Abstract
We investigated phylogeographic divergence among populations of Galápagos warbler finches. Their broad distribution, lack of phenotypic differentiation and low levels of genetic divergence make warbler finches an appropriate model to study speciation in allopatry. A positive relationship between genetic and geographical distances is expected for island taxa. Warbler finches actually showed a negative isolation by distance relationship, causing us to reject the hypothesis of distance‐limited dispersal. An alternative hypothesis, that dispersal is limited by habitat similarity, was supported. We found a positive correlation between genetic distances and differences in maximum elevation among islands, which is an indicator of ecological similarity. MtDNA sequence variation revealed monophyletic support for two distinct species. Certhidea olivacea have recently dispersed among larger central islands, while some Certhidea fusca have recently dispersed to small islands at opposite ends of the archipelago. We conclude that females have chosen to breed on islands with habitats similar to their natal environment. Habitat selection is implicated as an important component of speciation of warbler finches, which is the earliest known divergence of the adaptive radiation of Darwin's finches. These results suggest that small populations can harbour cryptic but biologically meaningful variation that may affect longer term evolutionary processes.
Keywords: dispersal, habitat selection, speciation, mtDNA, isolation by distance, phylogeography
1. Introduction
Islands that are closer together tend to have populations that are more similar to each other (Wallace 1880). Recent molecular genetic studies have provided evidence that this pervasive pattern in nature is caused by distance‐limited dispersal. Island populations of tortoises (Caccone et al. 2002), beetles (Finston & Peck 1995) and lizards (Wright 1983) in the Galápagos, and Drosophila (Desalle 1995), spiders (Hormiga et al. 2003) and crickets (Shaw 2002) in Hawaii tend to be genetically more closely related if they occur on neighbouring islands. The abundant evidence from nature has led to the assumption of distance-limited dispersal in models of island biogeography (MacArthur & Wilson 1967), geographical clines (Endler 1977; Kirkpatrick & Barton 1997), metapopulation dynamics (Hanski & Gilpin 1997) and speciation (Mayr 1942; Garcia-Ramos & Kirkpatrick 1997).
Evidence of distance-limited dispersal is commonly encountered in birds (Seutin et al. 1994; Clegg et al. 2002; Irwin et al. 2002). However, the greater vagility of birds compared with other organisms also implies an enhanced ability to sample different habitats during dispersal, which could cause deviations from distance-dependent dispersal. Birds are well known for selecting habitats after dispersing (Cody 1985). Their habitat preferences may be genetically based. Alternatively, habitat preferences may be acquired during development (Gruenberger & Liesler 1990; Teuschl et al. 1998), much like their songs and mate preferences (Grant & Grant 1996). Developmentally acquired preferences can reduce gene flow and accelerate divergence and speciation among populations, but evidence for this process in nature is limited (Price 1998; West-Eberhard 2003; Davis & Stamps 2004).
Interisland dispersal of Darwin's finches has traditionally been considered rare, allowing island populations to differentiate in allopatry (Lack 1947). A study of the founding of a population of large ground finches (Geospiza magnirostris) in 1982 suggested interisland movements may be more frequent (Grant et al. 2001). Founders of the now thriving Daphne Major population were a non random subset of potential immigrants from four other island populations, suggesting that birds sampled island habitats and chose to breed or continue moving. Such non random flow of genes among populations could lead to divergence and speciation over time (Epperson 2003). This can be investigated indirectly by quantifying population divergence in a very closely related species with older populations.
The Galápagos warbler finches (Certhidea) are an appropriate model for the study of population divergence and speciation in allopatry. They are the most widespread of all Darwin's finches, occurring on every major island. Warbler finch populations are phenotypically very similar and until recently were considered a single species (Lack 1947; Grant & Grant 2003). There is also a lack of evidence of premating isolation among populations based on song (Grant & Grant 2002). Thus, the discovery of large genetic differences among subsets of island populations of warbler finches was surprising (Freeland & Boag 1999a; Petren et al. 1999a) and suggested the presence of two allopatric species (Certhidea olivacea and Certhidea fusca).
Here, we present a comprehensive phylogeographic study of warbler finches using mtDNA (cytochrome b) sequence divergence among all 16 known populations. We use museum specimens to represent difficult to access and possibly extinct populations (Grant et al. 2005). We determine if there is support for two phylogenetically independent groups of warbler finches. We test the hypothesis that metapopulation structure is determined by distance-limited dispersal among islands, which predicts that genetic isolation will be correlated with geographical distance. Finally, we explore an alternative hypothesis, that genetic relatedness is determined by habitat-limited dispersal, and evaluate the evidence for recent dispersal.
2. Methods
(a) Sampling
We obtained tissue samples from 84 birds from all 16 known populations of warbler finches (table 1). Of these, 23 were museum specimens from the California Academy of Sciences, the American Museum of Natural History and the British Natural History Museum (Electronic Appendix Section 1). Museum specimens were sampled by cutting a small (approximately 3 ×2 mm) piece of tissue from the side of the largest toe pad using a sterile scalpel (Mundy & Woodruff 1997). Field-captured birds were sampled from different expeditions and different parts of islands where possible. A small drop of blood was taken by venipuncture and absorbed onto EDTA-soaked filter paper and dried (see Petren 1998; Petren et al. 1999b).
Table 1.
Numbers and sources of warbler finch specimens sequenced including partial haplotypes. (three approximately 300 bp regions; 864 bp total.)
| island | sourcea | date | n | 1st | 2nd | 3rd | all |
|---|---|---|---|---|---|---|---|
| Darwin | CAS | 1906 | 2 | 2 | 2 | 2 | 2 |
| Española | field | 1997 | 4 | 4 | 3 | 1 | 1 |
| Fernandina | AMNH | 1894 | 3 | 3 | 3 | 1 | 1 |
| field | 1997, 1999 | 4 | 4 | 3 | 1 | 1 | |
| Floreana | CAS | 1906 | 1 | 1 | 1 | 1 | 1 |
| Genovesa | BNHM | 1897 | 3 | 2 | 3 | 2 | 1 |
| fieldb | 1997 | 3 | 3 | 3 | 3 | 3 | |
| Hermanos | AMNH | 1902 | 4 | 4 | 4 | 3 | 3 |
| Isabela | fieldc | 1999 | 10 | 10 | 9 | 1 | 1 |
| Marchena | CAS | 1906 | 2 | 2 | 2 | 1 | 1 |
| field | 1988 | 2 | 2 | 2 | 1 | 1 | |
| Pinta | field | 1997 | 9 | 9 | 8 | 1 | 1 |
| Pinzón | CAS | 1906 | 4 | 3 | 2 | 3 | 1 |
| field | 2004 | 3 | 3 | 3 | 0 | 0 | |
| Rábida | field | 2002 | 1 | 1 | 1 | 1 | 1 |
| San Cristóbal | field | 1997, 1999 | 7 | 7 | 7 | 1 | 1 |
| Santa Cruz | CAS | 1906 | 2 | 1 | 1 | 2 | 1 |
| field | 1996/7 | 4 | 3 | 3 | 2 | 1 | |
| Santa Fé | field | 1999, 2004 | 6 | 6 | 6 | 1 | 1 |
| Santiago | field | 1996 | 8 | 8 | 7 | 1 | 1 |
| Wolf | CAS | 1898 | 1 | 1 | 1 | 0 | 0 |
| BNHM | 1897 | 1 | 1 | 1 | 1 | 1 | |
| totals | 84 | 80 | 74 | 30 | 25 |
Sources: AMNH, American Museum of Natural History; CAS, California Academy of Sciences; BNHM, British Natural History Museum; field, field captured. Museum accession numbers are given in Electronic Appendix 2.
Includes two sequences from Sato et al. (1999).
Includes individuals from Isabela volcanos Alcedo, Wolf and Darwin.
(b) Laboratory methods
DNA was extracted from tissue (Qiagen tissue kit) by digestion for 48 h at 55 °C, with twice the recommended amount of proteinase K (140 μg). Previously published sequences (Sato et al. 1999) were used to develop primers for an 864‐bp region of the cytochrome b gene, and internal primers for three shorter segments which were used for polymerase chain reaction (PCR) of lower quality DNA from museum specimens (Electronic Appendix Section 2). PCR was carried out for 33 cycles (30 s at 94 °C; 30 s at 57 °C; 90 s at 72 °C) using pre mixed components (ABI Amplitaq Gold mix). An early set of reactions showing evidence of contamination in a blank control was discarded. No further evidence of contamination was detected after stricter procedures were implemented, including the use of a spatially isolated facility dedicated to low copy number DNA extraction (Leonard et al. 2000). To facilitate the detection of contamination, individual birds from the same population were processed in different batches, and populations were intermingled within batches.
At least one complete haplotype was obtained per island, and additional complete haplotypes were obtained for any individuals whose partial haplotype revealed differences that could affect interpretation (greater then 0.5% divergence). Unique haplotypes based on bidirectional sequencing of 22 birds were subjected to the full analysis and posted in GenBank (accession numbers AY672044–AY672065).
(c) Genetic distance comparisons
We used Mantel correlations (Mantel 1967) to compare average genetic distances among populations with the physical features of islands on which they occur. Statistical significance was assessed with permutations (n=1000; Legendre & Vaudor 1991). Interisland distances (shore-to-shore) were from Hamilton & Rubinoff (1967). Maximum elevation of islands was used as an approximate indicator of habitat variation (figure 1). High islands create more local rainfall, thus several distinct vegetation zones are evident as one moves up an elevational gradient. Low islands are much more xeric and less heterogeneous, with few of the plant species characteristic of upland habitats (Wiggins & Porter 1971). We included island area in partial Mantel tests (Castellano & Balletto 2002) because area is expected to affect population size and levels of genetic variation, and because it is generally correlated with maximum elevation of islands.
Figure 1.
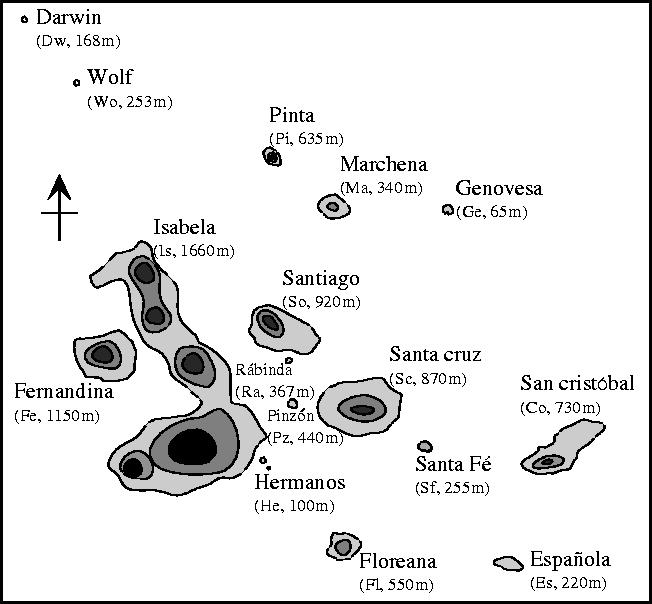
A map of the Galápagos with 300 and 600 m elevation contours. Abbreviated island names and maximum elevation in metres are given in parentheses.
(d) Nested clade analysis
We used nested clade phylogenetic analysis to test for differences in the geographical distribution of clades and haplotypes (Templeton 2004). This method can reveal biological processes that may have affected subsets of populations, including isolation by distance, range expansion, colonization, fragmentation and panmixia. Pairs of real and inferred haplotypes were grouped into clades based on an unrooted maximum parsimony network, and the process was repeated to create different levels of nesting clades. Clades containing variation in haplotypes and geographical distribution (n=12) were subjected to a contingency test permuted 10 000 times (GeoDis v.2.0; Posada et al. 2000). This method has performed well in recent evaluations (Templeton 2004), and the statistical tests are robust, but biological inferences made using the interpretation key must be made with caution (Knowles & Maddison 2002).
(e) Phylogeographic analysis
To assess support for monophyletic groupings, we used maximum parsimony and maximum likelihood (Swofford 1998), the latter with a ts/tv ratio of 2.0. Confidence in nodes was assessed using 1000 bootstrap replicates. We also employed Bayesian methods of inference with data partitions set to codon position, and a general time reversible model (Ronquist & Huelsenbeck 2003). Likelihood scores stabilized between generation 12 000 and 15 000 in successive runs using random starting trees, thus we discarded the first 20 000 generations. Over the following 30 000 generations, we saved one tree every 10 generations and estimated support for nodes from these 3000 trees. We used Tiaris obscura from Panama as the outgroup (Sato et al. 2001). Owing to uncertainty about which species are the most appropriate outgroup (Burns et al. 2002), we also determined if using other mainland species (T. olivacea, T. bicolor) or other Darwin's finches (Geospiza spp.) as the outgroup altered our results.
3. Results
(a) Patterns of genetic variation
The maximum amount of within-island divergence among haplotypes was 0.7%, or six changes out of 864 bp. Different haplotypes were found on the large islands of Santa Cruz and Fernandina and were included in the final analysis. For all other islands, haplotype divergence was less than 0.5%. Some complete haplotypes collected a century apart from Genovesa and Marchena were identical. The average genetic divergence among all complete haplotypes was 1.8% (0–3.8%).
(b) The hypothesis of distance‐limited dispersal
If dispersal is limited by distance, genetic and geographical distances should be positively correlated, producing a pattern of isolation-by-distance. The isolation by distance correlation is actually negative, contrary to the predicted direction. Therefore, the hypothesis of distance limited dispersal can be rejected. Populations generally fall into two major groups corresponding to C. olivacea and C. fusca, initially identified by microsatellites (Petren et al. 1999a). Genetic distances between these groups (; range, 2.3–3.8%) are nearly non overlapping with distances within groups regardless of the geographical distances among islands (figure 2). When C. olivacea and C. fusca are considered separately, the signs of the correlation coefficients remain negative but are not statistically significant (table 2; figure 2).
Figure 2.
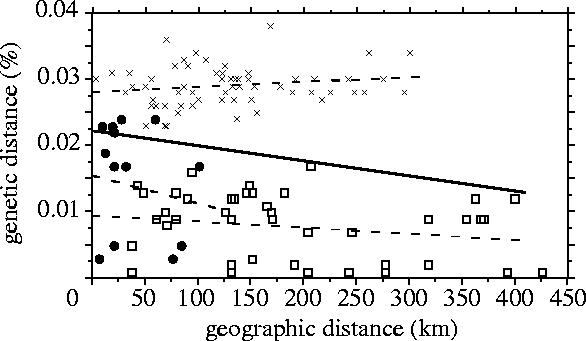
Uncorrected pairwise genetic distances of mitochondrial haplotypes plotted against interisland geographical distances. Least-squares regression lines are shown as a solid line for the entire dataset, and as dashed lines for Certhidea olivacea (circles) and Certhidea fusca (squares). Among-group distances (crosses) are greater and nearly non overlapping with within-group distances.
Table 2.
Mantel correlations (rM) of average genetic distance among populations and island characteristics. (Significant and marginally significant values are in boldface and negative correlations are underlined.)
| geographic distance | elevational (ecological) distance | |||||
|---|---|---|---|---|---|---|
| alone | partial, with area | |||||
| rM | p | rM | p | rM | p | |
| Certhidea all | −0.194 | 0.022 | 0.339 | 0.015 | 0.182 | 0.054 |
| Certhidea fusca only | −0.233 | 0.080 | 0.509 | 0.010 | 0.416 | 0.015 |
| Certhidea olivacea only | −0.135 | 0.412 | −0.489 | 0.010 | −0.164 | 0.416 |
(c) The hypothesis of habitat-mediated dispersal
If dispersers prefer to settle on islands with habitats similar to their natal environment, there should be a positive correlation of genetic distances between populations and habitat similarity. The habitat-mediated dispersal hypothesis is supported, as there is a significant positive correlation of genetic and elevational differences overall. The result remains the same when populations of C. fusca are considered separately (table 2). Controlling for island area in partial Mantel tests does not alter these results. The hypothesis is not supported statistically when C. olivacea populations are considered alone, perhaps because there are so few of them.
(d) Phylogeographic patterns of variation
We use nested clade analysis to distinguish among alternatives to the distance-dependent dispersal hypothesis. Four of the 12 clades tested (figure 3) show significant heterogeneity in the geographical distribution of subclades (table 3). For the entire cladogram, results indicate long distance dispersal in the main C. fusca clade (4.1), while the main C. olivacea clade (4.3) has a relatively limited distribution. Within the main C. olivacea clade (4.3), there is evidence of recent long-distance colonization, as clade 3.5 has a relatively broad distribution (Fernandina, Isabela, Pinzón, Rábida) compared with clade 3.6. We conclude that long-distance dispersal has occurred in both C. fusca and C. olivacea, and this was probably the underlying cause of our rejection of the distance limitation hypothesis. Non random patterns within two other C. fusca clades (4.1, 2.1) could not be attributed to any biological process other than panmixia.
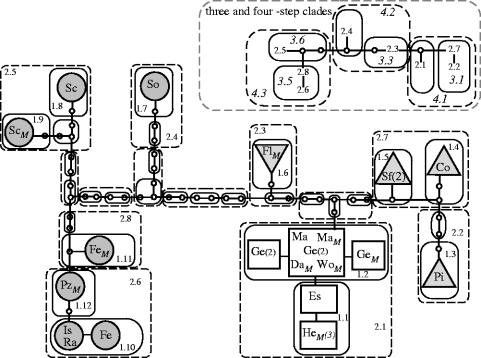
Figure 3.
A network of Certhidea mtDNA haplotypes showing minimum numbers of changes. Lines connecting real or inferred (open circles) haplotypes represent one base change. Abbreviations within haplotype symbols indicate the island where each haplotype was found. Unique haplotypes are indicated by circles for putative Certhidea olivacea, and triangles or squares for Certhidea fusca. Numbers in parentheses indicate more than one identical sequence. An ‘M’ indicates the haplotype is from museum specimens.
Table 3.
Nested clade phylogenetic analysis (Templeton 2004) results for four clades that showed significanta geographical structuring of the 12 clades tested.
| clade (subclade) | Dc(km) | Dn(km) | key codes and inference |
|---|---|---|---|
| total clade (χ2=54.0***) | [1, 2, 3, 5, 6, 7, 8] | ||
| (4.1, tip) | 138.2>* | 126.0>** | some long distance dispersal (clade 4.1) |
| (4.2, int.) | 60.0 | 78.7 | some restricted gene flow (clade 4.3) |
| (4.3, tip) | 48.8<** | 86.4<* | |
| I-T | −42.5 | −31.5 | |
| clade 4.3 (χ2=8.0) | [1, 2, 11, 12, 13] | ||
| (3.5, tip) | 82.3 | 66.5>* | range expansion, long distance colonization |
| (3.6, int.) | 0.0<* | 38.7 | |
| I-T | −82.3<* | −27.8 | |
| clade 4.1 (χ2=15.0**) | [0] | ||
| (3.1, tip) | 136.7 | 137.8 | panmixia |
| (3.2, int.) | 138.4 | 138.3 | |
| I-T | 1.7 | 0.5 | |
| clade 2.1 (χ2=12.0**) | [0] | ||
| (1.1, tip) | 92.0 | 141.1 | panmixia |
| (1.2, int.) | 137.0 | 136.8 | |
| I-T | 35.0 | −4.2 |
The direction of significant geographical differences relative to other clades is indicated by ‘<’ or ‘>’; p values are indicated by ‘*’ (<0.05), ‘**’ (<0.01) or ‘***’ (<0.001).
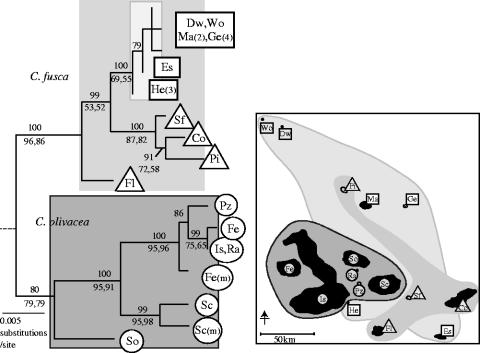
Phylogenetic reconstructions support the existence of two phylogenetically distinct species (figure 4, left). The geographical locations of clades reveal that neighbouring islands do not always harbour genetically similar populations. Instead, two striking patterns of interspersion of clades are revealed (figure 4, right). First, C. olivacea has a narrower range than C. fusca, but ranges overlap considerably, with C. fusca partly circumscribing C. olivacea. Second, a small clade of Certhidea haplotypes spans the entire length of the archipelago (figure 4, squares), and is distributed irregularly around the four remaining C. fusca haplotypes (triangles). This pattern is inferred to have become established recently, owing to the small numbers of base changes (less than or equal to 3) among all members of the widely dispersed clade. The geographical interspersion of clades contributes to the ambiguous interpretations of the nested clade analysis because it assumes that geographical overlap indicates panmixia.
Figure 4.
Phylogram of all unique mtDNA haplotypes (left), and the geographical locations of haplotypes (right). The maximum likelihood tree is shown, with horizontal branch lengths proportional to inferred evolutionary time. Certhidea olivacea (below, circles) and Certhidea fusca (above, triangles, squares) form phylogenetically distinct groups as indicated by strong support for reciprocal monophyly. A subset of C. fusca haplotypes (squares) are maximally 3 bp different, but span the entire archipelago (indicated by lighter shading). The topology shown differed from all other equally parsimonious trees (n=2) or trees with equivalent likelihood (n=1) only in the arrangement of branches within the recently diverged subclade of C. fusca (squares). Numbers above horizontal lines indicate support for each node based on Bayesian analysis (3000 trees). Numbers below lines are percentage bootstrap support for maximum parsimony (left, n=1000) and maximum likelihood (right, n=1000). Tiaris obscura was used as the outgroup, but substitution of different outgroup taxa did not affect results.
4. Discussion
Closely related populations of warbler finches that are ecologically, behaviourally and morphologically similar do not show isolation by distance patterns that reflect distance-dependent dispersal. Instead, genetic relatedness among populations is more closely associated with habitat similarity. For example, populations on islands at opposite ends of the archipelago with similar habitat, such as Darwin and Española, are genetically similar. Populations on neighbouring islands that differ in habitat, such as Isabela and Hermanos, belong to two distinct clades. The pattern is reflected in field-captured specimens so our results are not attributable to artefacts of degraded DNA (Hofreiter et al. 2001). We can also rule out the possibility of being misled by our single-locus mtDNA marker (Shaw 2002) or by insufficient sampling, because the division of C. fusca and C. olivacea populations is completely concordant with results of a nuclear microsatellite analysis (Petren et al. 1999a) of more loci (n=16) and more individuals per population (89 birds from six populations).
In our view, the evidence of recent long range dispersal implies that habitat choice is the most probable explanation for the positive association of genetic and habitat similarities among populations of warbler finches. An alternative explanation is that a recent wave of C. fusca dispersal was random, but immigrant haplotypes are now only detectable on small islands. This alternative assumes that population size is correlated with island size, but the correlation is far from perfect. For example, warbler finches are much less numerous on the medium-sized islands of Pinzón, Rábida and Santa Fé, where the widely dispersed haplotype is absent, than on Genovesa or Española where the haplotype is present (unpublished observation). Furthermore, the evidence shows that C. olivacea has recently dispersed among both large and small populations. This suggests that population size does not affect the detection of recent genetic movement, and a single wave of dispersal limited to C. fusca is unlikely. Reproductive surges that could lead to waves of dispersal are known in Darwin's finches, but have repeatedly been linked to recurring El Niño events (Grant et al. 2000), which affect the entire archipelago, not subsets of islands. Although it is difficult to completely rule out more complex alternatives, habitat choice is the most parsimonious explanation for the observed patterns. This conclusion is supported by results of another study (Grant et al. 2001) that suggested birds sample habitats, and choose not to breed on certain islands.
Other processes may have contributed to the interisland population structure of Certhidea. We briefly discuss two that are pertinent to recently diverged, vagile vertebrates. First, competition between C. olivacea and C. fusca may affect their current distributions. However, competition with C. olivacea does not prevent low island C. fusca from inhabiting the drier lowlands of larger islands, because C. olivacea is confined to moist upland forest on every island where they occur (personal observation). The absence of warbler finches from the lowlands of large islands is an unsolved problem that requires further ecological research, but it also suggests that there are indeed biological differences between these species. Second, mate choice can affect dispersal and influence speciation (Price 1998). Mate choice is related to song and morphology in Darwin's finches (Grant 1999) and it may have played a role in the settlement choices of immigrant G. magnirostris (Grant et al. 2001). However, the lack of evidence of premating barriers among Certhidea (Grant & Grant 2002), along with the absence of a connection with habitat, suggests mate choice is not a general explanation for Certhidea dispersal patterns.
The two warbler finches are the most genetically divergent phylogenetic species of the entire, and relatively young, adaptive radiation of Darwin's finches (Petren et al. 1999a; Freeland & Boag 1999a). Differences in cytochrome b sequence among C. olivacea and C. fusca is considerably less than the divergence between most sister species of North American passerines (; Klicka & Zink 1999). The divergence in habitat use of warbler finches contrasts in many ways with the rest of the adaptive radiation. Phenotypic differentiation among Certhidea populations is much more subtle than differences among populations of other Darwin's finch species commonly used to illustrate the early stages of speciation in allopatry (Lack 1947; Grant 1999; Grant & Grant 2003). Furthermore, other species of Darwin's finches share the same habitats, are not phylogenetically distinct (Freeland & Boag 1999b), but differ greatly in morphology, ecology and behaviour (Grant 1999), all in contrast to the warbler finches. Thus, although habitat selection may have helped to initiate the speciation process in this adaptive radiation, other processes must have shaped the rest of the radiation.
The divergence of Certhidea can be viewed as a case of cryptic speciation. Molecular methods have previously revealed cryptic speciation in birds, but it has typically been associated with geographical variation (e.g. Avise & Nelson 1989; Omland et al. 2000). Habitat choice has been implicated in speciation of other organisms, for instance, in insects that specialize on different hosts (Via 1999). Habitat use preceded divergence of other traits in Phylloscopus warblers (Richman & Price 1992). However, to our knowledge, selective dispersal among interspersed habitats has not previously been linked to speciation in birds. The short time-scale of divergence among warbler finches suggests that habitat imprinting may play a role in habitat choice and speciation (Davis & Stamps 2004), but with these data we cannot distinguish between acquired and innate habitat preferences.
Most spatial models of population structure assume that immigrants come from larger and closer source populations (Mayr 1942; Garcia-Ramos & Kirkpatrick 1997; Hanski & Gilpin 1997). Centrally located C. olivacea populations conform to this expectation, but peripheral C. fusca populations do not. The sources of genetic immigrants to low islands were not large or close islands but other low islands located further away. The role of habitat remained cryptic until this comprehensive analysis of 16 discrete island populations. This study provides an example of the need to carefully assess the biodiversity value of small populations (Blondel et al. 1999; Templeton et al. 2001). Small, peripheral populations that do not appear to be unique may nevertheless harbour biologically significant variation that may play an important role in evolution.
Acknowledgments
We thank the California Academy of Sciences, the American Museum of Natural History, and the British Natural History Museum for loaning valuable specimens. We thank the Galápagos National Parks for research permission; the Charles Darwin Research Station for logistical support; L. F. Keller and E. K. Monson for field assistance; D. J. Long, E. Bermingham, H. Vargas, M. Wikelski and M. Hau for lending samples; two anonymous reviewers for comments; and the National Science Foundation and the University of Cincinnati Research Council for financial assistance. We especially acknowledge the support and friendship of the late L. F. Baptista and T. C. Kane.
Footnotes
As this paper exceeds the maximum length normally permitted, the authors have agreed to contribute to production costs.
Supplementary Material
References
- Avise J.C, Nelson W.S. Molecular genetic relationships of the extinct dusky seaside sparrow. Science. 1989;243:646–648. doi: 10.1126/science.243.4891.646. [DOI] [PubMed] [Google Scholar]
- Blondel J, Dias P.C, Perret P, Maistre M, Lambrechts M.M. Selection-based biodiversity at a small spatial scale in a low-dispersing insular bird. Science. 1999;285:1399–1402. doi: 10.1126/science.285.5432.1399. [DOI] [PubMed] [Google Scholar]
- Burns K.J, Hackett S.J, Klein N.K. Phylogenetic relationships and morphological diversity in Darwin's finches and their relatives. Evolution. 2002;56:1240–1252. doi: 10.1111/j.0014-3820.2002.tb01435.x. [DOI] [PubMed] [Google Scholar]
- Caccone A, Gentile G, Gibbs J.P, Fritts T.H, Snell H.L, Betts J, Powell J.R. Phylogeography and history of giant Galápagos tortoises. Evolution. 2002;56:2052–2066. doi: 10.1111/j.0014-3820.2002.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Castellano S, Balletto E. Is the partial Mantel test inadequate? Evolution. 2002;56:1871–1873. doi: 10.1111/j.0014-3820.2002.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Clegg S.M, Degnan S.M, Kikkawa J, Moritz C, Estoup A, Owens I.P.F. Genetic consequences of sequential founder events by an island-colonizing bird. Proc. Natl Acad. Sci. USA. 2002;99:8127–8132. doi: 10.1073/pnas.102583399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody M.L, editor. Habitat selection in birds. Academic Press; Orlando: 1985. [Google Scholar]
- Davis J.M, Stamps J.A. The effect of natal experience on habitat preferences. Trends Ecol. Evol. 2004;19:411–416. doi: 10.1016/j.tree.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Desalle R. Molecular approaches to biogeographic analysis of Hawaiian Drosophilidae. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography: evolution on a hot spot archipelago. Smithsonian Institution Press; Washington: 1995. pp. 72–89. [Google Scholar]
- Endler J.A. Princeton University Press; Princeton, NJ: 1977. Geographic variation, species and clines. [Google Scholar]
- Epperson B.K. Princeton University Press; Princeton: 2003. Geographical genetics. [Google Scholar]
- Finston T.L, Peck S.B. Population structure and gene flow in Stomion: a species swarm of flightless beetles of the Galápagos Islands. Heredity. 1995;75:390–397. [Google Scholar]
- Freeland J.R, Boag P.T. Phylogenetics of Darwin's finches: paraphyly in the tree-finches and two divergent lineages in the Warbler Finch. Auk. 1999;116:577–588. [Google Scholar]
- Freeland J.R, Boag P.T. The mitochondrial and nuclear genetic homogeneity of the phenotypically diverse Darwin's ground finches. Evolution. 1999;53:1553–1563. doi: 10.1111/j.1558-5646.1999.tb05418.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramos G, Kirkpatrick M. Genetic models of adaptation and gene flow in peripheral populations. Evolution. 1997;51:21–28. doi: 10.1111/j.1558-5646.1997.tb02384.x. [DOI] [PubMed] [Google Scholar]
- Grant P.R. Princeton University Press; Princeton: 1999. Ecology and evolution in Darwin's finches. [Google Scholar]
- Grant B.R, Grant P.R. Cultural inheritance of song and its role in the evolution of Darwin's finches. Evolution. 1996;50:2471–2487. doi: 10.1111/j.1558-5646.1996.tb03633.x. [DOI] [PubMed] [Google Scholar]
- Grant B.R, Grant P.R. Lack of premating isolation at the base of a phylogenetic tree. Am. Nat. 2002;160:1–19. doi: 10.1086/339987. [DOI] [PubMed] [Google Scholar]
- Grant P.R, Grant B.R. Reversed sexual size dimorphism in the beak of a finch. Ibis. 2003;145:341–343. [Google Scholar]
- Grant P.R, Grant B.R, Keller L.F, Petren K. Effects of El Niño events on Darwin's finch productivity. Ecology. 2000;81:2442–2457. [Google Scholar]
- Grant P.R, Grant B.R, Petren K. A population founded by a single pair of individuals: establishment, expansion and evolution. Heredity. 2001;112/113:359–382. [PubMed] [Google Scholar]
- Grant, P. R., Grant, B. R., Petren, K. & Keller, L. F. 2005 Extinction behind our backs: the possible fate of one of the Darwin's finch species on Isla Floreana, Galápagos. Biol. Conserv.122, 499–503.
- Gruenberger S, Leisler B. Innate and learned components in the habitat selection of coal tits Parus ater. J. fuer Ornith. 1990;131:460–464. [Google Scholar]
- Hamilton T.H, Rubinoff I. On predicting insular variation in endemism and sympatry for the Darwin's finches in the Galápagos archipelago. Am. Nat. 1967;101:161–167. [Google Scholar]
- Hanski I, Gilpin M, editors. Metapopulation biology: ecology, genetics, and evolution. Academic Press; San Diego: 1997. [Google Scholar]
- Hofreiter M, Serre D, Poinar H.N, Kuch M, Pääbo S. Ancient DNA. Nat. Rev. Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- Hormiga G, Arnedo M, Gillespie R.G. Speciation on a conveyor belt: sequential colonization of the Hawaiian Islands by Osonwelles spiders (Araneae, Linyphiidae) Syst. Biol. 2003;52:70–80. doi: 10.1080/10635150390132786. [DOI] [PubMed] [Google Scholar]
- Irwin D.E, Bensch S, Price T.D. Speciation in a ring. Nature. 2001;409:333–337. doi: 10.1038/35053059. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N.H. Evolution of a species' range. Am. Nat. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- Klicka J, Zink R.M. Pleistocene effects on North American songbird evolution. Proc. R. Soc. B. 1999;266:695–700. [Google Scholar]
- Knowles L.L, Maddison W.P. Statistical phylogeography. Mol. Ecol. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Lack D. Cambridge University Press; Cambridge: 1947. Darwin's finches. [Google Scholar]
- Legendre P, Vaudor X. Université de Montréal; Montreal: 1991. The R package. [Google Scholar]
- Leonard J.A, Wayne R.K, Cooper A. Population genetics of ice age brown bears. Proc. Natl Acad. Sci. USA. 2000;97:1651–1654. doi: 10.1073/pnas.040453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur R.H, Wilson E.O. Princeton University Press; Princeton: 1967. The theory of island biogeography. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Mayr E. Columbia University Press; New York: 1942. Systematics and the origin of species. [Google Scholar]
- Mundy N.I, Woodruff D.S. Skin from feet of museum specimens as a non-destructive source of DNA for avian genotyping. Auk. 1997;114:126–129. [Google Scholar]
- Omland K.E, Tarr C.L, Boarman W.I, Marzluff J.M, Fleischer R.C. Cryptic genetic variation and paraphyly in ravens. Proc. R. Soc. B. 2000;267:2475–2482. doi: 10.1098/rspb.2000.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petren K. Microsatellite primers from Geospiza fortis and cross-species amplification in Darwin's finches. Mol. Ecol. 1998;7:1782–1784. doi: 10.1046/j.1365-294x.1998.00518.x. [DOI] [PubMed] [Google Scholar]
- Petren K, Grant B.R, Grant P.R. A phylogeny of Darwin's finches based on microsatellite DNA length variation. Proc. R. Soc. B. 1999;266:321–329. [Google Scholar]
- Petren K, Grant B.R, Grant P.R. Low extrapair paternity in the cactus finch (Geospiza scandens) Auk. 1999;116:252–256. [Google Scholar]
- Posada D, Crandall K.A, Templeton A.R. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol. Ecol. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Price T.D. Sexual selection and natural selection in bird speciation. Phil. Trans. R. Soc. B. 1998;353:251–260. [Google Scholar]
- Richman A.D, Price T. Evolution of ecological differences in the Old World leaf warblers. Nature. 1992;355:817–821. doi: 10.1038/355817a0. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sato A, O'hUigin C, Figueroa F, Grant P.R, Grant B.R, Tichy H, Klein J. Phylogeny of Darwin's finches as revealed by mtDNA sequences. Proc. Natl Acad. Sci. USA. 1999;96:5105–5106. doi: 10.1073/pnas.96.9.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tichy H, O'hUigin C, Grant P.R, Grant B.R, Klein J. On the origin of Darwin's finches. Mol. Biol. Evol. 2001;18:299–311. doi: 10.1093/oxfordjournals.molbev.a003806. [DOI] [PubMed] [Google Scholar]
- Seutin G, Klein N.K, Ricklefs R.E, Bermingham E. Historical biogeography of the bananaquit (Coereba flaveola) in the Caribbean region: a mitochondrial DNA assessment. Evolution. 1994;48:1041–1061. doi: 10.1111/j.1558-5646.1994.tb05292.x. [DOI] [PubMed] [Google Scholar]
- Shaw K.L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA. 2002;99:16 122–16 127. doi: 10.1073/pnas.242585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 1998. PAUP*. Phylogenetic analysis using parsimony and other methods, vol. 4. [Google Scholar]
- Templeton A.R. Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol. Ecol. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- Templeton A.R, Robertson R.J, Brisson J, Strasburg J. Disrupting evolutionary processes: the effect of habitat fragmentation on collared lizards of the Missouri Ozarks. Proc. Natl Acad. Sci. USA. 2001;98:5426–5432. doi: 10.1073/pnas.091093098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuschl Y, Taborsky B, Taborsky M. How do cuckoos find their host? The role of habitat imprinting. Anim. Behav. 1998;56:1425–1433. doi: 10.1006/anbe.1998.0931. [DOI] [PubMed] [Google Scholar]
- Via S. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution. 1999;53:1446–1457. doi: 10.1111/j.1558-5646.1999.tb05409.x. [DOI] [PubMed] [Google Scholar]
- Wallace A.R. Macmillan; London: 1880. Island life. [Google Scholar]
- West-Eberhard M.J. Oxford University Press; New York: 2003. Developmental plasticity in evolution. [Google Scholar]
- Wiggins I.L, Porter D.M. Stanford University Press; Stanford, CA: 1971. The flora of the Galápagos. [Google Scholar]
- Wright J.W. The evolution and biogeography of the lizards of the Galápagos archipelago: evolutionary genetics of Phyllodactylus and Tropidurus populations. In: Bowman R.I, Berson M, Leviton A.E, editors. Patterns of evolution in Galápagos organisms. AAAS Pacific Division; San Francisco: 1983. pp. 123–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.