Abstract
HIV-1 enters cells by membrane fusion, mediated by the trimeric viral envelope glycoprotein gp160, which is processed by a single proteolytic cleavage into stably associated gp120 and gp41. The gp120/gp41 trimer can be triggered to undergo an irreversible conformational change. Using a protein-based assay designed to mimic the gp41 conformational change, we screened for small molecules that prevent the formation of postfusion gp41. Several compounds were identified. One set of structurally related molecules inhibited formation of a postfusion-like assembly with an IC50 of ≈5 μM. The compounds also inhibited envelope-mediated membrane fusion in both cell–cell fusion and viral infectivity assays. Thus, our screen identifies effective fusion inhibitors. Tested against a panel of envelope proteins from primary HIV-1 isolates, the compounds inhibited fusion across a broad range of clades, including both M and T tropic strains. They bind in a highly conserved, hydrophobic pocket on the inner core of the gp41 trimer, a region previously identified as a potential inhibitor site.
Keywords: viral entry, antiviral drugs, high-throughput screen
Enveloped viruses such as HIV and influenza virus enter cells by membrane fusion (1). The gp160 envelope glycoprotein of HIV-1, its fusion protein, is synthesized as a single polypeptide chain with a C-terminal membrane anchor, and it requires a processing cleavage (to gp120 and gp41) for activation (2, 3). The fusion peptide, a glycine-rich, hydrophobic sequence required for interaction with the target-cell membrane, lies near the newly created N terminus of gp41 (4). Like other class 1 fusion proteins, such as influenza virus hemagglutinin, gp160 is a homotrimer, and its fusion-promoting fragment, gp41, is also trimeric (1).
The cleaved, mature trimer of gp120/gp41 can be triggered by receptor binding to undergo an irreversible conformational change (5–9). In effect, cleavage of the gp160 precursor produces a metastable conformation, but the barrier to its rearrangement is high. The image of a spring-loaded device has been used to describe this situation, and the triggering event then corresponds to releasing the catch on the spring (10).
The structure of the gp41 ectodomain in the postfusion conformation is a trimer of α-helical hairpins (11, 12). Its inner core is an α-helical coiled coil formed by N-terminal segments of the three polypeptide chains; the helices of its outer layer are the C-terminal segments of those chains. The 30-residue loop that connects the inner core and outer-layer helices [also called “heptad repeats 1 and 2” (HR1 and HR2, respectively)] is absent from the crystal structures; sequence conservation suggests that it resembles the loop seen in the ectodomain of the transmembrane (TM) subunit from Moloney murine leukemia virus (MoMuLV) and in the Gp2 ectodomain of Ebola virus (13, 14). A noteworthy feature of the contact between each outer-layer helix and the inner core is a hydrophobic pocket on the latter, containing residues Leu-57, Trp-60, and Lys-63, which receives the side chains of Trp-117, Trp-120, and Ile-124.
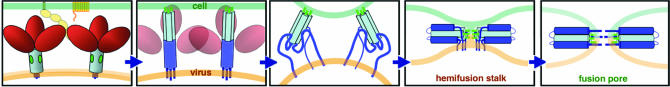
The sequence of molecular events during the fusion-promoting conformational rearrangement (refolding) of gp120/gp41 is thought to be as follows (Fig. 1) (1). The gp120 fragment dissociates, allowing the gp41 fragment to rearrange (2). An initial reorganization of the gp41 polypeptide chain pulls the fusion peptide from a buried position and projects it against the target-cell membrane. The result of this process is probably a conformation with significant half-life, the so called “prefusion intermediate” (3). In a subsequent step, each fusion-promoting fragment folds back on itself, probably by zippering up the outer layer along the surface of the inner core. This step brings together the transmembrane (TM) segment at the C terminus of the fusion-promoting fragment, anchored in the viral bilayer, and the fusion peptide at the N terminus, bound to the target-cell membrane. The two membranes are thus induced to approach each other and ultimately to merge.
Fig. 1.
Model for fusion by the HIV envelope glycoprotein. Binding of gp120 to CD4 and coreceptor (first panel) triggers a conformational change that releases the grip of gp120 (red) on gp41 (blue). The latter extends (second panel) so that its fusion peptide (green) inserts into the target-cell membrane (green bar at top). As the outer-layer region of gp41 (dark blue) zips up along the outside of the inner layer (light blue: a three-chain, α-helical coiled coil), the viral membrane (tan) and cell membrane (green) are drawn together (third and fourth panels). It is this step that is inhibited by T-20/enfuvirtide. The formation of a hemifusion stalk (fourth panel) and a fusion pore (fifth panel) completes the membrane-fusion reaction.
Even before the structure of the postfusion gp41 trimer was known, screening of peptides derived from gp160 had identified fusion inhibitors (15–19). The gp41 structure showed that some of these peptides represent inner-core segments and that others cover all or part of the outer helix (11, 12). The presumed mechanism of inhibition by the latter is the binding of the peptide to the inner-core portion of the prefusion intermediate, preventing completion of the fusion-promoting conformational rearrangement. The inhibitory peptide known as T-20 (now a licensed drug with the generic name enfuvirtide) contains residues 127–162 (16, 17, 20). Its sequence includes most, but not all, of the outer-layer helix and an additional tryptophan-rich region whose function is still unknown. It does not include Trp-117, Trp-120, or Ile-124, residues that would occupy the hydrophobic pocket near positions Trp-60 and Lys-63 on the inner core. Evidence suggests that this pocket would be a suitable site for an inhibitory drug (21). It was targeted by Eckert et al. (22) in their design of a 16-residue cyclic D peptide, which docks against the pocket and also inhibits fusion and viral infectivity. Ferrer et al. (23) used the pocket to select nonnatural binding elements tethered to a peptide derived from the outer helix. The conjugate was shown to inhibit envelope-mediated cell fusion. Molecular docking simulations against the pocket have also been performed (24). A quite different sort of fusion inhibitor is a single-chain model for five of the six helical segments in gp41, which presumably traps an incipient outer-layer helix (25).
Can small molecules, with more conventional drug-like properties, also inhibit the fusion-promoting refolding of gp41? This question is a particular case of the more generally debated issue: can small molecules block formation of protein–protein interfaces? Several small-molecule respiratory syncytial virus (RSV) inhibitors target the inner core of the F1 protein, a related class 1 fusion protein (26). We have found a set of compounds that interact with the gp41 inner core by using a screen based on blocking the association of outer-layer peptides with the inner core. These molecules reduce gp41-mediated cell–cell fusion and inhibit HIV-1 infectivity. We have narrowed down the target of binding to a hydrophobic pocket on the gp41 inner core. This pocket is highly conserved among the different clades of group M. Consistent with this conservation, we have shown that our compounds are active against a panel of HIV-1 primary isolates that includes both M and T tropic strains from different clades but are substantially less active against divergent strains from group O and simian immunodeficiency virus (SIV).
Results and Discussion
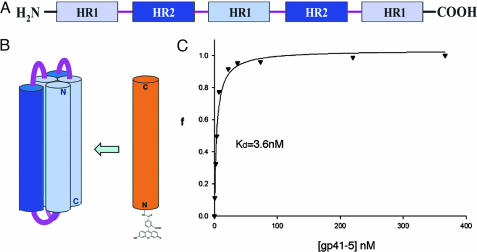
We designed and expressed a soluble, single-chain protein construct we call gp41-5 to provide a suitable target for a high-throughput assay (Fig. 2). This strategy takes advantage of the unusually simple structure of gp41 in its postfusion state, a trimer of hairpins, which can be linked into a single, covalent polypeptide that folds into a six-helix bundle. A related construct (called “five-helix”) was used as a fusion inhibitor, in studies mentioned above (25). Our gp41-5 contains three inner-core segments (residues 35–70), alternating with two outer-layer segments (residues 117–150). Short linkers connect the segments. After folding, the molecule contains five of the helices present in the six-helix bundle. It is relatively soluble, and it binds with high-affinity peptides that contain part or all of the sequence in the sixth helix. Moreover, molecules of any sort that can bind the groove exposed by the absence of the sixth HR region should also be able to bind the prehairpin intermediate.
Fig. 2.
Association of an outer-layer peptide with gp41-5 as a screening assay. (A) The sequence of a single-chain polypeptide that can fold into a model for five of the six helices in the postfusion form of the gp41 ectodomain. (B) Diagram illustrating that a fluoresceinated, outer-layer peptide can bind to gp41-5. N, N terminus; C, C terminus. (C) Fluorescence anisotropy binding curve for the association of the outer-layer peptide with gp41-5. The fraction bound, f, is plotted as a function of gp41-5 concentration (in nM), at a constant concentration of peptide (5 nM).
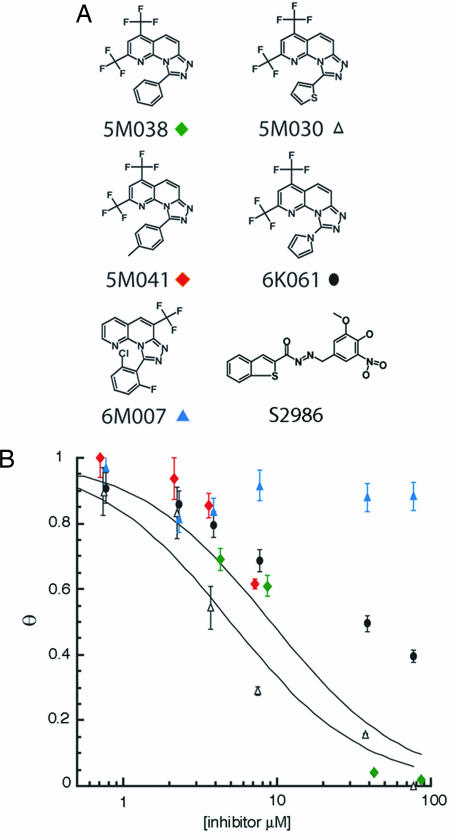
A peptide with the sequence of the missing outer helix (C38, residues 117–154) was labeled at its N terminus with fluorescein. Binding of the labeled peptide (C38*) to gp41-5 was monitored by fluorescence polarization (Fig. 2C). The assay is sensitive, enabling us to perform high-throughput screens. We screened several libraries of small molecules (Mr < 500), searching for compounds that interfered with the binding of C38*. As a positive control, C38 (6 μM) was added to one well on each plate. Of the 34,800 compounds screened, four (5M030, 5M038, 5M041, and S2986; Fig. 3A) completely blocked C38* binding to gp41-5 under our screening conditions (library compound concentration of 40 μM; C38* = 10 nM). Three contained a 2,4-bis(trifluoromethyl)[1,2,4] triazolo[4,3-a][1,8] naphthyridine ring, with different substituents in the 9 position. Follow-up analysis of structure activity relationships revealed two additional compounds structurally similar to 5M038 (Fig. 3A). Only compounds 5M038 and 5M030 had sufficient solubility and inhibitory activity to determine accurate IC50 values in our fluorescence polarization assay (≈5 and 9 μM, respectively; Fig. 3B). Compounds 5M041 and 6K061 also had significant inhibitory activity (30% and 40% inhibition, respectively, at 7.5 μM). Compound 6M007, which contains a similar ring system with the exception of a trifluoromethyl substituent in the 6 position rather than the 2 and 4 positions, showed no activity in the fluorescence polarization assay.
Fig. 3.
Results of high-throughput screen. (A) Compounds identified as strong inhibitors of outer-layer peptide binding (5M038, 5M030, 5M041, and S2986), as well as compounds in the chemical library related to compound 5M038. (B) Inhibition of peptide binding for five of the compounds shown in A. The fraction bound, θ, is plotted as a function of inhibitor concentration. Symbols correspond to those beneath the various compounds in A. The curves show optimal sigmoidal fits to the data for 5M030 and 5M038, the only two compounds that were sufficiently soluble to yield reliable plots. Compound S2986 had an IC50 of ≈5 μM in this assay (data not shown).
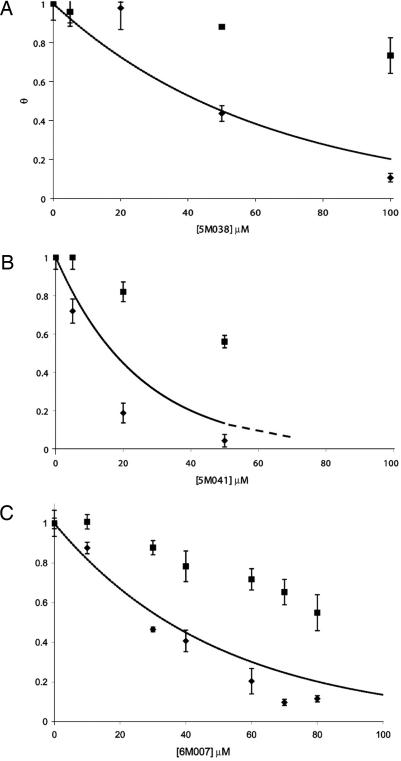
To determine the effects of these compounds on gp41-mediated membrane fusion, we used a cell–cell fusion assay (23, 27). Compound toxicities were initially determined by using trypan blue staining. In addition, to control for nonspecific effects of added compound, we prepared doubly transfected cells with plasmids encoding T7 polymerase and luciferase. Compounds 5M038 and 5M041 strongly inhibited gp41-mediated membrane fusion. At their solubility limits (90 μM for 5M038 and 50 μM for 5M041), both compounds reduced the level of cell–cell fusion by >80% (Fig. 4). Although compound 6M007 had little activity in the fluorescence polarization assay, it was as active as 5M038 against envelope-mediated membrane fusion. We discuss below this difference in assay response. Compounds 6K061 and 5M030 were too toxic to be tested properly.
Fig. 4.
Inhibition of cell–cell fusion by 5M038 (A), 5M041 (B), and 6M007 (C). The fractional degree of fusion, determined by the luciferase assay described in Materials and Methods, is plotted as a function of inhibitor concentration (diamonds). Squares show an experiment in which luciferase was expressed directly in the target cells, independent of fusion, as a control for toxicity and other side effects of the inhibitors. Curves are fractional inhibition, normalized for these toxicity effects. All points are the average of three experiments (error bars).
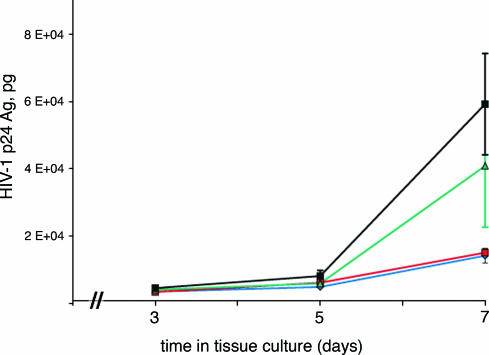
We determined the effects on HIV infectivity of the series of small molecules just described, by exposing peripheral blood mononuclear cells (PBMCs) to HIV-1 (isolate 2076) in the presence and absence of each compound. When cells were pretreated with a compound before HIV infection, compounds 5M038 and 5M041 blocked infection with IC50 values of 19 and 18 μM, respectively (Table 1). Compound 6M007 had a weaker effect, showing 50% inhibition at a concentration of >30 μM. To demonstrate that, like enfuvirtide, 5M038 can inhibit the cell-to-cell spread of virus, we compared the two directly over a 7-day period, as shown in Fig. 5. At a concentration as low as 30 μM, 5M038 nearly completely suppressed the appearance of p24 antigen over the course of the experiment.
Table 1.
Effects of 5M038, 5M041, and 6M007 on viral infectivity compared with their activities in other assays
| Compound | IC50 protein-based assay, μM | IC50 cell–cell fusion, μM | IC50 viral infectivity, μM |
|---|---|---|---|
| 5M038 | 10 | 38 | 19 |
| 5M041 | 10 | 18 | 18 |
| 6M007 | >100 | 39 | 38 |
Peripheral blood mononuclear cells were pretreated with virus for 1 h before addition of compound. After 5 days, viral levels were determined by the amount of p24 antigen present in the culture. [3H]thymidine incorporation was used to assay possible cytotoxic effects. The 50% toxicity concentration (TC50) values for 5M038, 5M041, and 6M007 were 43.2, 58, and 79.9 μM, respectively.
Fig. 5.
Effect of 5M038 during the time course of HIV infection. MT-2 cells were infected with HIV in the presence or absence of either 30 μM (blue) or 50 μM (red) 5M038. Time points were taken at 3, 5, and 7 days, and viral infection was measured by the amount of p24 antigen (p24 Ag) produced. Enfuvirtide, a potent peptidic viral entry inhibitor, was also tested (11 nM) and used for comparison (green). All points are the average of three experiments (bars represent standard error). The control (no added compound) is in black.
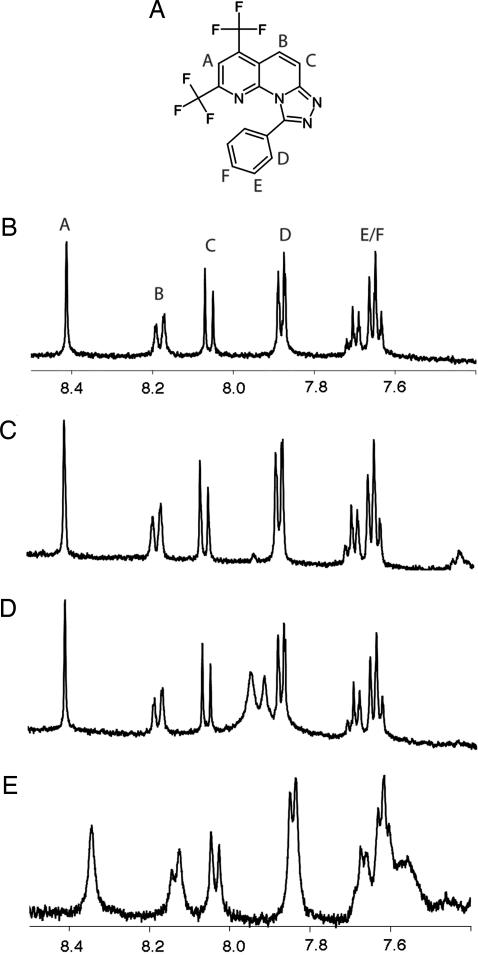
How do these molecules interact with gp41? Proton NMR experiments show that 5M038 gives significantly broadened proton resonances in the presence of gp41-5 (data not shown). To use this property to pinpoint the location of binding, we synthesized a set of three peptides containing 17-residue segments of the gp41 inner core attached to a 29-residue trimerization domain derived from the coiled coil of GCN4 (Fig. 6). The latter helps to solubilize and trimerize the short segment of the gp41 core and permits its use at concentrations suitable for NMR. Similar fusion proteins were used to crystallize the gp41 ectodomain (11), to crystallize the Ebola Gp2 ectodomain (14), and to develop cyclic D peptide fusion inhibitors (22). Proton NMR spectra were recorded from compound 5M038 (200 μM) in the presence of each of the three peptides (also at 200 μM). Resonances remained sharp in the presence of peptides 1 and 2, which contained residues 34–50 and 41–57, respectively. In the presence of peptide 3, which contained residues 54–70, the proton lines from 5M038 broadened significantly and shifted upfield. This segment of the inner core contains a deep cavity formed by residues surrounding Leu-57, Trp-60, and Lys-63 and occupied in the postfusion structure by Trp-117, Trp-120, and Ile-124 from the outer-layer helix. This pocket has been exploited previously (22, 23), and it seems likely that 5M038 binds it as well. To probe further the binding site for these molecules, we performed competition assays using peptides missing key residues that insert into the inner-core pocket. Whereas 5M038 was able to compete against a full-length peptide containing residues 117–154, it showed no activity against a shorter peptide containing residues 119–154 (data not shown). The latter lacks the key binding residue Trp-117; the result provides further evidence that 5M038 occupies the Trp-117 pocket. T-20/enfuvirtide does not include Trp-117, and 5M038 and enfuvirtide are indeed weakly synergistic in viral infectivity experiments (data not shown).
Fig. 6.
NMR measurements of the association of 5M038 with three-chain coiled coils containing segments of the inner core of postfusion gp41. (A) Chemical diagram showing the various protons detected in the spectra. (B) Spectrum of free 5M038. (C–E) Spectra of 5M038 in the presence of coiled coils containing residues 34–50 of gp41 (C), residues 41–57 (D), and residues 54–70 (E). There was significant broadening and shifting of the resonance peaks only in the last of the four spectra, showing that 5M038 associates selectively with the part of the gp41 inner core formed by residues 54–70.
The hydrophobic pocket formed by Leu-57, Trp-60, and Lys-63 is a very highly conserved region among the different isolates of HIV-1 group M (see Table 2). By targeting this region, we would expect this class of compounds to show activity across a broad range of isolates. Conversely, broad activity would be consistent with our identification of the site and mode of action. We selected a number of Env sequences derived from primary isolates and assayed them for cell–cell fusion in the presence of compound 5M038. The sequences include members from clades A, B, D, E, and F and include both T and M tropic strains. We also included two divergent strains, BCF03 from group O and SIV mac32H. The compound was active against all of the group M isolates tested while exhibiting much weaker activity toward the group O strain (only 40% inhibition at 100 μM) and SIV (35% inhibition at 100 μM).
Table 2.
Effect of 5M038 on cell–cell fusion mediated by envelope glycoproteins from various HIV and SIV isolates
| Isolate | IC50, μM | Clade | Coreceptor | Pocket sequence |
|---|---|---|---|---|
| HXB2 | 38 | B | X4 | LTVWGIKQLQARIL |
| 92ug024.2 | 30 | D | X4 | LTVWGIKQLQARVL |
| 93th976.10 | 40 | E | Unknown | LTVWGIKQLQARVL |
| JRFL | 40 | B | R5 | LTVWGIKQLQARVL |
| 93br029.2 | 50 | F | R5 | LTVWGITQLQARIL |
| 92ug037.8 | 50 | A | R5 | LTVWGIKQLQARVL |
| 92us716.6 | 70 | B | R5 | LTVWGIKQLQARVL |
| BCF03 | >100 | Group O | Unknown | LSVWGIRQLRARLQ |
| SIV mac32H | >100 | SIV | — | LTVWGTKNLQTRVT |
The pocket sequence is the amino acid sequence of residues 57–70 (HXB2 numbering) on the inner-core helix of gp41.
We used the fluorescence polarization assay to examine other inhibitors that target gp41. A peptide corresponding to the part of T-20 contained in gp41-5 (i.e., residues 127–154) and a peptidic entry inhibitor (23) both failed to compete with C38 at concentrations up to 100 and 150 μM, respectively, under our assay conditions ([*C38] = 5 nM), even though the peptidic entry inhibitor blocks cell–cell fusion. These results are similar to our observations with 6M007. The in vitro fluorescence polarization assay is an equilibrium measurement; the inhibition of fusion (cell–cell or virus–cell) is probably a kinetic one. Moreover, there are three potential sites per envelope trimer in the cellular assays, but only one in our gp41-5 measurement. For both these reasons, competitive inhibition of the binding of the outer helix to gp41-5 should be a more stringent criterion of interaction than inhibition of fusion, and the spectrum of potential small-molecule fusion inhibitors may be broader than the range detected in our present assay. The limited solubility of some of the compounds, such as 6M007, and its variation with buffer, ionic strength, or other factors may also account for differences between cell-based and protein-based assays.
The likelihood of finding small, drug-like compounds that can block protein association is currently a subject of considerable debate. The coiled coil is a relatively special kind of protein interface, but our results suggest that suitably designed, sensitive, structure-based assays can indeed detect such interference and that a screen of relatively modest extent can uncover compounds with IC50 values in the low micromolar range. Understanding properly how compounds such as 5M038 inhibit C38 association with gp41-5 and, hence, how they interfere with viral fusion will require structures of suitable bound complexes. Although efforts to cocrystallize 5M038 with gp41 have thus far failed, it is already clear that a structure-based assay like the one described here can lead to detection of refolding inhibitors in chemical libraries.
Materials and Methods
Gp41-5 Cloning, Expression, and Purification.
We have expressed a single-chain model for five of the six gp41 helices (Fig. 2A). The construct contains residues 35–70 of HR1 and residues 117–150 of HR2 from HXB2. The amino acid sequence for HR1 is SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL, and the sequence for HR2 is WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL. The C terminus of each of the first two inner-core segments is connected to the N terminus of the succeeding outer-layer helix by the 6-residue linker SGGRGG. The C terminus of each outer-layer helix is connected to the N-terminus of the succeeding inner-core segment by the 6-residue linker GGKGGS. The DNA fragment encoding gp41-5 was subcloned into the expression vector pRSET (Invitrogen, Carlsbad, CA) and transformed into Escherichia coli cells BL21 DE3/pUBS. For purification, cell pellets were dissolved in cold (4°C) glacial acetic acid and incubated on ice for 30 min. Cell debris was removed by centrifugation (39,000 × g, 30 min). The supernatant was diluted to 10% acetic acid with deionized water and loaded onto a reversed-phase C18 column (Vydac, Hesperia, CA). The column was eluted with an acetonitrile gradient (30–90%). The protein eluted at 50% acetonitrile; it was >90% pure as judged by SDS/PAGE.
Gp41-5 Refolding.
Lyophilized protein was dissolved in 6 M guanidine HCl at a concentration of 1 mg/ml and dialyzed successively against 100 mM glycine, pH 3.5, and PBS, pH 7.4. The precipitate was removed by centrifugation, and the protein (>98% pure as judged by SDS/PAGE) was used without further purification.
Peptide Synthesis.
All peptides were synthesized by using Fmoc chemistry on PAL (PE Biosystems, Warrington, U.K.) supports by using an Applied Biosystems (Foster City, CA) model 431 peptide synthesizer. Peptides were cleaved by using reagent R [TFA/thioanisole/1,2-ethanedithiol (EDT)/anisole, 90:5:3:2], precipitated into cold diethyl ether and purified by reversed-phase C18 HPLC (0.1% TFA/acetonitrile gradient). All peptides were characterized by electrospray mass spectrometry at the Mass Spectrometry Facility of the Department of Chemistry and Chemical Biology at Harvard University (Cambridge, MA). Labeling of peptides at the N terminus was achieved as follows. Synthetic peptide, still attached to the resin and with side chains protected but N terminus deprotected, was suspended in a small volume of NMP. 5-FAM (Molecular Probes, Carlsbad, CA) was dissolved in NMP and added to the peptide suspension followed by the addition of 10 μl of 4-methyl morpholine. The reaction was allowed to proceed under slow stirring for ≈2 days. Cleavage and deprotection of labeled peptides were performed as described above.
High-Throughput Screening.
All screens were performed at the Harvard Medical School Institute for Chemistry and Chemical Biology (ICCB). Gp41-5 at a concentration of 7.4 nM in PBS/Tween 20 was loaded into 384-well microtiter plates (30-μl volume per well), followed by the transfer of 0.1 μl of compound (5 mg/ml in DMSO). After incubation for 1 h at room temperature, fluorescein-labeled C38 was added to each well (final concentration of 5–10 nM). After a 90-min incubation period at room temperature, fluorescence polarization measurements were recorded on an Analyst HTS (Molecular Devices, Sunnyvale, CA). A 12-h time point revealed no major differences from the 90-min time point. Compounds that appeared to be active were retested independently. Commercial libraries screened included those from CEREP (Redmond, WA), Maybridge (Trevillet, Tintagel, Cornwall, U.K.), Bionet (Ryan Scientific, Mount Pleasant, SC), and ChemBridge (San Diego, CA). The hits described in this work were from Bionet (the 5M series) and CEREP (S2986).
Cell–Cell Fusion Assay.
Assays were performed as described previously (23, 27) with the following modifications. Target and effector cells were mixed briefly at equal concentrations before transfer to 96-well plates. All inhibitors were dissolved in DMSO and diluted 100-fold in the final assay. After the addition of inhibitor, cells were incubated at 30°C for 6 h, a time point previously determined to be in the linear range of the assay. Assays were stopped by aspiration of media followed by addition of reporter lysis buffer (Promega, Madison, WI) and transfer to −20°C.
Viral Infectivity Assay.
Peripheral blood mononuclear cells (PBMCs) were exposed to HIV-1 isolate 2076 [1,000 tissue culture 50% infective dose (TCID50) per 106 cells] for 1 h. Cells were washed once with medium, and 0.1 ml of HIV-infected PBMCs (2 × 105 cells) were added to each well of a 96-flat-well cell plate followed by the addition of compound. The final concentration of DMSO in each well was 0.5%. Supernatants were harvested after 5 days and assayed for HIV-1 p24 antigen production. For time course assays, 106 MT-2 cells were cultured in 1 ml of RPMI medium 1640 with 10% FCS in individual wells of a 24-well plate. In selected wells, 5M038 or enfuvirtide was added to each well to achieve final concentrations of 30 or 50 μM for 5M038 or 50 ng/ml (11 nM) enfuvirtide. Each control or experimental condition was conducted in triplicate. After 1 h, 500 TCID50 HIV-IIIB was added, and cells were incubated for 7 days at 37°C in 5% CO2. Supernatant fluid (100 μl) was removed from each well on days 3, 5, and 7 for HIV-1 p24 antigen quantification. The medium was replaced with culture medium with the appropriate concentrations of inhibitor. HIV-1 p24 antigen was quantified by using kits according to the manufacturer's instructions (Perkin–Elmer, Wellesley, MA).
Gp41 Chimeras.
All chimeras were synthesized by using Fmoc chemistry and purified as described above. The sequence for peptide 1 was Ac-RMKQIEDKIEEIESKQKKIENEIARIKKLLSQIVQQQNNLLRAIEA-NH2. The sequence for peptide 2 was Ac-RMKQIEDKIEEIESKQKKIENEIARIKKLQNNLLRAIEAQQHLLQL-NH2. The sequence for peptide 3 (IQN17) was Ac-RMKQIEDKIEEIESKQKKIENEIARIKKLLQLTVWGIKQLQARIL-NH2. All peptides were analyzed by CD spectroscopy and analytical ultracentrifugation. Peptides 1 and 3 were fully helical; peptide 2 was >60% helical. Peptides 1 and 3 gave apparent molecular weights consistent with a trimer, whereas peptide 2 gave a slightly lower apparent molecular weight (11,000 as compared with the expected 16,782). We interpreted this last result to indicate a mixture of monomer and trimer. Size exclusion chromatography supported this interpretation.
Proton NMR Experiments.
All assays were performed on a Varian (Palo Alto, CA) 400-MHz spectrophotometer with a triple resonance probe. Samples were prepared in PBS/2H2O. Compound stocks were prepared in DMSO-d6 at 10 mg/ml (≈26 mM) and diluted in PBS/2H2O to a final concentration of 200 μM (final DMSO, <1%). Spectra were in the presence or absence of protein, also at 200 μM (based on Mr of trimer).
Acknowledgments
We thank Genfa Zhou (FusoGen Pharmaceuticals, Tianjin, China), who designed the original gp41-5 construct; Rebecca Ward, who facilitated all initial contacts with the Harvard Medical School Institute for Chemistry and Cell Biology; Tim Strassmeier and Daniel Oprian (Brandeis University, Waltham, MA) for help in the early stages of this work; Beatrice Hahn (University of Alabama, Birmingham, AL) for DNA encoding envelopes from a panel of HIV and SIV isolates; Caroline Shamu for advice on screening; and Jon Clardy for comments on the manuscript. The screens were carried out at the Harvard Medical School Institute for Chemistry and Cell Biology. This work was supported by Ruth L. Kirschstein National Research Service Award AI52859 (to G.F.), National Institutes of Health Program Project Grant GM39589 (to S.C.H.), and Centers for AIDS Research Grant AI36214 (to X.-Q.Z. and R.T.S.). S.C.H. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- HR
heptad repeat
- SIV
simian immunodeficiency virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harrison SC. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 3.Veronese FD, DeVico AL, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 4.Gallaher WR. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- 5.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 6.Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway SR, Jr, Leary J, Bugelski PJ. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sattentau QJ, Moore JP. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuta RA, Wild CT, Weng Y, Weiss CD. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 9.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh WC, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 10.Carr CM, Kim PS. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 12.Chan DC, Fass D, Berger JM, Kim PS. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 13.Fass D, Harrison SC, Kim PS. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 14.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 15.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild C, Greenwell T, Matthews T. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 17.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S, Lin K, Strick N, Neurath AR. Biochem Biophys Res Commun. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 19.Jiang S, Lin K, Strick N, Neurath AR. Nature. 1993:365–113. doi: 10.1038/365113a0. (lett) [DOI] [PubMed] [Google Scholar]
- 20.Kilby JM, Eron JJ. N Engl J Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 21.Chan DC, Chutkowski CT, Kim PS. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 23.Ferrer M, Kapoor TM, Strassmaier T, Weissenhorn W, Skehel JJ, Oprian D, Schreiber SL, Wiley DC, Harrison SC. Nat Struct Biol. 1999;6:953–960. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 24.Debnath AK, Radigan L, Jiang S. J Med Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- 25.Root MJ, Kay MS, Kim PS. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 26.Cianci C, Langley DR, Dischino DD, Sun Y, Yu KL, Stanley A, Roach J, Li Z, Dalterio R, Colonno R, et al. Proc Natl Acad Sci USA. 2004;101:15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nussbaum O, Broder CC, Berger EA. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]