to eliminate RMA-S cells (Fig. 2C). However, pretreatment of scid mice with anti-NK1.1 abrogated this activity, indicating that NK1.1+ CD3− cells were responsible for the reconstituted tumor clearance and other cell types including macrophage and neutrophils had no such activity. The reconstitution experiments formally establish a significant role of NK1.1+ CD3− cells in tumor elimination in vivo. Collectively, the data strongly suggest that the functional defect in Tg mice is due to a selective deficiency in NK1.1+ CD3− cells and that these cells represent NK cells.
Role of NK Cells in Suppression of Tumor Metastasis and Outgrowth.
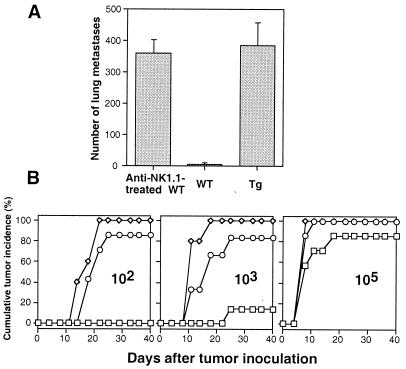
Taking advantage of the selective NK cell deficiency in Tg mice, we evaluated the role of NK1.1+ CD3− NK cells in tumor rejection using two established in vivo tumor models (37). When B16 cells were i.v. inoculated, dramatically increased numbers of tumor foci were found in Tg lungs after 2 wk (Fig. 3A). The Tg mice had >60-fold higher numbers of lung metastases than WT mice, indicating a critical role of NK cells in inhibition of tumor metastasis. To assess the relative contribution of NK/T cells, mice were depleted of both NK and NK/T cells by anti-NK1.1 treatment as a control group. Repeated treatments of WT mice with anti-NK1.1 resulted in development of increased lung metastases but not greater than those in Tg mice. Interestingly, the anti-NK1.1-treated WT mice also had a greater number of liver metastases whereas Tg mice had as few metastases as WT mice (data not shown), suggesting a redundant role of NK/T cells in this process as previously reported (38). Given the presence of NK/T cells in Tg mice, however, our data suggest a predominant role of NK cells in prevention of lung metastases in this experimental system using these tumor cells and doses.
Figure 3.
Control of experimental lung metastases and tumor outgrowth is impaired in Tg mice. (A) Mice were injected i.v. with 3 × 104 B16 cells. The numbers of macroscopic tumor metastases in the lungs were counted on day 14. Anti-NK1.1-treated mice received three i.p. injections (200 μg/injection) of anti-NK1.1 on days −2, +2, and +7. Results are represented as the average number ± SD from four mice per group. (B) Varying numbers of RMA-S cells as indicated were inoculated s.c. into the flanks of WT (seven mice per group, □), Tg (six to seven mice per group, ○) and anti-NK1.1-treated WT (three to five mice per group, ⋄) mice, respectively. Anti-NK1.1-treated mice received five i.p. injections (200 μg/injection) of anti-NK1.1 on days −4, −2, +7, +14, and +21. Palpable tumors were scored twice weekly.
In tumor outgrowth assays, Tg mice failed to reject as few as 100 RMA-S cells inoculated s.c. whereas no palpable tumors developed in WT mice over a 40-d interval (Fig. 3B). Nearly 100-fold more tumor cells were needed for similar levels of tumor incidence in WT mice. Repeated treatments of WT mice with anti-NK1.1 resulted in increased tumor incidence slightly higher than that observed in Tg mice. Again, given the presence of NK/T cells in Tg mice, our data suggest a predominant role for NK cells. Therefore, studies with our Tg mice, together with reconstitution studies, provide direct compelling evidence that NK cells play a significant role in tumor rejection in vivo.
Contribution of NK Cells to IFNγ Production Induced by LPS or IL-12.
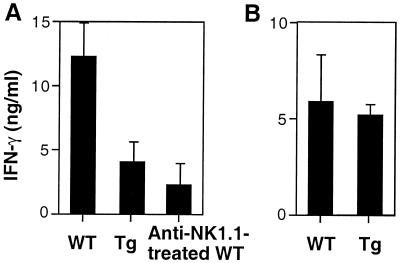
Many physiological functions of NK cells are probably mediated by their production of cytokines. In many infections, NK cells have been proposed as an important early source of IFNγ (reviewed in ref. 17). However, the specific contribution of NK cells to IFNγ production and the regulatory mechanisms in vivo remain to be further established. To gain an insight into these issues, we examined in vivo production of IFNγ after systemic exposure to bacterial endotoxin (LPS). Upon i.p. injection of 200 μg of LPS, WT mice produced significant amounts of IFNγ, reaching peak serum levels at 7 hr (Fig. 4A). However, Tg mice produced markedly lower serum levels. With lower doses of LPS, there were more pronounced reductions in Tg mice (data not shown). Compared to Tg mice, anti-NK1.1-treated WT mice produced slightly lower levels of IFNγ. These results indicated that NK cells are the predominant source of acute IFNγ production in response to LPS and suggest that NK/T cells have a redundant but minor effect. The predominant contribution of NK cells to IFNγ production in response to LPS suggests an important role for NK cells in innate immunity upon bacterial infection.
Figure 4.
IFNγ production in response to LPS but not to IL-12 is impaired in Tg mice. (A) Mice were injected i.p. with 200 μg LPS. Sera were collected after 7 hr and assayed for IFNγ by ELISA. Results are expressed as the average ± SD from five to seven mice per group. (B) Mice were injected i.p. with 1 μg IL-12, and sera were collected after 24 hr. Results are expressed as the average ± SD from five mice per group.
Surprisingly and by contrast, NK cells do not contribute significantly to the in vivo IFNγ response to IL-12. Despite the early description of IL-12 as having potent capacity to induce NK cell-mediated IFNγ production in vitro (39), Tg mice produced IFNγ at levels comparable to those produced by WT mice upon injection of IL-12 (Fig. 4B). This observation fits with previous studies suggesting that NK/T cells are the primary source of IFNγ in response to IL-12 in vivo and express higher levels of IL-12 receptors compared to NK cells (22, 23). In addition, a more recent study has demonstrated that even dendritic cells and macrophages are more efficient producers of IFNγ than NK cells in response to IL-12 in vitro (40). Taken together, our data suggest that NK cells become potent producers of IFNγ under the physiologic conditions where other costimulatory cytokines such as tumor necrosis factor-α are produced together with IL-12 (41). Our data may also provide an explanation for why NK cells are not often required for IL-12-mediated effects in many studies (21, 42).
The Role of NK Cells in Antibody Response to T Cell-Independent Antigen and the Generalized Shwartzman Reaction.
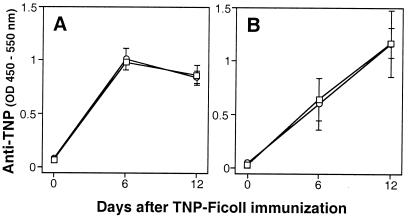
NK cells have been long suspected to have regulatory effects on B cell differentiation and Ig secretion (43). In particular, studies using in vitro models of TI type 2 Ab responses have suggested that NK cells, probably through their production of IFNγ, play an important role in this response (44, 45). To test this possibility in vivo, we examined Ab responses of WT and Tg mice to TNP-Ficoll, a prototypic TI type 2 antigen (43). Analysis of serum levels of TNP-specific Abs indicated that following IgM response (Fig. 5A), WT mice developed predominant IgG3 response (Fig. 5B). Unexpectedly, Tg mice produced virtually identical levels of TNP-specific IgM and IgG3. In addition, there were no apparent differences in production of other IgG isotypes between WT and Tg mice (data not shown). These data indicated that Tg mice developed normal TI type 2 Ab responses including class switching. Similarly, pretreatment of WT mice with anti-NK1.1 had no apparent effect on these responses (data not shown). Thus, the data suggest that NK cells are not required for the induction of in vivo TI type 2 humoral immunity.
Figure 5.
Normal Ab responses to TI type 2 antigen in Tg mice. WT (□) and Tg (○) mice were immunized i.p. with 10 μg of TNP-Ficoll. Sera were collected at the indicated dates, diluted 500-fold, and TNP-specific IgM (A) and IgG3 (B) were measured by ELISA. Results are expressed as the average optical density ± SD from three mice per group.
Mice succumb to lethal shock upon two (priming and challenging) deliberate injections of LPS, termed the generalized Shwartzman reaction (46). Previously, administration of Abs reactive with NK cells rendered mice resistant to this reaction (19). Here we show that under conditions in which WT mice succumbed to lethal shock, Tg mice also died (Table 1). By contrast, most of WT mice that received two previous injections of ascites fluid containing high concentration of anti-NK1.1 survived as previously reported (19). Interestingly, however, no protective effect was observed when WT mice were treated once with 200 μg of anti-NK1.1, a dose that was capable of depleting >90% of NK, NK/T cells, and natural killing activity in splenocytes (data not shown). Thus, our data suggest that NK cells are not required for the induction of the generalized Shwartzman reaction.
Table 1.
Normal induction of the generalized Shwartzman reaction in Tg mice
| LPS, μg
|
Anti-NK1.1-treated WT† | WT (dead/tested) | Tg |
|---|---|---|---|
| Challenging* | |||
| 100 | 1 /9 | 0 /5 | |
| 200 | 1 /6 | 3 /6 | 4 /6 |
| 300 | 2 /6 | 15 /16 | 14 /17 |
Mice were first primed with the footpad injection of 5 μg of S. marcescens-derived LPS 24 hr before i.v. injection of indicated challenging doses of LPS.
† Mice were treated twice with 100 μl of anti-NK1.1 ascites fluid 4 and 1 days before the priming injection.
One explanation for the difference in susceptibility of Tg mice and anti-NK1.1-treated mice is that NK/T cells are essential for the shock response. In this regard, previous work has suggested that NK/T cells are required for the induction of a modified Shwartzman reaction elicited by LPS following IL-12 injection (23). Another possible explanation is that NK and NK/T cells have redundant function. We favor this explanation because NK cells are the predominant source of IFNγ in response to LPS as demonstrated above and this cytokine plays a critical role in the LPS-induced lethal shock (46). Studies with NK/T cell-deficient mice such as CD1-deficient mice should aid in addressing this issue.
In conclusion, the current study describes a mouse model with a profound and selective defect involving NK1.1+ CD3− NK cells. Studies with this mouse model provide direct evidence for significant in vivo roles of NK1.1+ CD3− NK cells in tumor rejection and IFNγ production. In addition, comparative studies with anti-NK1.1-treated mice have suggested contributions of NK/T cells to these immune responses. Importantly, our data warrant caution in interpretation of observations obtained with the Ab administration strategy alone. Our mouse model will be useful for studies of specific NK cell functions in primary tumor induction, chronic infection, and autoimmunity.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Mike White, Jacque Mudd, Debra Rateri, and Kim Marlotte. These studies were supported by an National Institutes of Health grant to W.M.Y. K.I. was supported by a fellowship from the Missouri Chapter of the Lupus Foundation. W.M.Y. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- NK
natural killer
- WT
wild type
- Tg
transgenic
- TI
T cell-independent
- LPS
lipopolysaccharide
- TNP
2,4,6,-trinitrophenyl
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050588297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050588297
References
- 1.Trinchieri G. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama W M. In: Fundamental Immunology. Paul W E, editor. New York: Lippincott-Raven; 1999. pp. 575–603. [Google Scholar]
- 3.Dorshkind K, Pollack S B, Bosma M J, Phillips R A. J Immunol. 1985;134:3798–3801. [PubMed] [Google Scholar]
- 4.Talmadge J E, Meyers K M, Prieur D J, Starkey J R. Nature (London) 1980;284:622–624. doi: 10.1038/284622a0. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa M D, Nguyen Q A, Tchernev V T, Ashley J A, Detter J C, Blaydes S M, Brandt S J, Chotai D, Hodgman C, Solari R C, et al. Nature (London) 1996;382:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scharton-Kersten T M, Sher A. Curr Opin Immunol. 1997;9:44–51. doi: 10.1016/s0952-7915(97)80157-4. [DOI] [PubMed] [Google Scholar]
- 7.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen K J, Podack E R, Zinkernagel R M, Hengartner H. Nature (London) 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 8.Heusel J W, Wesselschmidt R L, Shresta S, Russell J H, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 9.Georgopoulos K, Bigby M, Wang J H, Molnar A, Wu P, Winandy S, Sharpe A. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 10.Cao X, Shores E W, Hu-Li J, Anver M R, Kelsall B L, Russell S M, Drago J, Noguchi M, Grinberg A, Bloom E T, et al. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H, Duncan G S, Takimoto H, Mak T W. J Exp Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohteki T, Yoshida H, Matsuyama T, Duncan G S, Mak T W, Ohashi P S. J Exp Med. 1998;187:967–972. doi: 10.1084/jem.187.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton K, Muthusamy N, Fischer C, Ting C N, Walunas T L, Lanier L L, Leiden J M. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 14.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Nature (London) 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka K, Chaplin D D, Wang Y, Wu Q, Pegg L E, Yokoyama W M, Fu Y X. Proc Natl Acad Sci USA. 1999;96:6336–6340. doi: 10.1073/pnas.96.11.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Biron C, She J, Higgins K, Sunshine M J, Lacy E, Lonberg N, Terhorst C. Proc Natl Acad Sci USA. 1994;91:9402–9406. doi: 10.1073/pnas.91.20.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott P, Trinchieri G. Curr Opin Immunol. 1995;7:34–40. doi: 10.1016/0952-7915(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 18.Mond J J, Lees A, Snapper C M. Annu Rev Immunol. 1995;13:655–692. doi: 10.1146/annurev.iy.13.040195.003255. [DOI] [PubMed] [Google Scholar]
- 19.Heremans H, Dillen C, van Damme J, Billiau A. Eur J Immunol. 1994;24:1155–1160. doi: 10.1002/eji.1830240522. [DOI] [PubMed] [Google Scholar]
- 20.Bendelac A. Curr Opin Immunol. 1995;7:367–374. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 21.Cui J Q, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura T, Takeda K, Mendiratta S K, Kawamura H, Van Kaer L, Yagita H, Abo T, Okumura K. J Immunol. 1998;160:16–19. [PubMed] [Google Scholar]
- 23.Ogasawara K, Takeda K, Hashimoto W, Satoh M, Okuyama R, Yanai N, Obinata M, Kumagai K, Takada H, Hiraide H, Seki S. J Immunol. 1998;160:3522–3527. [PubMed] [Google Scholar]
- 24.Arase H, Arase N, Saito T. J Exp Med. 1996;183:2391–2396. doi: 10.1084/jem.183.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Yokoyama W M. Cell Immunol. 1998;183:106–112. doi: 10.1006/cimm.1998.1252. [DOI] [PubMed] [Google Scholar]
- 26.Aguila H L, Hershberger R J, Weissman I L. Proc Natl Acad Sci USA. 1995;92:10192–10196. doi: 10.1073/pnas.92.22.10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama W M, Jacobs L B, Kanagawa O, Shevach E M, Cohen D I. J Immunol. 1989;143:1379–1386. [PubMed] [Google Scholar]
- 28.Roland J, Cazenave P A. Int Immunol. 1992;4:699–706. doi: 10.1093/intimm/4.6.699. [DOI] [PubMed] [Google Scholar]
- 29.Karlhofer F M, Ribaudo R K, Yokoyama W M. Nature (London) 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi M, Ogasawara K, Takeda K, Hashimoto W, Sakihara H, Kumagai K, Anzai R, Satoh M, Seki S. J Immunol. 1996;156:2436–2442. [PubMed] [Google Scholar]
- 31.Watanabe T, Kawamura T, Kawamura H, Haga M, Shirai K, Watanabe H, Eguchi S, Abo T. J Immunol. 1997;158:5805–5814. [PubMed] [Google Scholar]
- 32.Hackett J, Jr, Bennett M, Kumar V. J Immunol. 1985;134:3731–3738. [PubMed] [Google Scholar]
- 33.Tutt M M, Kuziel W A, Hackett J, Jr, Bennett M, Tucker P W, Kumar V. J Immunol. 1986;137:2998–3001. [PubMed] [Google Scholar]
- 34.Held W, Cado D, Raulet D H. J Exp Med. 1996;184:2037–2041. doi: 10.1084/jem.184.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahlen L, Khoo N K, Daws M R, Sentman C L. Eur J Immunol. 1997;27:2057–2065. doi: 10.1002/eji.1830270833. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimoto T, Paul W E. J Exp Med. 1994;179:1285–1295. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herberman R B, Gorelik E. In: Functions of the Natural Immune System. Reynolds C W, Wiltrout R H, editors. New York: Plenum; 1989. pp. 3–37. [Google Scholar]
- 38.Seki S, Hashimoto W, Ogasawara K, Satoh M, Watanabe H, Habu Y, Hiraide H, Takeda K. Immunology. 1997;92:561–566. doi: 10.1046/j.1365-2567.1997.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan S H, Perussia B, Gupta J W, Kobayashi M, Pospisil M, Young H A, Wolf S F, Young D, Clark S C, Trinchieri G. J Exp Med. 1991;173:869–879. doi: 10.1084/jem.173.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohteki T, Fukao T, Suzue K, Maki C, Ito M, Nakamura M, Koyasu S. J Exp Med. 1999;189:1981–1986. doi: 10.1084/jem.189.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wherry J C, Schreiber R D, Unanue E R. Infect Immun. 1991;59:1709–1715. doi: 10.1128/iai.59.5.1709-1715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunda M J, Luistro L, Warrier R R, Wright R B, Hubbard B R, Murphy M, Wolf S F, Gately M K. J Exp Med. 1993;178:1223–1230. doi: 10.1084/jem.178.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snapper C M, Yamaguchi H, Moorman M A, Sneed R, Smoot D, Mond J J. J Immunol. 1993;151:5251–5260. [PubMed] [Google Scholar]
- 44.Snapper C M, Yamaguchi H, Moorman M A, Mond J J. J Immunol. 1994;152:4884–4892. [PubMed] [Google Scholar]
- 45.Vos Q, Snapper C M, Mond J J. Int Immunol. 1999;11:159–168. doi: 10.1093/intimm/11.2.159. [DOI] [PubMed] [Google Scholar]
- 46.Ozmen L, Pericin M, Hakimi J, Chizzonite R A, Wysocka M, Trinchieri G, Gately M, Garotta G. J Exp Med. 1994;180:907–915. doi: 10.1084/jem.180.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]