Abstract
Background and aim: The Krüppel-like transcription factor KLF6 is a novel tumor-suppressor gene. It was inactivated in human prostate cancer and other tumors tissue, as the result of frequent mutation and loss of heterozygosity (LOH). However, there is no data reporting the levels of KLF6 both mRNA and protein in hepatocellular carcinomas (HCCs). We therefore detected mutations and expression of KLF6 in HCC tissues and further observed the effect of it on cell growth in HCC cell lines. Methods: We analyzed the exon-2 of KLF6 gene by direct DNA sequencing, and detected the expression of KLF6 by RT-PCR and Western blot in 23 HCC tissues and corresponding nontumorous tissues. Loss of growth suppressive effect of the HCC-derived KLF6 mutant was characterized by in vitro growth curves plotted, flow cytometry and Western blotting. Results: KLF6 mutations were found in 2 of 23 HCC tissues and one of mutations was missense. Expression of KLF6 mRNA or protein was down-regulated in 8 (34.7%) or 9 (39.1%) of 23 HCC tissues. Wild-type KLF6 (wtKLF6) inhibited cellular proliferation and prolonged G1-S transition by inducing the expression of p21WAF1 following stable transfection into cultured HepG2 cells, but tumor-derived KLF6 mutant (mKLF6) had no effects. Conclusion: Our findings suggest that KLF6 may be involved in pathogenesis of HCC.
Keywords: Tumor suppressor gene, Krüppel-like factor 6 (KLF6), Mutation, Gene expression, Hepatocellular carcinoma
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common cancers and a leading cause of cancer patients’ death in the world, especially in areas such as Eastern Asia and Saharan Africa. Although it is well known that HCC is associated with chronic infection with hepatitis B virus and C virus, alcohol consumption, chronic exposure to the mycotoxin or aflatoxin B1 (AFB1), cirrhosis of the liver, the precise molecular mechanism of HCC is not well-understood (Bruix et al., 2004). It is generally believed that genetic alterations in oncogenes and tumor suppressor genes contribute to each step of human multistage carcinogenesis. In the past years, inactivation of several tumor suppressor genes including p53 (Staib et al., 2003) and Rb (Feitelson et al., 2002), p16INK4 (Hui et al., 1996) has been observed in HCCs. However, inactivation of these suppressor genes is not observed in all HCC tissues. For example, down-regulation of p53 is present in 30%~40% cases, and mutation frequency of p53 in HCC tissues is no more than 30% (Staib et al., 2003). Rb gene mutations are found in 20% to 25% HCCs as a single gene mutation (Feitelson et al., 2002). So it is possible that other tumor suppressors exist and play a role in hepatocarcinogenesis.
Krüppel-like factor 6 (KLF6; zf9, core promoter binding protein), a ubiquitously expressed zinc finger transcription factor, was found to be inactivated in prostate cancer as the result of frequent mutations and LOH, and might be a candidate tumor suppressor gene (Narla et al., 2001). Mutations and LOH of KLF6 gene have also been identified in other human cancers, including colorectal cancer (Reeves et al., 2004), malignant glioma (Jeng and Hsu, 2003), nasopharyngeal carcinoma (Chen et al., 2002) and primary hepatocellular carcinoma (Kremer-Tal et al., 2004a; Wang et al., 2004). It was recently reported that KLF6 is frequently down-regulated and induced apoptosis in not small cell lung cancer cells (Ito et al., 2004). Furthermore, KLF6 inhibited cell proliferation partly through transactivating p21WAF1, which encodes a cyclin-dependent kinase inhibitor of the cell cycle in a p53-independent pathway (Narla et al., 2001). So KLF6 may play a role in cancer pathogenesis. However, the relation of KLF6 with HCCs is little known. So in this study we detected the frequency of KLF6 mutation and the expression of KLF6 in 23 HCC tissues, and further observed its effects on cell growth in HCC cell lines.
MATERIALS AND METHODS
Tumor sample and cell lines
Twenty-three HCC tissues and their corresponding noncancerous tissues originated from patients hospitalizing at the First Affiliated Hospital, Zhejiang University. Completely informed consent was obtained from all the patients before tissue collection. There were 19 men and 4 women in this series, with a mean age of 58.3 years (range, 27~77). All the tumor tissue samples and non-tumor tissue samples were identified by pathological examination and frozen immediately after surgical resection, then stored at −80 °C until use. Hepatoblastoma cell line HepG2 (ATCC HB 8065) was obtained from the American Type Culture Collection.
DNA preparation
Genomic DNA from liver tissues was extracted using a “GENOME DNA isolation kit” (NNIQ-10 Sangon, China), according to the manufacturer’s instructions.
Sequencing analysis of the KLF6 gene
The whole coding region of KLF6 gene exon-2 was amplified by PCR with primers ex2-S (5′-caatcacgtgccttctctgg-3′) and primers ex2-AS (5′-gagaaagtgaggatttgtctg-3′). The PCR condition was 94 °C (30 s), 56 °C (40 s) and 72 °C (30 s) for 35 cycles, and a final extension of 72 °C (5 min) after the initial denaturation step (94 °C for 5 min). PCR products were directly sequenced after purification using QIAquick PCR purification Kit (Qiagen, Germany). Direct DNA sequencing was performed on an ABI Prism 3730 Automated Sequencer. All mutations were identified by sequencing in both directions and further confirmed by repeat experiments using PyrBest DNA polymerase (Takara, Japan).
RNA preparation and RT-PCR
Total RNA was isolated from 23 paired HCCs and corresponding non-tumor liver tissue using trizol (Invitrogen, USA) according to the manufacture’s protocol. After reverse transcription, the cDNA was used as a template for PCR amplification. The primer sequences were as follows: sense (S) (5′ cgagccctgctatgtttc 3′) and antisense (AS) (5′ catcgccatttcccttgt 3′). The product size was 437 bp. Human β-actin was amplified as an internal control using the forward primer (5′ cgctgcgctggtcgtcgaca 3′) and the reverse primer (5′ gtcacgcacgatttcccgct 3′) under the same conditions. Each PCR product was loaded directly onto 1.5% agarose gels, stained with ethidium and visualized under UV illumination.
Western blot analyses
Tissues or cells were treated with lysis buffer containing 50 mmol/L HEPES (pH 7.0), 250 mmol/L NaCl, 0.1% Nonidet P-40, 5 mmol/L ethylenediaminetetraacetate (EDTA), 1 mmol/L phenylmethylsulfonyl fuoride (PMSF), 1 mmol/L dithiothreitol (DTT), and protease inhibitor cock tail (EMD Biosciences, Inc., USA), and protein extraction was performed. Total protein (30 μg) was separated on 12% SDS-polyacrylamide gel electrophoresis gel, and transferred to PVDF membrane. The membranes were blotted with 5% milk, washed and then probed with rat polyclonal antibodies against KLF6 (Santa Cruz, CA, at 1:250 dilution), or p21WAF1 (Santa Cruz, CA, at 1:250 dilution) and β-actin (Cell Signaling, USA, at 1:1000 dilution). After washing, the membrane was then incubated with the appropriate secondary antibody and detected by the enhanced chemiluminescence ECL kit procedure (Santa Cruz Biotechnology, Santa Cruz, CA), and band intensity was measured by densitometry using a Kodak digital science image station 440CF. The immunoblots were quantitated by densitometry and protein levels were normalized with respect to β-actin level.
Plasmid constructions
The open reading frame of KLF6 gene was generated by RT-PCR using cDNA synthesized from normal liver tissues and the forward primer was 5′ agcgaattcgacatggacgtgctccccatg 3′ (italicized nucleotides indicate EcoRI site) and the reverse primer was 5′ agcggatccgaggtgcctcttcatgtg 3′ (italicized nucleotides indicate artifical BamHI site). After being double-digested with EcoRI and BamHI, the PCR product was cloned into the EcoRI/BamHI restriction sites of pcDNA3.1(-)A (Invitrogen, USA). The entire insert was confirmed by sequencing in both directions, and the construction was named pcDNA3.1(-)A/wtKLF6. The mutation confirmed in the coding region of KLF6 gene exon-2 in this study was introduced into the pcDNA3.1(-)A/wtKLF6 by PCR directed mutagenesis. We named the construction pcDNA3.1(-)A/mKLF6. The construction was sequenced to confirm mutation of the appropriate nucleotide.
Cell culture, transfection, and establishment of KLF6-transfected HepG2 cells
HepG2 cells were maintained in Dulbecco’s MEM (DMEM) with 10% fetal bovine serum at 37 °C in a humid atmosphere of 5% CO2. Cells in 6-well plates were transfected with plasmid pcDNA3.1(-)A/wtKLF6, plasmid pcDNA3.1(-)A/mKLF6 or empty plasmid pcDNA3.1(-)A using Lipofectamine 2000 reagent (Invitrogen, USA) respectively, according to the manufacturer’s instructions. At 48 h after transfection, the cells were cultured in DMEM medium containing 1000 μg/ml G418 (Gibco-BRL). After 3 weeks in selective medium, individual G418-resistant colonies were isolated and amplified.
Growth curves
The parent cells not transfected with vector, mock clones transfected with plasmid pcDNA3.1(-)A alone, wtKLF6-overexpressing clones and mKLF6-overexpressing clones were planted on 24-well plates at a density of 1×104 well−1. At 1, 2, 3, 4 and 5 d after planting, cells were removed from the culture wells by short exposure to trypsin and EDTA. The total cells in each well were counted by trypan blue staining under microscope and growth curves were plotted.
Cell cycle analysis
Exponentially growing control cells, wtKLF6-overexpressing clones and mKLF6-overexpressing clones were trypsinized and collected, respectively. Cell pellets were fixed in 70% ethanol and stored at −20 °C overnight. Then, fixed cells were washed with PBS and resuspended in 50 mg/ml propidium iodide containing 125 U/ml RNase A. After 30 min incubation, DNA contents were analyzed by flow cytometry using the Cell-FIT software (Becton-Dickinson Cell-cycle analysis). The cells percentage in different phases of cell cycle was determined with the ModFit 2.0 computer program (Verity Software).
Statistical analysis
To determine statistical significance, data were analyzed by Student’s t test or χ 2 test (SPSS 10.0 for Windows, 1999, SPSS Inc., USA). P<0.05 was considered statistically significant.
RESULTS
Mutation analysis of the KLF6 gene
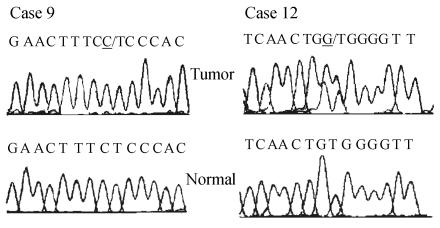
To detect mutations of the KLF6 gene in HCC, direct sequencing of exon-2 in KLF6 gene in 23 cases of HCC tissues and corresponding nontumor tissues was performed, because the majority of KLF6 mutations previously found in tumors, including HCC, were localized to this exon. We found that 2 of 23 (8.7%) HCCs harbored KLF6 mutations, whereas none of the nontumorous tissue had the KLF6 mutation. One (case 12) of the two mutations was a missense variant caused by single-nucleotide substitutions (TGG-GGG), which result in single amino acid substitution of tryptophan (W) by glycine (G) at codon 162 (W162G). The other one (case 9) was silent mutation (Fig.1).
Fig. 1.
Sequence analysis reveals KLF6 mutations in HCCs. The case number, nucleotide substitution and sequence chromatogram are shown for paired and tumor DNA for each mutation described. The mutated alleles are underlined
Expression of the KLF6 mRNA
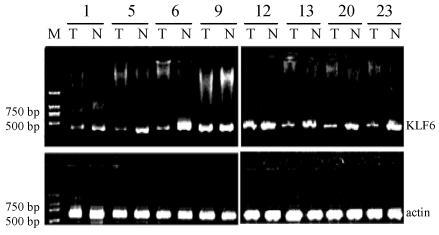
The expression of KLF6 mRNA in HCC was examined by RT-PCR. The amplified fragment of 437 bp DNA was detected in all of the 23 primary HCC tissues and corresponding non-cancerous tissues. The PCR product was identified to be the corresponding fragment of KLF6 cDNA by direct sequencing. The level of each KLF6 mRNA was measured with the Digital Imaging System from Alpla Innotech Corporation (UK) and normalized by β-actin cDNA level. In 8 of 23 (34.7%) cases, the KLF6 cDNA level of HCC was lower than their normal counterpart, and in 7 of the 8 cases, the T/N ratios (the relative ratio of normalized KLF6 cDNA of a tumor to that of the corresponding nontumorous tissue from the same patient) were less than 0.5 (Fig.2).
Fig. 2.
The representative results of RT-PCR analysis of KLF6 cDNA in human HCC tissues (T) and corresponding nontumorous tissues (N). RT-PCR-amplified 437 bp fragments was found in any speciman, including tumor and nontumorous tissues. The amplification of β-actin (607 bp) was shown as a positive control for cDNA synthesis
Expression of the KLF6 protein
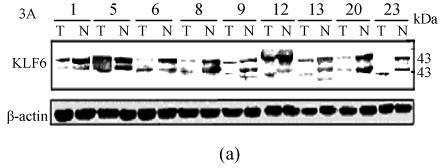
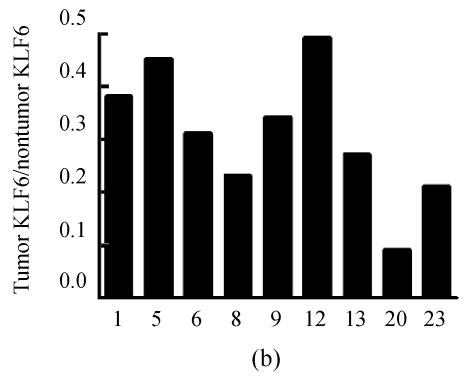
To determine KLF6 protein expression in HCC, we examined the KLF6 protein in 23 HCC tissues and corresponding noncancerous tissues using Western blot assay. Two bands within a size range of 35~43 kD were detected, which is likely due to various phosphorylation states of KLF6 as described by others (Slavin et al., 2004). We found that KLF6 protein levels in HCCs varied dramatically compared to corresponding noncancerous tissues (Fig.3), but with the exception of sample 8, KLF6 mRNA levels correlated with KLF6 protein levels. In 9 of 23 (39.1%) HCCs, KLF6 protein expression levels were lower than their normal countparts and decreased by more than 50%. Interestingly, we found a pattern of KLF6 isoform with the lower molecular weight was displayed in 5 of 23 (21.7%) HCC but not in noncancerous tissues.
Fig. 3.
The representative results of Western blot analysis of KLF6 protein in human HCC tissues (T) and corresponding nontumorous tissues (N). (a) KLF6 protein was shown as two bands within a size range of 35~43 kD. Case 23 had only a band for KLF6 protein (upper panel). β-actin protein was detected as a control for sample loading (lower panel); (b) Densitometric measurements from Western blot were used to calculate KLF6 protein levels (KLF6 level/β-actin level) in tumor tissue relative to KLF6 protein levels (KLF6 level/β-actin level) in corresponding nontumorous tissue
Overexpression of KLF6 in human hepatoma cells
KLF6 protein was detected by Western blot assay using established stable wKLF6-overexpressing clones and mKLF6-overexpressing clones. The results showed that KLF6 protein levels increased approximately 2.2- and 2.5-fold, respectively, compared to the mock clones which had been transfected with an empty plasmid (Fig.4).
Fig. 4.
Western blot analysis of KLF6 expression in KLF6-overexpressing HepG2 cells
Lane 1: Mock; Lane 2: wtKLF6 clones; Lane 3: mKLF6 clones
Effects of KLF6 on cellular growth and cell-cycle distribution
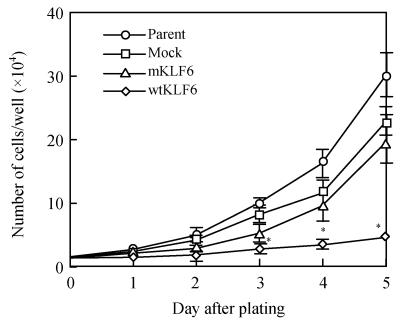
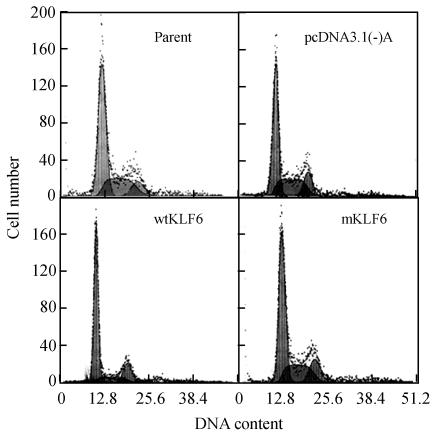
Analysis of cell number revealed strong inhibition of cell growth in wtKLF6-overexpressing clones but no remarkable inhibition of cell growth in mKLF6-overexpression clones, compared with the mock clones (Fig.5). To determine the mechanisms of the reduced rate of cellular proliferation, we then conducted cell cycle analysis of control cells and KLF6-transfectants using flow cytometry. As shown in Fig.6 and Table 1, wtKLF6 overexpression increased the percentage of the total cell population in the G0-G1 phase, decreased the percentage of cells in the S phase, but, had no consistent effect on the percentage of cells in the G2-M phase, compared to the controls (P<0.01). Thus, wtKLF6-overexpressing cells were arrested at the G0-G1 phase. But percentage of G0-G1 phase cells or S phase cells in mKLF6-overexpressing HepG2 cell lines did not significantly differ from the control (P>0.05).
Fig. 5.
Effect of KLF6 overexpression on cell growth. The total cell number was counted under microscopy at the times indicated. Data represent means±SD of three experiments performed in triplicate
* P<0.01; Parent: Cells not transfected with vector; Mock: Cells tranfected with vector alone
Fig. 6.
Cell cycle profile of HepG2 cells stably transfected with KLF6 gene. Cell cycle distribution of HepG2 cells transfected with wtKLF6 or mKLF6 gene was analyzed by flow cytometry
Table 1.
Phase of the cell cycle analyzed by flow cytometry in KLF6-overexpressing HepG2 cells
| Groups | G0-G1 (%) | S (%) | G2-M (%) |
| Parent | 55.7±6.0 | 33.6±5.6 | 10.7±1.5 |
| Mock clones | 57.6±4.1 | 30.9±3.7 | 11.5±2.3 |
| wtKLF6 clones | 59.6±4.6* | 29.4±4.3 | 11.0±1.9 |
| mKLF6 clones | 71.9±5.2** | 13.8±4.7 | 14.3±3.6 |
Note: Values in the table show percentage of the total population at the indicated phase of the cell cycle; data represent the means±SD of three experiments performed in triplicate. Parent: cells not transfected with vector; Mock clones: cells transfected with vector alone
P>0.05
P<0.01
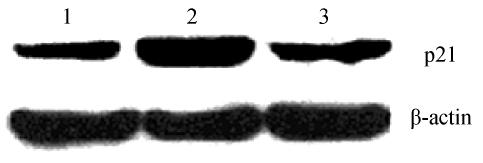
KLF6 induce expression of p21WAF1
The levels of p21WAF1 protein were detected by Western blot assay in established stable wtKLF6-overexpressing clones and mKLF6-overexpressing clones. The results showed that wtKLF6 markedly increased the levels of p21WAF1 protein compared to control cells, whereas mKLF6 (W162G) failed to do it (Fig.7).
Fig. 7.
Western blot analysis of p21WAF1 in KLF6-overexpressing HepG2 cells
Lane 1: Mock; Lane 2: wtKLF6 clones; Lane 3: mKLF6 clones. wtKLF6 transfection increased p21 expression. In contrast, mKLF6 transfection failed
DISCUSSION
The present study demonstrated that missense mutation of KLF6 gene and decreased KLF6 gene expression was found in HCC tissues, and wild-type KLF6 (wtKLF6) not cancer-derived KLF6 mutant (mKLF6) inhibited cellular proliferation and prolonged G1-S transition by inducing the expression of p21WAF1. These data suggest that KLF6 might be involved in pathogenesis of HCC.
Mutation of KLF6 gene was found in 2 of 23 HCC tissues with sequencing analysis. The frequence mutation of KLF6 in HCCs was consistent with a previous report by Kremer-Tal et al.(2004a) which demonstrated that mutations of KLF6 was detected in 6 of 41 HCCs. Another report also showed mutations of KLF6 in 6 of 27 HCCs (Wang et al., 2004). In prostate adenocarcinoma cells, wild-type KLF6 was shown to suppress tumor growth, possibly due to up-regulation of p21WAF1 in a p53-independent manner, whereas tumor-derived KLF6 mutants did not (Narla et al., 2001). In this study, we first identified W162G mutant of KLF6 in HCC tissues and found that it also inactivated the KLF6 function, hence, showing that mutations of KLF6 may induce uncontrolled cell proliferation and cancer formation. On the other hand, a recent study failed to find mutations of KLF6 in 71 HCCs (Boyault et al., 2005). So the highly variable frequency of KLF6 mutations may be caused by differences in methodology and sample sets. For example, some reports showed that KLF6 mutations are absent in prostate cancers (Mühlbauer et al., 2003) and human astrocytic gliomas (Koivisto et al., 2004; Köhler et al., 2004; Montanini et al., 2004), while other authors reported high frequencies of KLF6 mutations in prostate cancer (55%) (Narla et al., 2001) and astrocytic gliomas (9%) (Jeng and Hsu, 2003). Therefore, extensive investigation in both HCC and other human cancers is needed to determine the frequency of KLF6 mutation and its role in hepatocarcinogenesis.
Downregulation or loss of KLF6 expression in other human cancers including not small cell lung cancer cells (Ito et al., 2004) and glioblastoma multiforme (GBM) (Kimmelman et al., 2004) has been reported. Our results also demonstrated decreased expression of KLF6 mRNA was found in 8 of 23 (34.7%) HCC tissues and a low level of KLF6 protein was detected in 9 of 23 (39.1%) HCC tissues compared to corresponding nontumorous tissue. These results were supported by Kremer-Tal et al.(2004b) that KLF6 mRNA expression was reduced in the large majority of HCC tumors, and decreased more than 50% in almost 2/3 of tumors compared to normal liver.
In our study, we found shorter KLF6 band by Western blot in 5 of 23 tumour tissues, but did not find in corresponding nontumourous tissues. The variant form of KLF6 protein could be generated from a spliced KLF6 mRNA, or be due to a post-translational event such as truncation and/or enhanced proteolytic degradation in tumor samples. Nevertheless, we recently confirmed the short version of KLF6 protein generated from a splicing of KLF6 mRNAs in these tumour tissues (data not published). We speculated that generation of some alternative splice forms of KLF6 might be involved in carcinogenesis including HCCs and other cancers. The speculation was supported by two recent reports. Narla et al.(2005) reported alternative splicings of KLF6 were associated with increased prostate cancer risk and confirmed that the above splice variant proteins antagonize wtKLF6 function, leading to decreased p21 expression and increased cell growth, although the functional significance of the novel splicing variant expression remains to be evaluated. Furthermore, our results showed that all of those HCCs harboring mutations or spliced form of KLF6 had concomitantly decreased KLF6 expression at RNA and protein level. This suggests that gene mutation or alteration in posttranscriptional level could contribute to downregulation of KLF6 expression aside from the transcriptional silencing by promoter hypermethylation reported by Yamashita et al.(2002).
Overexpression of the wtKLF6 gene, not the KLF6 mutant derived from tumor tissues, significantly inhibited cell proliferation of HepG2, and led to G1-S arrest in these cells accompanied by increased expression of p21WAF1. The data accorded with previous reports on other cancers. Recently, KLF6 has been shown to induce apoptosis in NSCLC (non-small cell lung cancer) cells (Ito et al., 2004), and inhibit a number of key oncogenic signaling pathways including the cyclin-dependent kinase complex CDK-cyclinD1 and c-jun (Slavin et al., 2004; Benzeno et al., 2004). Furthermore, expression of KLF6 can revert to the tumorigenic phenotype in glioblastoma cell lines in culture and in vivo (Kimmelman et al., 2004). All these functions and features are consistent with KLF6’s role as a tumor suppressor.
In summary, in this study we found KLF6 was mutated and frequently down-expressed in HCCs, and exogenously induced wtKLF6 inhibited tumor cell growth. These results indicated KLF6 may be involved in the pathogenesis of HCC and may be an attractive candidate for gene therapy approaches. Of course, further studies are needed to elucidate the role of other mechanisms of KLF6 inactivation in HCCs.
References
- 1.Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Didhl JA, Germain D, Freedman SL. Cyclin dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64(11):3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- 2.Boyault S, Herault A, Balabaud C, Zucman-Rossi J. Absence of KLF6 gene mutation in 71 hepatocellular carcinomas. Hepatology. 2005;41(3):681–682. doi: 10.1002/hep.20588. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5(3):215–219. doi: 10.1016/S1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen HK, Liu XQ, Lin J, Chen TY, Feng QS, Zeng YX. Mutation analysis of KLF6 gene in human nasopharyngeal carcinomas. Ai Zheng. 2002;21(10):1047–1050. (in Chinese) [PubMed] [Google Scholar]
- 5.Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan JB, Lian ZR. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21(16):2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 6.Hui AM, Sakamoto M, Kanai Y, Ino Y, Gotoh M, Yokota J, Hirohashi SL. Inactivation of p16INK4 in hepatocellular carcinoma. Hepatology. 1996;24(3):575–579. doi: 10.1002/hep.510240319. [DOI] [PubMed] [Google Scholar]
- 7.Ito G, Uchiyama M, Kondo MS, Mori S, Usami N, Maeda O, Kawabe T, Hasegawa Y, Shimokata K, Sekido Y. Krüppel-like factor 6 (KLF6) is frequently downregulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64(11):3838–3843. doi: 10.1158/0008-5472.CAN-04-0185. [DOI] [PubMed] [Google Scholar]
- 8.Jeng YM, Hsu HC. KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer. 2003;105(5):625–629. doi: 10.1002/ijc.11123. [DOI] [PubMed] [Google Scholar]
- 9.Kimmelman AC, Qiao RF, Narla G, Banno A, Lau N, Bos PD, Rodriguez NN, Liang BC, Guha A, Friedman SC, et al. Suppression of glioblastoma tumorigenicity by the Krüppel-like transcription factor KLF6. Oncogene. 2004;23(29):5077–5083. doi: 10.1038/sj.onc.1207662. [DOI] [PubMed] [Google Scholar]
- 10.Köhler B, Wolter M, Blaschke B, Reifenberger G. Absence of mutations in the putative tumor suppressor gene KLF6 in glioblastomas and meningiomas. Int J Cancer. 2004;111(4):644–645. doi: 10.1002/ijc.20302. [DOI] [PubMed] [Google Scholar]
- 11.Koivisto PA, Zhang X, Sallinen SL, Sallinen S, Helin HJ, Dong JT, van Meir EG, Haapasalo H, Hyytinen ER. Absence of KLF6 gene mutations in human astrocytic tumors and cell lines. Int J Cancer. 2004;111(4):642–643. doi: 10.1002/ijc.20301. [DOI] [PubMed] [Google Scholar]
- 12.Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Usage P, Martignetti JA, et al. Frequent inactivation of the tumor suppressor Krüppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40(5):1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 13.Kremer-Tal S, Narla G, Banck M, Difeo AV. Significantly decreased expression of the tumor suppressor KLF6 in 85% of HCCs contributes to enhanced growth and reduced differentiation. Hepatology. 2004;40:520A. [Google Scholar]
- 14.Montanini L, Bissola L, Finocchiaro G. KLF6 is not the major target of chromosome 10p losses in glioblastomas. Int J Cancer. 2004;111(4):640–641. doi: 10.1002/ijc.20303. [DOI] [PubMed] [Google Scholar]
- 15.Mühlbauer KR, Grone HJ, Ernst TE, Grone E, Tschata R, Hergenhahn M, Hollstein M. Analysis of human prostate cancers and cell lines for mutations in the TP53 and KLF6 tumour suppressor genes. Br J Cancer. 2003;89(4):687–690. doi: 10.1038/sj.bjc.6601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294(5551):2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 17.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65(4):1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 18.Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kermer S, et al. Krüppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126(4):1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, Chatton B, Bocco JL. A new role for the Krüppel-like transcription factor KLF6 as an inhibitor of c-jun proto-oncoprotein function. Oncogene. 2004;23(50):8196–8205. doi: 10.1038/sj.onc.1208020. [DOI] [PubMed] [Google Scholar]
- 20.Staib F, Hussain SP, Hofseth LJ, Wang X, Harris CC. TP53 and liver carcinogenesis. Human Mutation. 2003;21(3):201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 21.Wang SP, Chen XP, Qiu FZ. A candidate tumor suppressor gene mutated in primary hepatocellular carcinoma: Krüppel-like factor 6. Zhonghua Wai Ke Za Zhi. 2004;42(20):1258–1261. (in Chinese) [PubMed] [Google Scholar]
- 22.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–495. doi: 10.1016/S1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]