Abstract
HIV-1 coreceptor usage plays a critical role in virus tropism and pathogenesis. A switch from CCR5- to CXCR4-using viruses occurs during the course of HIV-1 infection and correlates with subsequent disease progression. A single mutation at position 322 within the V3 loop of the HIV-1 envelope glycoprotein gp120, from a negatively to a positively charged residue, was found to be sufficient to switch an R5 virus to an X4 virus. In this study, the NMR structure of the V3 region of an R5 strain, HIV-1JR-FL, in complex with an HIV-1-neutralizing antibody was determined. Positively charged and negatively charged residues at positions 304 and 322, respectively, oppose each other in the β-hairpin structure, enabling a favorable electrostatic interaction that stabilizes the postulated R5 conformation. Comparison of the R5 conformation with the postulated X4 conformation of the V3 region (positively charged residue at position 322) reveals that electrostatic repulsion between residues 304 and 322 in X4 strains triggers the observed one register shift in the N-terminal strand of the V3 region. We posit that electrostatic interactions at the base of the V3 β-hairpin can modulate the conformation and thereby influence the phenotype switch. In addition, we suggest that interstrand cation-π interactions between positively charged and aromatic residues induce the switch to the X4 conformation as a result of the S306R mutation. The existence of three pairs of identical (or very similar) amino acids in the V3 C-terminal strand facilitates the switch between the R5 and X4 conformations.
Keywords: 447-52D, gp120, NMR
The third variable (V3) region of the HIV type 1 (HIV-1) envelope glycoprotein gp120 binds to chemokine receptors CCR5 and CXCR4, which are involved in HIV-1 infection. The amino acid sequence of V3 determines whether the virus binds to CCR5 (“R5 viruses”) and infects predominantly macrophages or to CXCR4 (“X4 viruses”) and infects mostly T cells (1). The presence of a basic residue at V3 positions 306 or 322 is associated with X4 and dual-tropic, X4R5 viruses, whereas the presence of a negatively charged residue and a neutral residue at positions 322 and 306, respectively, is correlated with R5 viruses (the “11/25 rule”) (2). Numerous investigations have confirmed that mutation of a negatively charged residue at position 322 to a positively charged one converts an R5 strain into an X4 strain (2–4).
To gain insight into the structure of the V3 region and the mechanism for phenotype conversion, we used solution NMR spectroscopy to study the conformation of synthetic V3 peptides in complex with V3-specific anti-gp120 antibodies. An assumption underlying this approach is that the native conformation of V3 is induced in linear V3 peptides upon binding to V3-directed antibodies that were elicited against the entire gp120 protein. We studied two V3-specific antibodies. The first, murine mAb 0.5β, is a potent strain-specific HIV-1-neutralizing antibody that was raised against a full-length gp120 protein of the X4 virus HIV-1IIIB (5). The second antibody, human mAb 447-52D, was derived from B cells of an HIV-1-infected donor and neutralizes a broad spectrum of R5 and X4 viruses. The phenotype of the virus that elicited the production of the 447-52D antibody is unknown.
Our previous NMR studies consistently revealed a β-hairpin conformation in V3 peptides bound to HIV-1-neutralizing antibodies. Among the V3 β-hairpin structures observed by NMR, two distinct N-terminal β-strand conformations were found that differed by 180° in side chain orientation (6) and by a one-register shift in the residues occupying the hydrogen bond-forming positions. We suggested that these structures represent two alternative conformations of the N-terminal V3 β-strand that create surfaces with different topology. These two conformations, together with possible involvement of the residues at positions 306 and 322 in coreceptor interactions, determine the selectivity of binding of gp120 to either CCR5 or CXCR4. Because 0.5β was raised against an X4 virus (5), we concluded that the conformation recognized by 0.5β represented the X4 conformation of V3 whereas 447-52D recognized the R5 conformation. Furthermore, a search of the structural database (www.rcsb.org/pdb) revealed a clear analogy between the alternative V3 conformations of R5 and X4 viruses and the β2,β3-hairpin of the CCR5 and CXCR4 chemokines, respectively (6). Similar to the X4 structure of V3 peptides, the conformation of the β2,β3-hairpin of stromal cell-derived factor 1 (SDF-1), a CXCR4 chemokine, differs from that of the corresponding hairpin in the CCR5 chemokines by a one-register shift in the pairing of residues resulting in a 180° difference in side-chain orientation, and a one-register shift in the hydrogen bond positions at the N-terminal strand. This homology between V3 structures and either CCR5 or CXCR4 chemokines supports our contention that the different V3 structures are relevant to virus selectivity.
The crystal structure of the gp120 core with the entire V3 region was solved recently by x-ray crystallography (7). Whereas the base and tip of the V3 loop adopt well defined β-strand and β-turn conformations, respectively (the latter of which agrees with results of our NMR studies), a large central segment of the C-terminal strand, as well as a short central segment of the N-terminal strand, is flexible. Residue 322 was found to be separated from residues 304 and 306 by Cα distances of 13 Å and 17 Å, respectively. These three residues, important for HIV-1 phenotype conversion, were located in flexible segments in the crystal structure.
The present NMR study of a V3JR-FL peptide (representing the consensus R5 sequence) bound to 447-52D reveals a well defined structure for residue E322 that is critical for phenotype conversion. In contrast to the recent crystal structure, the part of the V3JR-FL stem that is included in the NMR structure (most of the V3 stem) is not flexible, and the hairpin conformation places E322 directly opposite R304, suggesting a role for a cross-strand electrostatic interaction in determining the V3 conformation and phenotype conversion. Comparison of the V3JR-FL structure with that of a V3MN peptide bound to the same antibody suggests the involvement of interstrand diagonal cation-π interactions in the conformational switch of V3 when residue 306 is mutated to arginine.
Results
The Structure of the Bound V3JR-FL Peptide.
To determine the structure of the V3JR-FL peptide in complex with the Fv fragment of mAb 447-52D, the chemical shifts of 96% and 97% of the backbone and side chain atoms, respectively, were assigned by using multidimensional NMR techniques. NOE interactions characteristic of a β-hairpin conformation were detected between backbone atoms of the N- and C-terminal halves of the peptide and between the side chains of opposing residues (Table 2, which is published as supporting information on the PNAS web site). Five NOE interactions between the α, β, and γ protons of E322 and the α and β protons of R304 were unambiguously assigned, providing clear evidence that these two residues are close in space and allowing definition of the structure of a V3 segment that includes these two residues. 3JHNHα coupling constants >7.7 Hz, typical of a β-strand, were measured for T303, S306, I309, R315, F317-T319, and I323.
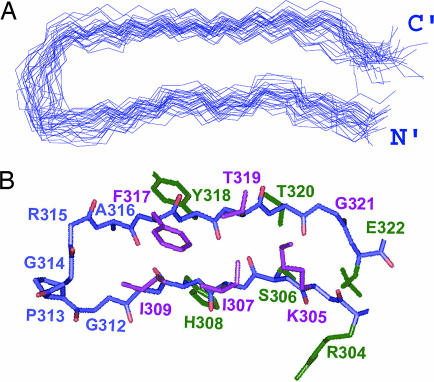
The structure of the V3JR-FL peptide bound to 447-52D Fv (PDB ID codes 2ESX and 2ESZ) was determined by using 308 NMR-derived distances, 2 hydrogen bond constraints, and 25 dihedral angles (Table 1). The backbone superposition of the 30 lowest energy structures of the bound peptide that satisfy the experimental restraints with no NOE violation >0.5 Å defines a β-hairpin consisting of two strands encompassing residues R304-I309 on the N side and F317-E322 on the C side (Fig. 1A). Most of these residues are in an extended but not ideal β-strand conformation. However, two short well defined anti-parallel β-strands are formed by residues S306-I307 and T319-T320. The side chains of residues R304, S306, H308, Y318, T320, and E322 form the lower face of the β-hairpin, whereas the side chains of K305, I307, I309, F317, and T319 form the upper face (green versus purple, respectively, in Fig. 1B). The structure of residues G312-R315 at the turn of the β-hairpin is not as well defined as the flanking strands. The rmsd values for the entire V3 epitope (R304-E322) were 0.85 Å and 1.82 Å for the backbone and all heavy atoms, respectively. The statistical data for the final set of structures are presented in Table 1. The Ramachandran plot (Fig. 6, which is published as supporting information on the PNAS web site) of the mean structure of the V3JR-FL peptide bound to 447-52D Fv shows that the ϕ and ψ angles of all non-glycine peptide residues except R304 occupy allowed regions.
Table 1.
NMR constraints and structural statistics for the refined structures of the 447–52D Fv-bound V3JR-FL peptide (30 structures)
| NMR distance constraints | |
| Total | 308 |
| Intra-residue | 180 |
| Sequential | 55 |
| Medium- and long-range | 73 |
| Dihedral angle | 25 |
| Hydrogen bond | 2 |
| NOE violations | |
| Maximum individual violation, Å | 0.5 |
| rmsd of NOE violation | 0.0108 ± 0.0031 |
| Deviation from ideal covalent geometry | |
| Bond lengths, Å | 0.0011 ± 0.0002 |
| Bond angles, ° | 0.3496 ± 0.0127 |
| Improper angles, ° | 0.1202 ± 0.0341 |
| Mean rmsd values, Å | |
| All backbone atoms | 0.85 |
| All heavy atoms | 1.82 |
Fig. 1.
Solution structure of a V3JR-FL peptide bound to the 447-52D Fv. (A) Backbone superposition of the 30 lowest-energy structures of 304–322gp120JR-FL. (B) Stick representation of 304–322gp120JR-FL bound to the 447-52D Fv. Side chains pointing out from the page are colored purple, side chains pointing inward are colored green, and side chains of the loop residues are colored blue.
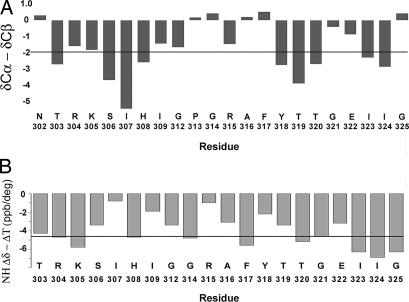
The presence of a β-hairpin conformation is additionally supported by the deviations of the Cα and Cβ chemical shifts from their random coil values, ΔCα and ΔCβ, respectively. To obtain a value that is independent of the reference chosen to calibrate the 13C chemical shift, we calculated the difference ΔCα − ΔCβ (8). This difference was more negative than 2 ppm for T303, S306, I307, and H308 at the N-terminal strand and for Y318, T319, T320, I323, and I324 at the C-terminal strand (Fig. 2A), indicating that these two strands are mostly in an extended conformation. Some deviations are observed in the N-terminal (R304 and K305) and C-terminal (G321 and E322), strands probably the result of the kink at G321 observed in the structure of the V3JR-FL peptide bound to 447-52D (Fig. 1). The two strands are linked by the five residue loop constituting G312-A316. The deviations of the Cα and Cβ chemical shifts of T303, I323, and I324 from random coil values and the large 3JNH coupling constants of T303 and I323 residues indicate that the backbone of the V3 is probably in an extended structure also for the residue preceding R304 and the two residues following E322.
Fig. 2.
The difference between the deviations of δCα and δCβ from random coil values (ΔCα-ΔCβ) and temperature coefficients for each of the V3JR-FL residues. (A) (ΔCα-ΔCβ) are represented by vertical bars. The horizontal line denotes a threshold value of −2 ppm. Consecutive residues with values less than −2 ppm indicate existence of a β-strand conformation (8). (B) The temperature coefficients of each residue are displayed as bars. The horizontal line represents a threshold value of −4.6 ppb/K. Values greater than −4.6 ppb/K are indicative of protection from solvent exchange. Residues N302 and P313 are not included in B.
Hydrogen Bonds.
Very low temperature coefficients (more positive than −2 ppb/K) were measured for the amide protons of I307, I309, and R315, providing strong evidence that these form hydrogen bonds (Fig. 2B). Such hydrogen bonds would be consistent with those found in the crystal structure of a V3MN peptide in complex with 447-52D Fab (9). The V3JR-FL and V3MN peptides share the same N-terminal strand conformation when bound to 447-52D (see below). In the crystal, MNI307 and MNI309 formed hydrogen bonds with residues of the antibody CDR3, creating an intermolecular β-sheet, and MNR315 amide-hydrogen bonded with the carbonyl oxygen of MNG312, stabilizing the reverse-turn formed by the GPGR segment. Several other amide protons exhibit temperature coefficients more positive than −4.6 ppb/K, including S306, G312, A316, Y318, T319, and E322, indicative of partial protection from the solvent and possible involvement in hydrogen bonds.
Comparison Among the Structures of V3JR-FL, V3IIIB, and V3MN.
The structures of V3JR-FL and V3IIIB bound to 447-52D (the latter of which we previously postulated was the R5 conformation of V3) (10) are similar in the pairing of the β-hairpin residues, the side chain orientation, and the register of the hydrogen bond-forming positions in both the N- and C-terminal strands (Fig. 3A and B). Intriguingly, the overall structure of V3JR-FL bears greater similarity to that of V3IIIB bound to 447-52D Fv than to V3MN bound to the same Fv (structure not shown). This finding is surprising, given the 93% sequence identity between V3JR-FL and V3MN in K305-T320 vs. only 71% sequence identity between V3JR-FL and V3IIIB in the same segment and the two-residue insertion in V3IIIB in comparison with V3JR-FL and V3MN. Whereas the structures of the N-terminal strands of both V3JR-FL and V3MN bound to 447-52D are similar, the two peptides differ by a one register shift in the pairing of the residues, a 180° difference in the side chain orientation and in the hydrogen bond positions in the C-terminal strand. As a result, the backbone rmsd between V3JR-FL and V3IIIB bound to 447-52D is 1.28 Å when the segments K305-I309 and F317-T320 are compared and 1.95 Å when the corresponding segments in V3JR-FL and V3MN bound to 447-52D are compared.
Fig. 3.
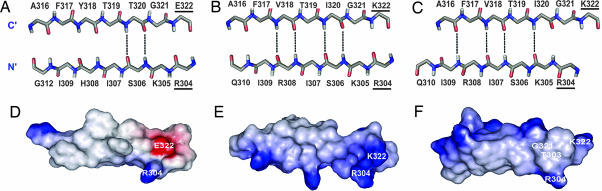
The influence of the sequence and residue-pairing in the V3 β-hairpin on the electrostatic potential at the base of the hairpin. (A) The residue pairing and hydrogen bonds in the schematic structure of V3JR-FL bound to 447-52D Fv. (B) The residue pairing and hydrogen bonds in the schematic structure of the V3IIIB bound to the 447-52D Fv. (C) The residue pairing and hydrogen bonds in V3IIIB bound to 0.5β Fv. Hydrogen bonds are indicated by black dashed lines. (D–F) Electrostatic potential map calculated by the DELPHI program in different V3 structures: V3JR-FL bound to 447-52D Fv (D) and V3IIIB bound to 447-52D Fv (E). Residues R304 and K322 were modeled according to the conformation of V3JR-FL bound to 447-52D Fv (the R5 conformation of V3). (F) V3IIIB bound to 0.5β Fv; residues T303 and K322, not included in the NMR structure, were modeled. The charged residues involved in the switch between the R5 and X4 conformation are underlined. Positive potential is shown in blue, and negative potential is shown in red.
Electrostatic Potential of V3 Conformations.
The region of the Fv-bound V3JR-FL for which we obtained a well defined structure includes residues R304 and E322, the latter of which is known to be critical for phenotype conversion. These residues were not ordered in any of the three structures of antibody-bound V3 peptides that we previously reported (6, 10, 11). The short distance, 4.6 Å, between the positively charged guanidinium proton of R304 and the negatively charged oxygen of the E322 carboxyl suggests the existence of an electrostatic interaction between these two residues. The electrostatic attraction between R304 and E322 in V3JR-FL is most likely responsible for a well defined structure for these residues in the V3JR-FL/447-52D Fv complex despite the absence of any detected interactions between these residues and 447-52D Fv.
To analyze the impact of the electrostatic interaction between residues 304 and 322 on the conformation of V3, we calculated the electrostatic potential of (i) V3JR-FL bound to 447-52D Fv, (ii) V3IIIB bound to 447-52D Fv, and (iii) V3IIIB bound to 0.5β Fv, using the DELPHI module of InsightII (Accelrys, San Diego, CA). The first two V3 structures correspond to the postulated R5 conformation, even though in terms of sequence, only V3JR-FL has an R5 V3 sequence (whereas V3IIIB has an X4 sequence). The third V3 structure is the postulated X4 structure of this region of gp120. Because the NMR structure of V3IIIB bound to 447-52D (10) did not include either IIIBK322 or IIIBR304 and the NMR structure of V3IIIB bound to 0.5β Fv (11) did not include IIIBT303 and IIIBK322, we “extended” the structure of V3IIIB bound to 447-52D and that of V3IIIB bound to 0.5β by one residue at each strand by modeling V3IIIB residues R304 and K322 in the first complex (Fig. 3 B and E) and V3IIIB residues T303 and K322 in the second complex (Fig. 3 C and F).
Inspection of these structures reveals that, when an R5 V3 sequence adopts an R5 conformation, as found in the present study, a favorable electrostatic interaction can occur between R304 and E322, which oppose each other in the R5 β-hairpin conformation (Fig. 3 A and D). This electrostatic interaction can stabilize the β-hairpin conformation and dictate the residue-pairing across the two β-strands. However, when V3IIIB, an X4 peptide, is in the R5 conformation, as shown in Fig. 3 B and E, the two modeled positively charged residues IIIBR304 and IIIBK322 oppose and electrostatically repel each other. The repulsion between R304 and K322 in V3IIIB bound to 447-52D probably resulted in increased flexibility and poor definition of the structure of these residues in the V3IIIB/447-52D Fv complex. In contrast, when V3IIIB adopts the postulated X4 V3 conformation (Fig. 3C), because of the one-register shift in the N-terminal strand, IIIBR304 opposes IIIBG321, and IIIBK322 opposes IIIBT303 (data not shown in Fig. 3C), partially alleviating the electrostatic repulsion between the side chains of the positively charged residues. This reduction in electrostatic repulsion is aided by these side chains pointing in different directions (Fig. 3F).
Electrostatic Potential in Chemokines and the Conformational Switch.
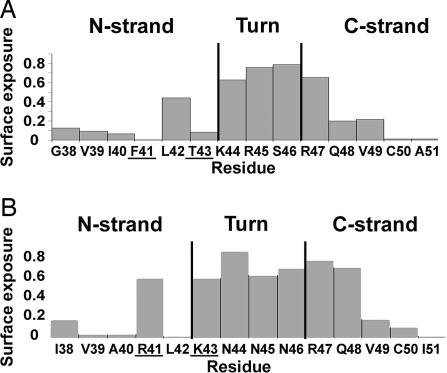
Although the CCR5 chemokines macrophage inflammatory protein (MIP) 1α, MIP-1β, and RANTES (regulated upon activation, normal T cell expressed and secreted) and the CXCR4 chemokine SDF-1 share very similar 3D structures, they differ in the conformation of the β2,β3-hairpin in analogy with the dual β-hairpin conformations of the V3 region (6). We suggest that the difference between the CCR5 chemokines and SDF-1 is mostly due to replacement of F41 and T43 in the CCR5 chemokines with arginine and lysine, respectively, in SDF-1. F41 and T43 are buried in the CCR5 chemokines (Fig. 4A). To prevent the occlusion of the two positively charged residues at these positions in SDF-1, a one-register shift occurs in the N-terminal strand with respect to the C-terminal strand that results in a 180° difference in the orientation of the side chains, exposing the side chains of R41 and K43 in SDF-1 (Fig. 4B). Interestingly, A40, R41, and L42 undergo significant changes in chemical shift when SDF-1 binds to a peptide corresponding to an N-terminal segment of CXCR4, suggesting that these residues of SDF-1 are involved in interactions with CXCR4 (12). Thus, it is likely that, in the chemokines as well as in gp120, the change in specificity is contributed both by mutations in interacting residues as well as a topological change in a region involved in receptor binding.
Fig. 4.
Surface exposure of the β2-β3 hairpin in macrophage inflammatory protein (MIP) 1α (A) and SDF-1 (B). Surface exposure was calculated by using the 3D structures of these proteins [PDB ID codes 1B52 and 1SDF, respectively (20, 21)].
Discussion
Molecular Switch for Phenotype Conversion.
The NMR structure of the V3JR-FL peptide bound to 447-52D reveals the structures of residues R304 and E322. Unequivocal NOE connectivities show that the side chains of these two residues are in close proximity. This finding contrasts with the crystal structure of a V3-containing gp120 core in complex with CD4 and the anti-gp120 X5 antibody-Fab, which showed the Cα atoms of these two residues to be separated by 13 Å, vs. 4.8 Å in the NMR structure. Our structural findings led us to suggest that an electrostatic interaction between the residue at position 322 and the conserved arginine at position 304 is responsible for the switch between the R5 and X4 V3 conformations. Thus, HIV-1 phenotype conversion arises not only from mutations of residues that possibly interact with the coreceptor but also from a switch in the V3 conformation mediated by mutation to positively charged residues at position 322 that is known to be critical for viral selection (2).
The electrostatic attraction between R304 and E322 in V3JR-FL stabilizes the β-hairpin conformation and determines the pairing of the residues in the V3 loop. Indeed, recent NMR measurements of β-hairpin stability have shown that a salt bridge between lysine and glutamate contributes 1.2–1.3 kJ·mol−1 to the stability of β-hairpins in short synthetic peptides dissolved in water (13). Molecular dynamic simulations of two β-hairpins from protein G showed that a salt bridge between their termini provides stabilization of 5.5 kJ·mol−1 (14). Moreover, in a site-directed mutagenesis study of the stability of parallel β-strands, the greatest stabilization came from electrostatic interactions (15).
Mutation from a negatively to a positively charged residue at position 322, known to be sufficient for phenotype conversion (3), will cause repulsion between residues 304 and 322 and destabilization of the postulated R5 conformation. This repulsion is alleviated by the one-register shift of the N-terminal strand relative to the C-terminal strand that we observed in the postulated V3 X4 conformation manifested by the structure of a V3IIIB peptide bound to the 0.5β Fv (Fig. 3 C and F).
Phenotype Switch by the S306R Mutation.
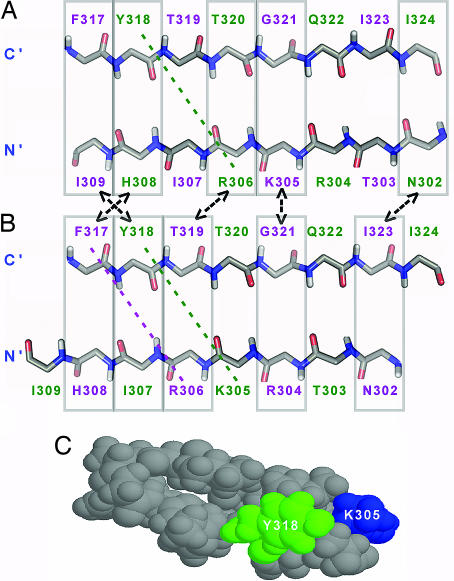
The 11/25 rule suggests that simultaneous mutations of S306 to arginine and E322 to a neutral residue result in a shift in HIV-1 phenotype (2). These two mutations create a segment of three consecutive positively charged residues (R304, K305, and R306) in the N-terminal strand of the V3 β-hairpin (Fig. 5A and B) with no obvious negative charges on the C-terminal strand that could form stabilizing electrostatic interactions. However, it is well known that the π-electrons of the aromatic amino acids tryptophan, tyrosine, and phenylalanine can interact with cationic centers in lysine and arginine. A one-register shift of the N-terminal strand as occurs in the postulated X4 conformation would enable two such diagonal cation-π interactions between K305 and Y318 and between R306 and F317 (Fig. 5B), versus only one cation-π interaction that is possible in the R5 conformation (between R306 and Y318)(Fig. 5A), thereby stabilizing the X4 conformation and favoring the phenotype conversion. Diagonal cross-strand interactions are between residues whose side chains point in the same direction but are not directly opposite each other. Such cation-π interactions have been found to contribute significantly to the stability of the β-hairpin conformation in designed β-hairpins (16, 17) and are possible as a result of the right handed twist that is usually observed in β-hairpins and β-sheets (16).
Fig. 5.
A V3 conformational switch that enhances interstrand cation-π interactions explains phenotype conversion associated with the S306R mutation. Schematic representations of the two suggested β-hairpin structures of R5 (A) and X4 (B) are shown. The presented structures have two point mutations, S306R and E322Q, that according to the 11/25 rule switch the R5 to an X4 conformation (2). Possible diagonal cation-π interactions in the suggested conformations are marked by diagonal colored dashed lines. Similar interstrand pairs are marked by rectangles connected by two-headed arrows. Residues pointing out of the page are purple; residues pointing inward are colored green. (C) CPK representation of the structure of the V3MN peptide bound to the 447-52DFv (6) showing diagonal side-chain–side-chain interactions between residue Y318 (green) and K305 (blue).
Support for the above explanation comes from the structure of V3MN bound to 447-52D, which, surprisingly, does not adopt the postulated R5 conformation at the C-terminal strand despite 93% sequence identity with V3JR-FL in the epitope recognized by 447-52D (gp120305–320). The only difference between V3JR-FL and V3MN in this segment is the mutation S306R in V3MN. Previous NMR findings on the V3MN peptide bound to 447-52D (6) included an NOE interaction between the side chains of K305 and Y318, supporting their proximity in the structure of the V3MN peptide bound to 447-52D Fv (Fig. 5C). This interaction may indicate a cation-π interaction between the lysine and tyrosine residues. Existence of cation-π interaction involving residue 306 would explain the surprising difference in the C-terminal strand conformation between V3MN and V3JR-FL bound to the same antibody.
Pairs of Identical or Similar Residues in the C-Terminal Strand.
Frequently the sequence of β-strands is characterized by the motif PHPH in which P represents a polar residue and H represents a hydrophobic residue (18). This motif exists in the N-terminal strands of many V3 regions as the sequence SIHI. However, the consensus sequence of the C-terminal strand of R5 viruses contains two pairs of identical residues (T319 and T320, I323 and I324) and one pair of similar residues (F317 and Y318) (these residues have a high propensity to assume β-sheet structures and likely favor β-strand formation by this region of V3). As a result of the residue repeats in the V3 primary sequence, 62% of the pairs of opposing residues in the X4 and R5 conformations of V3 are identical or very similar, e.g., Y318/H308, T320/R306, G321/K305, I324/N302, and F317/I309 in the R5 conformation, versus F317/H308, T319/R306, G321/R304, I323/N302, and Y318/I307 in the X4 conformation (Fig. 5 A versus B). Although the order of the pairs differs, the overall identity or similarity of cross-strand interactions in the two topologies of the β-hairpin may result in a small energy difference between the R5 and the X4 conformations of V3, especially when residues 306 and 322 are neutral, and thereby facilitate interconversion between the two conformations. The flexibility of the GPGR turn, because of the two glycine residues, further facilitates the conformational switch in the V3 β-hairpin. This sequence-dictated conformational flexibility may help explain the existence of dual tropic viruses.
Cross-Reactivity of 447-52D with the R5 and X4 Conformation.
A question arises as to how 447-52D can interact with both R5 and X4 V3 regions if these adopt two different β-hairpin conformations. We suggest that the conformational flexibility of V3 may enable 447-52D to impose the R5 conformation on an X4 virus as long as the V3 sequence contains the conserved triad K305, I307, and I309, which interact extensively with this antibody (10), as is the case for V3JR-FL (Fig. 7, which is published as supporting information on the PNAS web site). The absence of 447-52D interactions with residue 322 (Fig. 7) means that this antibody should be “insensitive” to the nature of the amino acid at this position. In addition the existence of five consecutive tyrosine residues in the third complementarity determining region (CDR3) of the heavy chain of 447-52D leads to similar backbone/backbone and side-chain/side-chain interactions with both the R5 and X4 β-hairpin conformation.
Comparison with the Crystal Structure of a gp120-core That Includes V3.
The NMR structure of the V3JR-FL peptide differs from that in the crystal structure reported by Huang et al. (7). Most notably, residues R304 and E322 were found to be separated by a Cα distance of 13 Å in the crystal structure (7) vs. a Cα distance of 4.8 Å in the NMR structure of V3JR-FL peptide bound to 447-52D Fv. This discrepancy may reflect the flexibility of the V3 stem, which is most pronounced for the C-terminal segment Y318-E322, as manifested by high B values reported for this region of the crystal structure (7). This flexibility could result from the remarkable protrusion of the long V3 region from the gp120-core molecules lacking the V1 and V2 regions and almost all carbohydrates (which constitute 50% of the gp120 molecule). As a result of the V3 flexibility in the system used in the crystallographic studies, the observed involvement of V3 in crystal packing interactions with the X5 Fab fragment and with adjacent molecules in the crystal lattice (7) could easily affect the V3 conformation.
Antibody 447-52D was elicited in the course of natural infection and most likely recognizes a native conformation of the V3 region. Whereas most probably 447-52D was generated against a gp120 conformation before CD4 binding, the conformation of gp120 relevant for interaction with the coreceptor is induced only after CD4 binding. However, the conformation of the V3 recognized by anti-V3 antibodies must be one that remains similar before and after CD4 binding because CD4 does not inhibit binding of such antibodies to gp120. On the contrary, in a few cases, CD4 binding enhanced the binding and neutralization of HIV-1 by V3-specific antibodies by increasing exposure of V3. Thus, we conclude that the NMR structure of the R5 V3 in complex with 447-52D Fv is relevant both for the rational design of HIV-1 immunogens and for understanding the mechanism underlying the phenotype switch. The idea that the alternative V3 conformations observed by NMR correspond to the R5 and X4 conformations of V3 and the β-hairpin structure of V3 enables us to explain the role of residues 306 and 322 in phenotype conversion and the 11/25 rule (2). Specifically the proximity of R304 and E322 in the NMR structure suggests an electrostatic interaction that stabilizes the R5 conformation of the V3 region. Mutation to a positively charged residue at position 322 (V3 position 25) will trigger the switch to the X4 V3 conformation to alleviate the repulsion between the positively charged residues at positions 304 and 322. In the case of the S306R mutation (at V3 position 11), the conformational switch will enhance interstrand cation-π interactions.
Experimental Procedures
The 23-residue peptide, NNTRKSIHIGPGRAFYTTGEIIG, corresponding to residues N301–G325 of the gp120 of HIV-1JR-FL strain and the consensus V3 sequence of clade-B R5 viruses, was expressed as a fusion protein in Escherichia coli, and purified as described (6). The 447-52D Fv was expressed in the BL21(DE3)pLysS strain as described (19). The peptide-Fv complex was prepared as described (10). All samples contained 10 mM d4-acetic acid buffer at pH 5, and 0.02% NaN3 was added as preservative (some of the samples contained 0.005% Thimerosal instead of NaN3). For measurements in D2O, the lyophilized peptide/Fv complex was dissolved in 99.99% D2O. NMR experiments and structure calculations were done as described.
Supplementary Material
Acknowledgments
We thank Dr. Zolla-Pazner (New York University, New York, NY) for a most fruitful and exciting collaboration and for the 447-52D cell line, Dr. Naama Kessler for the expression of 447-52D Fv, Dr. Jordan Chill for help with the NMR experiments, Mrs. Rina Levy and Mr. Yehezkiel Hayek for excellent technical assistance, Dr. Fred Naider for critical reading of the manuscript, and Dr. Sandy Livnat for editorial assistance. This research was supported by National Institutes of Health Grant GM 53329 (to J.A.). J.A. is the Joseph and Ruth Owades Professor of Chemistry.
Abbreviation
- SDF-1
stromal cell-derived factor 1.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2ESX (average structure) and 2ESZ (ensemble)].
References
- 1.Dittmar MT, McKnight A, Simmons G, Clapham PR, Weiss RA, Simmonds P. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 2.Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, Miedema F, Schuitemaker H. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resch W, Hoffman N, Swanstrom R. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 5.Matsushita S, Maeda H, Kimachi K, Eda Y, Maeda Y, Murakami T, Tokiyoshi S, Takatsuki K. AIDS Res Hum Retroviruses. 1992;8:1107–1115. doi: 10.1089/aid.1992.8.1107. [DOI] [PubMed] [Google Scholar]
- 6.Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Structure (London) 2003;11:225–236. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 7.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, et al. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Eghbalnia HR, Bahrami A, Markley JL. J Biomol NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- 9.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structure (Cambridge, MA) 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Rosen O, Chill J, Sharon M, Kessler N, Mester B, Zolla-Pazner S, Anglister J. Biochemistry. 2005;44:7250–7258. doi: 10.1021/bi047387t. [DOI] [PubMed] [Google Scholar]
- 11.Tugarinov V, Zvi A, Levy R, Hayek Y, Matsushita S, Anglister J. Structure Fold Des. 2000;8:385–395. doi: 10.1016/s0969-2126(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 12.Gozansky EK, Louis JM, Caffrey M, Clore GM. J Mol Biol. 2005;345:651–658. doi: 10.1016/j.jmb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Ciani B, Jourdan M, Searle MS. J Am Chem Soc. 2003;125:9038–9047. doi: 10.1021/ja030074l. [DOI] [PubMed] [Google Scholar]
- 14.Tsai J, Levitt M. Biophys. Chem. 2002;101–102:187–201. doi: 10.1016/s0301-4622(02)00198-9. [DOI] [PubMed] [Google Scholar]
- 15.Merkel JS, Sturtevant JM, Regan L. Structure Fold Des. 1999;7:1333–1343. doi: 10.1016/s0969-2126(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 16.Syud FA, Stanger HE, Gellman SH. J Am Chem Soc. 2001;123:8667–8677. doi: 10.1021/ja0109803. [DOI] [PubMed] [Google Scholar]
- 17.Tatko CD, Waters ML. Protein Sci. 2003;12:2443–2452. doi: 10.1110/ps.03284003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenberg D, Weiss RM, Terwilliger TC. Proc Natl Acad Sci USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler N, Zvi A, Ji M, Sharon M, Rosen O, Levy R, Gorny M, Zolla-Pazner S, Anglister J. Protein Expr Purif. 2003;29:291–303. doi: 10.1016/s1046-5928(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 20.Czaplewski LG, McKeating J, Craven CJ, Higgins LD, Appay V, Brown A, Dudgeon T, Howard LA, Meyers T, Owen J, et al. J Biol Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 21.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.