Abstract
Solution phase combinatorial chemistry holds an enormous promise for modern drug discovery. Much needed are direct methods to assay such libraries for binding of biological targets. An approach to encoding and screening of solution phase libraries has been developed based on the conditional photorelease of externally sensitized photolabile tags. The encoding tags are released into solution only when a sought-for binding event occurs between the ligand and the receptor, outfitted with an electron-transfer sensitizer. The released tags are analyzed in solution revealing the identity of the lead ligand or narrowing the range of potential leads.
Keywords: dithiane tag, photoinduced externally sensitized fragmentation, solution library encoding and screening
High-throughput combinatorial chemistry is critical for modern drug discovery, with the most prolific adaptation being the split-and-mix synthesis of libraries immobilized on polymeric beads (1, 2). These one-bead–one-compound libraries are assayed for binding of biological targets through fluorescence-guided mechanical segregation of the winning beads. The required mechanical manipulations impose a lower limit on the size of the particle used as a solid support. It is further recognized that solution phase combinatorial chemistry holds even greater promise, as it is compatible with both divergent and convergent multistep synthetic schemes, and not constrained to linear synthesis (3). However, its immense potential has not yet been fully realized, partly due to the complexity of assaying solution mixtures for binding. Although various iterative deconvolution methods (4) have been developed to synthesize and screen soluble sublibraries, all of them require redundant synthetic steps and are time consuming and expensive.
We have developed a methodology for direct screening of solution phase libraries encoded with photolabile tags that does not depend on mechanical manipulation or iterative deconvolution.
Results and Discussion
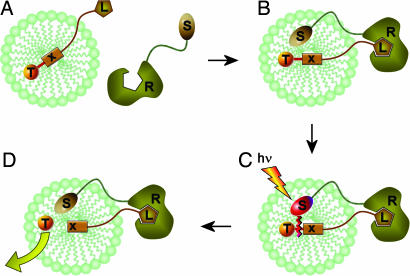
Our tagging methodology is based on dithiane adducts of carbonyl compounds, which are capable of efficient photoinduced fragmentation, but only in the presence of an external electron transfer (ET) sensitizer. At the core of this approach is the concept we term photolabile scaffolds for molecular recognition: binary molecular systems in which the sensitizer and the dithiane-based photocleavable fragments are each tethered to the respective components of a host–guest molecular recognition pair (5–7). In these pairs, photoinduced fragmentation is contingent on a molecular recognition event, which is necessary to arm the system, making it photolabile. This general concept has now been developed into a methodology for screening of encoded libraries, both unsupported or immobilized on nanosized carriers too small for mechanical handling. Scheme 1 gives a general outline for screening the micelle-solubilized library of ligands L, encoded with the tethered tags T. The receptor R is outfitted with an ET-sensitizer S, and incubated with the library (A). The host–guest binding brings the sensitizer into the vicinity of the adduct (B), which, upon irradiation (C), triggers the release of dithiane tags (D).
Scheme 1.
Our strategy is to encode individual molecules one tag at a time. The kth library member Lk can be encoded with a set of tags {T}k, for example, Tk1, Tk2, and Tk3, in the following fashion: one fraction of Lk molecules is encoded with Tk1, another fraction is encoded with Tk2, and yet another is encoded with Tk3, such that Lk is present in the solution as three subpopulations: Lk–tether–Tk1, Lk–tether–Tk2, and Lk–tether–Tk3. In this case, irradiation yields the desired result due to the fact that all of the tags encoding individual molecules of the bound ligand are collectively released into the solution, revealing the identity of the lead compound.
The following considerations were taken into account in the designing of tags. The tags should (i) be amendable for detection at very low concentrations by ubiquitous analytical techniques; (ii) not possess any functional groups implicated in biological interactions; (iii) not interfere with synthetic steps; (iv) be easy to separate from the aqueous screening environment, for example, due to significant hydrophobicity; and (v) be accessible via a simple and straightforward synthetic approach. We will demonstrate that 2-alkyl substituted dithianes satisfy these requirements and can be used for encoding as binary digits, i.e., 1 indicates that free dithiane is present in the solution and 0 indicates that no dithiane is found.
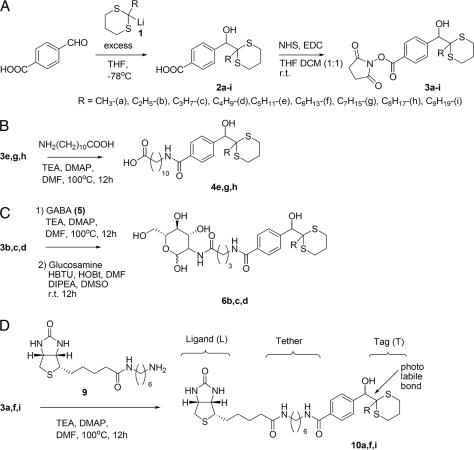
The ability to detect tags at very low levels is critical for any successful library encoding scheme. We have found that dithianes are particularly suitable for subpicomolar detection by gas chromatography with mass-selected detector (GCMS) with electron impact ionization. A better than 10:1 signal-to-noise ratio is achieved at a level of 500 femtomoles of alkyl dithianes per injection. In theory, for a library of one million tagged compounds, averaging 1–2 kDa, only a few milligrams are needed for the binding assay. The adducts of 2-alkyl dithianes and 4-formylbenzoic acid were chosen as tagging modules because alkyl dithianes add readily to nonenolizable aromatic aldehydes and the carboxylate serves as a practical handle to tether the tag to a ligand. A series of N-hydroxysuccinimide esters 3a–i were synthesized as shown in Scheme 2A (for experimental details, refer to Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Scheme 2.
To prove the concept we chose the archetype host-guest system, biotin-avidin, and synthesized a model minilibrary comprising three members (Scheme 2B-D), each encoded with a set of three dithiane tags: a carboxylate 4e,g,h encoded with 2-pentyl, 2-heptyl and 2-octyl dithianes (decimal 208; binary 11010000), a sugar 6b,c,d (ethyl-, propyl-, and butyl; 14; 1110), and biotin 10a,f,i (methyl-, hexyl-, and nonyl; 289; 100100001). In this model library, the encoding of individual members was intended not to overlap, to accurately assess the validity of the method and ensure unambiguous interpretation of the screening results.
The receptor, ImmunoPure avidin (Pierce, Rockford, IL), was outfitted with xanthone as an ET-sensitizer. The N-hydroxysuccinimide ester of 11-(xanthone-2-carboxamido)undecanoic acid was coupled to avidin in a 20 mM sodium phosphate buffer according to a described procedure (8), with subsequent purification on a Sephadex G-25 column. The degree of immobilization was quantified by UV spectroscopy to be 0.77, indicating that, on average, each tetramer of avidin was carrying approximately three tethered xanthone carboxylates. Tethering the xanthone-based sensitizer to avidin is, in a way, no different from outfitting a receptor with a fluorophore for the conventional on-the-bead fluorescence-guided assays.
To ensure fidelity of the method we compartmentalized the screening volume with micellar detergent, dodecyl phosphocholine (DPC), preventing indiscriminant collisional quenching of avidin-tethered xanthone by unbound molecules and thus limiting the ET sensitization exclusively to the bound host–guest complex. Besides the fact that amphiphiles displaying phosphocholine are common in biological settings, utilization of DPC micelles offers a number of additional benefits: (i) it ensures that the screening is always compatible with aqueous media, regardless of aqueous solubility of the tested libraries; (ii) it allows us to center the design of the tagging system around readily available hydrophobic alkyl dithianes, which can be selectively extracted after irradiation with hexane or other nonpolar solvents for unobstructed GCMS analysis; (iii) it spatially segregates the photofragmentation chemistry from molecular recognition, eliminating potential interference between them; (iv) it restricts the photochemistry to the micelle interior improving quantum efficiency of fragmentation, as it is known to increase in the nonpolar environment; and (v) the micelle-assisted design offers an option of solubilizing certain target proteins, which are not water soluble. Although not an issue with avidin, this functionality may prove critical in assaying insoluble membrane proteins.
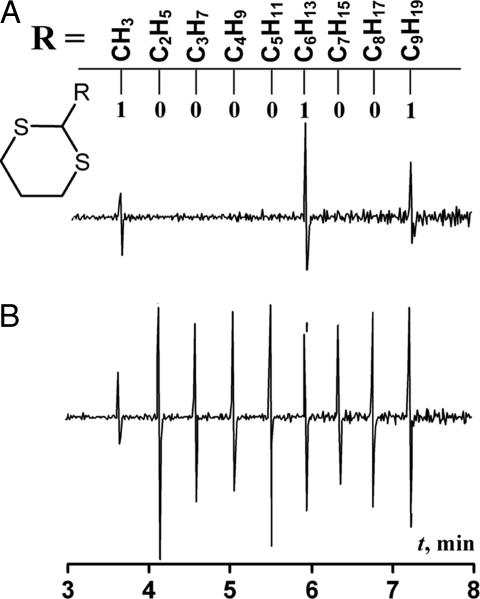
A typical screening procedure involved solubilization of the minilibrary, ≈0.5 mg per tagged compound, in a 0.6-ml aqueous solution containing 60 mg of DPC. To this clear micellar solution, 0.5 ml of avidin–xanthone conjugate was added, so the final concentrations were 0.7 mM of each library member carrying one tag (6.3 mM total), 23 μM protein, and 155 mM DPC. The micelle-embedded molecules had apparent translational diffusion coefficients of 7 × 10−7 cm2·s−1, as measured with spin-echo pulse field gradient (PFG) 1H NMR. This corresponds to the hydrodynamic radius of ≈3 nm, indicating that the occupied DPC micelles did not deviate much from their original 5- to 5.5-nm size. The resulting micellar solution was incubated in an orbital shaker for 1 h, purged with argon for 45 min, and irradiated for 4 h by using a 335-nm long pass filter. Then the mixture was extracted with 0.5 ml of hexane, concentrated to 100 μl and analyzed by GCMS. Fig. 1 clearly shows that only the biotin-encoding tags, namely methyl-, hexyl-, and nonyl-dithianes (binary 100100001, read from left to right), were detected in the chromatogram. The other six dithiane tags encoding glucosamine and aminoundecanoic acid were not discernible at all, attesting to the high fidelity of the assay.
Fig. 1.
The first derivative GCMS single ion monitoring (SIM) traces. (A) The trace encoding biotin in binary 100100001, obtained after the photolytic assay. (B) The trace for all nine alkyl dithianes at 1 pmol per injection.
The integrated intensity of dithiane peaks in most experiments was comparatively uniform within 30–50%. The quantum yield of alkyldithiane photo-release from ketone adducts increases in small increments of 2–3% in a homologous series, leveling off for the higher alkyls (9). The GCMS sensitivity of dithianes detection also varies insignificantly. If needed, these small variations can be offset by adjusting the quantities of individual tags used for encoding of each step.
To demonstrate that the compartmentalization requirement is not necessarily strict, one micelle in the described screening experiment contained on average two tagged library molecules (assuming that the aggregation number of DPC is 50–60; ref. 10). In theory, if the protein-tethered sensitizer indiscriminately releases both tags from the two occupants of the bound micelle, the integrated intensity of the false peaks in the chromatogram should constitute more than one third of the correct peak's intensity. Experimentally we did not detect any false tags, with the signal to noise ratio of the single-ion monitoring current exceeding 20:1. This shows that either the sensitizer discriminates between the bound and nonbound occupants of the micelle, preferentially triggering the release of the bound tag, or the micelles containing two biotin molecules bind much better than the micelles containing only one biotin, in which case false release is not at all possible. It is also conceivable that both factors operate concurrently, improving the fidelity of the method. If needed, a one-molecule–one-micelle compartmentalization can be readily achieved in practice by increasing the detergent concentration.
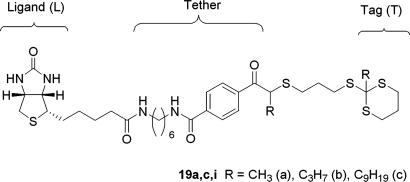
The generality of this approach is not limited to the tagging assemblies based on dithiane–aldehyde adducts. Potentially, any externally sensitized fragmentation reaction can be used in such binding assays. We achieved similar results with different tags comprised of thio ortho esters, 2-alkylthio-dithianes (11). The model minilibrary was tagged with nine thio ortho esters (19), three tags per library member, with biotin encoded by a different decimal 261, binary 100000101 (Fig. 2). The DPC micellar solution of the library was incubated with the avidin–xanthone conjugate, irradiated and extracted with hexane. The GCMS trace again showed only the dithianes encoding biotin. None of the other six dithianes were detected in the hexane extract after irradiation.
Fig. 2.
Thio ortho esters-encoded biotin.
In order for this screening methodology to afford maximal information content, the encoding strategy is to tag each reagent used at each step of combinatorial synthesis, so that every library member is encoded with the same total number of tags, equal to the number of synthetic steps. As an example, a library of pentapeptides with 7 possible amino acid residues, containing 75 = 16,807 members, would need 5 × 7 = 35 tags for encoding. The same 35 tags, of course, encode a library of 57 = 78,125 heptapeptides with five possible residues per position. The one-tag-per-reagent encoding is much more informative and less ambiguous than the conventional binary scheme suitable for the libraries immobilized on polymeric beads (12, 13). For example, tags T1 through T7 each encode the first-step reagents A1 through A7; tags T8 through T14 encode the second position, and so on. In the best case scenario, when the assayed library contains a lead ligand with much higher binding affinity than the next closest competitor, our photoassay identifies such a ligand in just one screening. In cases when a library contains several high-affinity ligands with very similar KD values, analysis of multiple GCMS peaks within each individual range, i.e., tags 1–7, tags 8–14, etc., should dramatically reduce the number of possibilities, providing statistically significant information about the most preferred components/reagents at each position/synthetic step. If needed, a much smaller sublibrary can be synthesized and the lead compound determined.
Of more importance is the actual binding, not the mathematics of deconvolution. In a typical case of fluorescence-guided on-the-bead assays, the multiple hits are not due to the fact that the respective KD values are nearly identical, but rather because the receptor is overloaded and free to saturate several binding equilibria. Subsequent dilution of the receptor to very low concentrations should help limit the number of the identified winners, with the constraint being the dithiane detection limit. As we demonstrated, we can confidently detect subpicomolar amounts of dithianes per injection by using conventional analytical instrumentation. Extracted dithianes can be further preconcentrated before GC injection, allowing for handling the full physiological range of KD values, down to subnanomolar.
In an early classical example by Lam et al. (14), a large pentapeptide library of 2,476,099 members was synthesized and screened against a monoclonal antibody, known to bind the β-endorphin epitope (YGGFL). The authors identified four peptide ligands with KD values under 37 nM: YGGFQ (15.0 nM), YGGFL–native ligand (17.5 nM), YGGFA (32.9 nM), and YGGFT (36.9 nM). The KD for the next best ligand, YGGLS, was 20 times greater (726 nM). In a case like this, our photo-assay methodology, with a sensitizer-tethered antibody, would produce one peak in each range of tags encoding positions 1–4, YGGF, and four peaks in the range of tags encoding position 5. Thus, in a single solution-phase screening, all four winners can be unambiguously identified.
Dynamic encoding of libraries, prepared by the split-pool approach, is normally achieved with a coupling scheme orthogonal to the used synthetic steps. The tagging synthons, esters 3, can be coupled with the library molecules via tethers containing primary amino groups, as in conventional polypeptide encoding (15, 16), which involves the appropriate cycling of the BOC or FMOC protection. Alternatively, the esters 3 can be used for dynamic tagging in conjunction with other efficient coupling reactions, such as Staudinger ligation (17–19) or Sharpless' click chemistry (20).
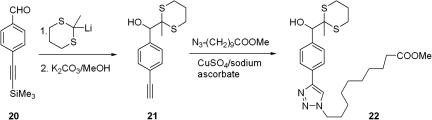
To this end we coupled 3a with propargyl amine, providing the acetylenic tagging component S20 for the azide-acetylene click pair. Another readily available tag series is prepared from commercially available acetylenic benzaldehyde as shown in Scheme 3. Whereas the Staudinger ligation produces the benzamide linkage, found in compounds 4, 6, and 10, for which we have proven the concept, acetylene–azide click chemistry yields triazoles, which needed further testing to ascertain their compatibility with photoinduced fragmentation and the release of dithiane tags. Methyl 10-azidodecanoate, emulating an azide-tethered library member, was “clicked” onto adduct 21, forming triazole 22, which upon benzophenone sensitization released methyl dithiane with a quantum efficiency very similar to the parent (unsubstituted) benzaldehyde adduct. Triazole 22 was unchanged after prolonged irradiation in the absence of the ET-sensitizer, showing no self-cleavage at wavelengths >330 nm. This was a critically important finding because premature self-cleavage is detrimental to screening, as it produces false positives.
Scheme 3.
In conclusion, we have described here a methodology for encoding and screening of combinatorial libraries based on externally sensitized photolabile tags. The approach does not depend on mechanical handling of the beads, allowing for the screening of unsupported solution phase libraries and, potentially, libraries immobilized on nano-carriers, including dendrimers. This methodology was tested on a model minilibrary, for which the high-fidelity of screening was demonstrated. We also have prepared acetylene-bearing tags for encoding via click chemistry and have outlined a general approach to dynamic tagging of libraries. Our findings have the potential to fundamentally transform combinatorial encoding and screening of solution phase libraries, as currently there are no direct methods to assay solution mixtures for binding.
Materials and Methods
Common reagents were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. THF was refluxed over and distilled from potassium benzophenone ketyl before use. 1H and 13C NMR spectra were recorded at 25°C on a Varian Mercury 400 MHz instrument, in CDCl3, DMSO-d6, or CD3OD using tetramethylsilane as an internal standard. Pulsed Field Gradient NMR studies were carried out with the Varian Performa I PFG module and a four-nuclei autoswitchable PFG probe. Column chromatography was performed on silica gel, 70–230 mesh. The UV-visible spectra were recorded on a Beckman DU-640 spectrophotometer. Irradiations were carried out in a carousel Rayonet photo reactor (Southern New England Ultra Violet, Branford, CT), equipped with RPR-3500 UV lamps (300- to 400-nm spectral distribution of irradiance density with a maximum at 350 nm) and a 335-nm long-pass solution filter. Gas chromatography was performed on a Varian Saturn 2100 T Ion-Trap instrument with electron ionization. Selective ion monitoring (m/z 119, 74) was used to detect dithiane tags after fragmentation.
Dithiane–Aldehyde Adducts 2(a–i).
Dithiane–aldehyde adducts 2(a–i) were synthesized according to the general procedure: n-BuLi (14.6 ml, 23.3 mmol, 1.6 M solution in THF) was added at −25°C to a solution of 2-alkyl-1,3-dithiane (23.3 mmol) in dry THF (40 ml) under nitrogen atmosphere. The solution was stirred for 2 h, the temperature was reduced to −78°C, and 4-formylbenzoic acid (0.5 g, 3.33 mmol) in 20 ml of THF was added. The solution was stirred at −78°C for an additional 2 h and then allowed to warm to room temperature. The reaction was quenched with saturated aqueous ammonium chloride (20 ml) and extracted twice with 20 ml of ethyl acetate. The aqueous layer was acidified with 5% HCl and extracted with ethyl acetate (100 ml), and the extract was dried over Na2SO4. The solvent was removed in vacuum. The crude product was recrystallized from toluene to furnish adducts 2a–i in 90–96% yields.
Tagging of biotin, glucosamine and aminoundecanoic acid with the encoding pendants 2a–i was carried out according to modified literature procedures by using EDC or DIPEA coupling reagents. All members of the two model minilibraries (10a–i, 16f,g,h, 18b,d,e, and 19a,c,i) were individually purified and characterized by 1H and 13C NMR. For detailed synthetic procedures and spectra, see Supporting Materials and Methods.
Xanthone Conjugate of Avidin.
100 μl of 13 (5 mg/ml in DMSO) was added to a solution of avidin (Pierce, Rockford, IL) (10 mg, 0.147 μmol) in 1 ml of 20 mM sodium phosphate buffer. The mixture was gently shaken for 2 h at ambient temperature, followed by purification on a Sephadex G-25 column. The degree of immobilization was quantified spectroscopically to be 0.77 as described in Supporting Materials and Methods.
Photochemical Screening.
In a typical screening experiment, 0.5 mg each of 10a, 6b, 6c, 6d, 4e, 10f, 4g, 4h, and 10i were added to a solution of DPC (Anatrace, Maumee, OH) (60 mg) in 0.6 ml of D2O. The mixture was stirred for 24 h at ambient temperature until a clear micellar solution was obtained, and 0.5 ml of the avidin–xanthone conjugate was added. This solution was incubated with shaking for 1 h, degassed with argon for 45 min, and irradiated for 4 h by using Rayonet reactor (RPR-3500 lamps) with a 335-nm long-pass solution filter. The micellar solution was extracted with 0.5 ml of hexane, concentrated to 0.1 ml, and analyzed by GCMS with single ion monitoring.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant CHE314344 and National Institutes of Health Grant GM067655.
Abbreviations
- ET
electron transfer
- GCMS
gas chromatography mass spectrometry
- EI
electron impact ionization
Footnotes
The authors declare no conflict of interest.
References
- 1.Lam KS, Lebl M, Krchňák V. Chem Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 2.Lam KS, Liu R, Miyamoto S, Lehman AL, Tuscano JM. Acc Chem Res. 2003;36:370–377. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]
- 3.Baldino CM. J Combinatorial Chem. 2000;2:89–103. doi: 10.1021/cc990064+. [DOI] [PubMed] [Google Scholar]
- 4.Konings DAM, Wyatt JR, Ecker DJ, Freier SM. J Med Chem. 1997;40:4386–4395. doi: 10.1021/jm970503o. [DOI] [PubMed] [Google Scholar]
- 5.Mitkin OD, Kurchan AN, Wan Y, Schiwal BF, Kutateladze AG. Org Lett. 2001;3:1841–1844. doi: 10.1021/ol015933u. [DOI] [PubMed] [Google Scholar]
- 6.Mitkin O, Wan Y, Kurchan A, Kutateladze A. Synthesis. 2001;7:1133–1142. [Google Scholar]
- 7.Wan Y, Angleson JK, Kutateladze A. G. J Am Chem Soc. 2002;124:5610–5611. doi: 10.1021/ja016874i. [DOI] [PubMed] [Google Scholar]
- 8.Wilchek M, Bayer EA, editors. Methods Enzymol. Vol. 184. 1990. p. 176. [DOI] [PubMed] [Google Scholar]
- 9.Gustafson TP, Kurchan AN, Kutateladze AG. Tetrahedron. 2006;62:6574–6580. [Google Scholar]
- 10.Brown LR, Bösch C, Wüthrich K. Biochim Biophys Acta. 1981;642:296–312. doi: 10.1016/0005-2736(81)90447-8. [DOI] [PubMed] [Google Scholar]
- 11.Valiulin RA, Kottani R, Kutateladze AG. J Org Chem. 2006;71:5047–5049. doi: 10.1021/jo060780f. [DOI] [PubMed] [Google Scholar]
- 12.Ohlmeyer MHJ, Swanson RN, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borchardt A, Still WC. J Am Chem Soc. 1994;116:373–374. [Google Scholar]
- 14.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 15.Franz AH, Liu R, Song A, Lam KS, Lebrilla CB. J Comb Chem. 2003;5:125–137. doi: 10.1021/cc020083a. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Marik J, Lam KS. J Am Chem Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 17.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 18.Saxon E, Armstrong JI, Bertozzi CR. Org Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson BL, Kiessling LL, Raines RT. Org Lett. 2000;2:1939–1941. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]
- 20.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.