Abstract
Lysine-specific demethylase 1 (LSD1) was recently identified as the first histone demethylase that specifically demethylates monomethylated and dimethylated histone H3 at K4. It is a component of the CoREST and other corepressor complexes and plays an important role in silencing neuronal-specific genes in nonneuronal cells, but the molecular mechanisms of its action remain unclear. The 2.8-Å-resolution crystal structure of the human LSD1 reveals that LSD1 defines a new subfamily of FAD-dependent oxidases. The active center of LSD1 is characterized by a remarkable 1,245-Å3 substrate-binding cavity with a highly negative electrostatic potential. Although the protein core of LSD1 resembles other flavoenzymes, its enzymatic activity and functions require two additional structural modules: an N-terminal SWIRM domain important for protein stability and a large insertion in the catalytic domain indispensable both for the demethylase activity and the interaction with CoREST. These results provide a framework for further probing the catalytic mechanism and the functional roles of LSD1.
Keywords: histone modification, flavoenzyme, catalysis
Histone proteins are subject to a variety of posttranslational modifications, including acetylation, methylation, phosphorylation, and ubiquitination, and it is these histone modifications that function as the molecular switches that alter the state of compaction of chromatin to allow gene activation or repression (1–3). Some histone modifications (e.g., acetylation and phosphorylation) are highly dynamic, whereas others (e.g., methylation) have been regarded as “permanent” chromatin marks. However, the discoveries of lysine-specific demethylase 1 (LSD1) and jumonji domain C (JmjC) domain-containing histone demethylase 1 (JHDM1) have changed this picture (4, 5). Shi and colleagues demonstrated that LSD1 is a histone lysine demethylase that specifically demethylates monomethylated and dimethylated histone H3 at K4 (4). More recently, we and others have shown that many JmjC domain-containing proteins are capable of demethylating dimethylated and trimethylated histone proteins (4–10). These findings suggest that histone methylation is a reversible modification and can be regulated under similar enzymatic control as other histone modifications (11–13).
LSD1 is a component of a number of transcriptional corepressor complexes, such as CoREST, CtBP, and HDAC complexes, and plays an important role in silencing neuronal-specific genes in nonneuronal cells (14–19). The C-terminal two-thirds of LSD1 contains an amine oxidase-like (AOL) domain, which shares extensive sequence homology to FAD-dependent oxidases (Fig. 1A and C) (20–22). In addition, LSD1 also contains an N-terminal SWIRM domain (Fig. 1A), which has been recently identified as a conserved motif often found in chromatin remodeling and modifying complexes with unknown function (23). Recent studies suggest that the specificity and activity of LSD1 can be modulated by its interacting factors (4, 24–26). However, the molecular mechanism by which this regulation is achieved remains unclear.
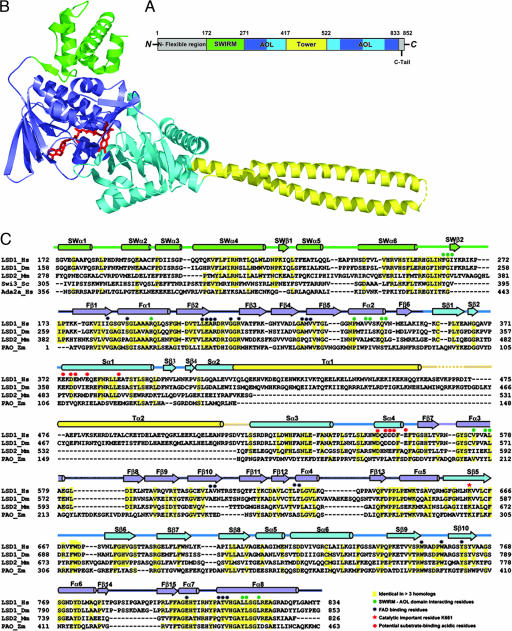
Fig. 1.
Overview of the LSD1 structure. (A) Domain organization of LSD1. The SWIRM domain is shown in green, the AOL domain is in blue (the FAD-binding subdomain) and cyan (the substrate-binding subdomain), and the Tower domain is in yellow. The N-terminal flexible region and the C-terminal tail that are not included in the structure determination are colored in gray. (B) Ribbon diagram of the LSD1 structure. The molecule is colored as in A. FAD is in ball-and-stick representation and is colored in red. (C) Structural-based sequence alignments of the SWIRM and AOL domains of LSD1 and its homologs. Secondary structure assignments from the LSD1 crystal structure are shown as cylinders (α-helices) and arrows (β-strands) above the aligned sequences and colored as in B.
Here we report the crystal structure of LSD1 at 2.8-Å resolution (Fig. 1). The x-ray analysis reveals that the overall structure of LSD1 closely resembles those of flavin-dependent oxidases (22) and contains a marked large catalytic cavity with a highly negative electrostatic potential. We further demonstrate that a large insertion in the catalytic domain is indispensable both for the demethylase activity of LSD1 and the interaction between LSD1 and CoREST.
Results
The Overall Structure of LSD1.
A recombinant protein of human LSD1 with an N-terminal deletion, LSD1ΔN (residues 172–833) (Fig. 1A), was prepared by overexpression and purification from Escherichia coli. LSD1ΔN was chosen for structural study because the N-terminal region of LSD1 was confirmed to be dispensable for LSD1 activity and is an unstructured region (27). For simplicity, we hereafter use LSD1 to represent LSD1ΔN unless stated otherwise. The purified LSD1 was crystallized in space group P6122, and the structure was determined by multiwavelength anomalous dispersion method. The crystallographic refinement resulted in a model with Rwork and Rfree values of 23.0% and 28.3%, respectively (Table 1, which is published as supporting information on the PNAS web site).
The structure of LSD1 contains three domains. Two of them, the SWIRM (residues 172–270) and AOL (residues 271–417 and 523–833) domains, pack together through extensive interactions resulting in a globular structure (Fig. 1 A and B). The SWIRM domain consists mostly of α-helices and is a newly identified structural module often found in chromatin-associated proteins (Fig. 1 B and C) (28–30). The AOL domain folds into a compact structure that exhibits a topology found in several flavin-dependent oxidases (Fig. 1B) (22, 31). The AOL domain contains a large insertion (residues 418–522) that forms an additional domain and adopts a tower-like structure (Tower domain) protruding away from the AOL domain by 75 Å (Fig. 1 B and C). The Tower consists of a pair of long helices that adopts a typical antiparallel coiled-coil conformation (Fig. 1C).
The SWIRM Domain.
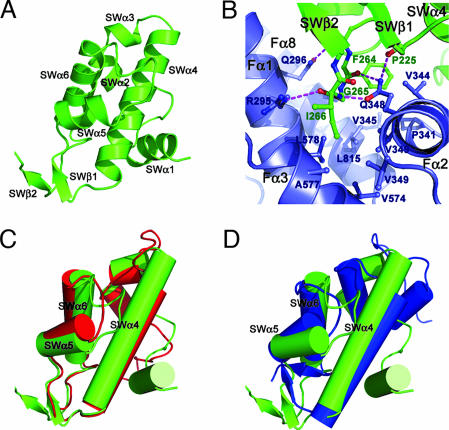
The SWIRM domain of LSD1 reveals a six-helical bundle architecture characterized by a long helix, SWα4, in the center surrounded by five other helices (Fig. 2A). One unique feature of the LSD1 SWIRM domain is an additional two-stranded β-sheet formed between the SWα4–SWα5 loop and the C terminus of the SWIRM domain (Figs. 1C and 2A). This short β-sheet helps anchor the interactions between the SWIRM and AOL domains by forcing the SWα6–SWβ2 loop to protrude out into a hydrophobic pocket formed by helices Fα1, Fα2, Fα3, and Fα8 of the AOL domain (Fig. 2B). The LSD1 SWIRM domain is structurally most similar to those of the chromatin-remodeling proteins Swi3 and ADA2α (Fig. 2 C and D). The three SWIRM domains have very similar hydrophobic core packings. The differences among the three structures are primarily confined to SWα1 and SWα2, which are not conserved and are positioned further away from the hydrophobic core (Fig. 2 C and D).
Fig. 2.
The structure of SWIRM domain and its interactions with the AOL domain. (A) Ribbon diagram of the SWIRM domain. (B) A three-amino-acid motif (F264G265I266) of the SWIRM domain mediates the interactions between the SWIRM and AOL domains. The SWIRM and AOL domains are colored in green and blue, respectively. Side chains of residues important for the interactions are shown explicitly. The hydrogen bonds are shown as dotted purple lines. (C and D) Superposition of the LSD1 SWIRM domain (green) with those of Swi3 (red) and Ada2α (blue).
The SWIRM–AOL Domain Interaction.
The SWIRM domain packs closely against the AOL domain and is located far away from both the FAD-binding site and the catalytic center of LSD1 (Fig. 1B). A three-amino-acid motif (F264G265I266) between SWα6 and SWβ2 anchors the binding through extensive interactions with the AOL domain, involving both hydrophobic contacts and hydrogen-bonding interactions (Fig. 2B). The side chains of F264 and I266 and the main chain Cα atom of G265 stack against a hydrophobic pocket from the AOL domain. These hydrophobic contacts are buttressed by extensive hydrogen-bonding interactions between the backbone of this tri-residue motif and the side chains of R295 and Q348. The SWIRM–AOL packing buries a total surface area of 1,115 Å2, and the residues that form the interface are highly conserved across species (Fig. 1C). Based on these observations, we conclude that the prearrangement of the SWIRM and AOL domains is important for LSD1 protein functions. Although the SWIRM domain can be expressed as a separate protein with native conformation (28), efforts to prepare the AOL and Tower domains of LSD1 yielded insoluble protein (data not shown), suggesting that it requires an interface with the SWIRM domain for stability.
The AOL Domain.
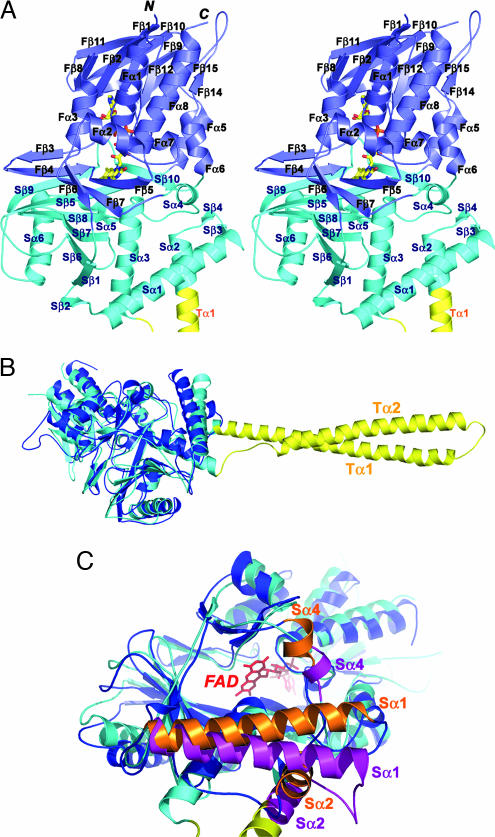
The AOL domain of LSD1 folds into two well defined subdomains, an FAD-binding subdomain and a substrate-binding subdomain, with their sequences intermingled together (Figs. 1C and 3A). The FAD-binding subdomain has three fragments (residues 271–356, 559–657, and 770–833) and adopts a mixed α–β structure (Fig. 3A). As is evident from the high degree of sequence conservation, the binding mode and the conformation of the FAD in the LSD1 structure closely resemble those observed in the enzymes sharing the dinucleotide-binding fold (Fig. 6, which is published as supporting information on the PNAS web site) (22, 32–34). The substrate-binding subdomain comprises three fragments (residues 357–417, 523–558, and 658–769) and is characterized by a six-stranded mixed β-sheet flanked by six α-helices (Fig. 3A). The two subdomains create a big cavity that defines the enzyme activity center at their interface (Fig. 3 A and C).
Fig. 3.
The structure of the AOL domain. (A) Stereo ribbon diagram of the AOL domain of LSD1. The molecule is colored as in Fig. 1B. The FAD is in ball-and-stick representation with carbon colored yellow, nitrogen colored blue, oxygen colored red, and phosphorus colored orange. (B) Superposition of LSD1 (the AOL and the Tower domains) on the structure of PAO. LSD1 is in cyan, and PAO is in blue. The Tower domain of LSD1 is colored in yellow. (C) Superposition of the AOL domain of LSD1 on the structure of PAO highlighting the differences in the catalytic cavity. LSD1 is in cyan and purple, and PAO is in blue and orange. The Tower domain of LSD1 is colored in yellow. The FAD, in red and ball-and-stick representation, lies in the back of the cavity. The positional change of three α-helices (Sα1, Sα2, and Sα4) of PAO relative to those of LSD1 significantly reduces the volume of the catalytic cavity.
The AOL domain of LSD1 is most similar to the structure of maize polyamine oxidase (PAO). The two structures can be superimposed with a 2.9-Å rmsd in the positions of 419 of 462 Cα atoms (Figs. 1C and 3B). Although the similarity is evident in the FAD-binding subdomain, the structure of LSD1 substrate-binding subdomain differs from that of PAO in several aspects. First, LSD1 comprises a long insertion between Sα2 and Sα3 compared with PAO (Fig. 3B). Second, LSD1 contains a large catalytic cavity between the FAD- and substrate-binding subdomains with a volume of 1,245 Å3 (Figs. 3C and 4A). In contrast, the catalytic center of PAO is located in a narrow U-shaped tunnel with a significantly smaller volume (716 Å3) (32, 35). The difference in the shape of the substrate-binding site reflects the different substrate specificities of the two enzymes; the substrates of LSD1 are methylated histone peptides, which are bulkier than the linear-shaped substrates of PAO, like spermidine (35). Taken together, these data reinforce the conclusion that, although PAO and the AOL domain of LSD1 share a similar overall folding topology, LSD1 defines a new subfamily of FAD-dependent oxidases with unique substrate specificity (4).
Fig. 4.
The catalytic center of LSD1. (A) Stereo ribbon diagram of the catalytic center of LSD1. The LSD1 and FAD are colored as in Fig. 3A. The modeled Cξ atom (green sphere) is located 3.6 Å from the N5 atom of the FAD. The side chain of the catalytically important K661 is shown in ball-and-stick representation. The conserved water molecule, which makes hydrogen-bonding interactions with both K661 and FAD, is shown as a red sphere. The hydrogen-bonding interactions are represented by dotted purple lines. Also highlighted are the side chains of the residues that may be involved in substrate recognition. (B) Electrostatic surface representation of the substrate-binding pocket. The view is the same as in A. The 3D coordination system denotes the orientation relative to that in C. (C) Electrostatic surface representation of LSD1 shows an acidic surface formed by helices Sα1 and Sα3 at the entrance of the catalytic cavity. The 3D coordination system denotes the orientation relative to that in B. (D) Histone demethylase activity assay of LSD1 and its mutants. Approximately equal amounts (2 μg) of wild-type and mutant recombinant LSD1 were incubated with equal amounts of 3H-labeled H3K4 methylated substrates. Released 3H-formaldehyde from each reaction was quantified by scintillation counting (cpm).
The Catalytic Center.
The catalytic center of LSD1 consists of a remarkable large cavity that occupies the center of the AOL domain at the interface between the substrate- and FAD-binding subdomains (Fig. 4A). The cavity is framed by many structural elements that surround the catalytic center; a six-stranded β-sheet (Sβ1 and Sβ6–Sβ9) and a long helix (Sα3) form the left side and the bottom surfaces, respectively, whereas a short one-turn helix (Sα4), a two-stranded β-sheet (Fβ5–Fβ7), and several loops with variable lengths cover the rest of the cavity (Fig. 4A). There is a marked contrast in the chemical nature of different areas on the inner surface of the catalytic cavity. The left side is a flat surface and is lined mainly by hydrophobic residues (Fig. 4 A and B). In contrast, the right side of the cavity presents mostly the acidic side chains and the backbone carbonyl oxygen atoms on the concave-shaped surface (Fig. 4 A and B). Notably, point mutations of the residues on both sides of the cavity abolished the LSD1 demethylase activity (Fig. 4D). The cavity goes deep into the protein, extending for a length of ≈23 Å from its entrance to the core of the catalytic site where the flavin ring is located. K661 is a crucial element characterizing the innermost section of the catalytic center (Fig. 4A). It is hydrogen-bonded to the N5 atom of the FAD via a conserved water molecule (Fig. 4A). The fact that a point mutation, K661A, completely abolished the demethylation activity of LSD1 suggests that K661 plays an essential role in flavin reduction as its counterpart, K300, in the case of PAO (25, 36).
Compared with other flavin-dependent oxidases, LSD1 has another unique structural feature: the two diagonally interacting helices, Sα1 and Sα3, form a highly acidic flat surface at the entrance of the catalytic cavity, serving as an additional binding site for the basic histone H3 tail (Fig. 4 A and C). Most of the residues forming this surface and the catalytic cavity are highly conserved not only among LSD1 orthologs in different species but also in LSD2, a close homolog of LSD1 (Fig. 1C) (4). Thus, it is likely that LSD2 also contains a similar catalytic center with a large cavity. This finding is consistent with the observation that the two proteins share the same substrate specificity (unpublished data).
The Tower Domain Contains an Antiparallel Coiled Coil and Is Indispensable for the Demethylase Activity of LSD1.
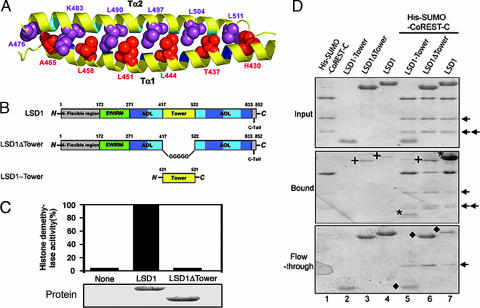
The overall structure of the LSD1 Tower domain is a typical antiparallel coiled coil, with two extended α-helices (Tα1 and Tα2) that pack together in a left-handed superhelix (Figs. 1B and 5A). Tα1 is the extension of helix Sα2 of the AOL domain (Figs. 1B and 3A). The sequence of the Tower domain reveals a repeating pattern of seven residues, (abcdefg)n, the characteristic of coiled-coil proteins, with hydrophobic amino acids predominating at positions a and d of the heptad repeat (Fig. 5A) (37).
Fig. 5.
The Tower domain of LSD1 is an antiparallel coiled coil and is indispensable for both the demethylase activity and the interaction between LSD1 and CoREST. (A) Ribbon diagram of the coiled coil of the Tower domain. The amino acids at the d positions of the heptad repeat of the two helices are in space-filling representation and colored in red (Tα1) and purple (Tα2), respectively. (B) Schematic diagrams of LSD1 and the LSD1 deletion mutants. The domains are colored as in Fig. 1A. (C) Histone demethylase activity assay of LSD1 and its deletion mutant LSD1ΔTower. Approximately equal amounts (2 μg) of wild-type and mutant recombinant LSD1 (Lower) were incubated with equal amounts of 3H-labeled H3K4 methylated substrates. Released 3H-formaldehyde from each reaction was quantified by scintillation counting (cpm) (Upper). (D) The Tower domain of LSD1 interacts with CoREST. Comparable wild-type and deletion mutants of LSD1 were incubated with Ni2+ beads in the presence or absence of His-SUMO-CoREST-C. The bound proteins (Middle) and the flow-through proteins (Bottom) were analyzed by SDS/PAGE. The input proteins are shown in Top. The asterisks in Middle indicate the bound LSD1 and LSD1-Tower, and the diamonds in Bottom indicate the unbound LSD1 (lane 7) and LSD1 mutants (lanes 5 and 6) after incubation with His-SUMO-CoREST-C. LSD1-Tower did not bind the Ni2+ beads (no LSD1-Tower band in Middle). The crosses indicate the nonspecifically bound LSD1 and LSD1ΔTower (lanes 3, 4, and 6 in Middle). The arrows indicate two degradation products of the His-SUMO-CoREST-C protein.
The Tower protrudes from the catalytic center of LSD1 and makes no obvious contact with the rest of the protein (Fig. 1B). This arrangement raises the question of whether the Tower domain plays a role in the histone demethylase activity. To address this issue, a deletion mutant of LSD1 (LSD1ΔTower), in which the Tower domain was deleted and replaced by a pentaglycine loop, was expressed and purified from E. coli (Fig. 5B). LSD1 was able to reduce the methylation level at K4, whereas LSD1ΔTower failed to do so even at a much higher protein concentration (Fig. 5C and data not shown). An effect of this deletion mutation on the structural stability was ruled out, owing to the fact that its biophysical and structural properties are similar to those of the wild-type LSD1 protein as assessed by gel-filtration chromatography analysis (data not shown). These results support the notion that the Tower domain is indispensable for the histone demethylase activity of LSD1.
The Tower Domain Mediates the Interaction Between LSD1 and CoREST.
Besides being important for the LSD1 demethylase activity, the Tower domain of LSD1 may also act as an adaptor to recruit other proteins. LSD1 was identified as a component of the putative LSD1 complex containing HDAC1/2, CtBP1, CoREST, HBC80, and BRAF35, among others (17, 25). Growing evidence showed that LSD1-interacting proteins regulate its activity and substrate specificity (24–26). One of these proteins, CoREST, endows LSD1 with the ability to demethylate nucleosomal substrates and protects LSD1 from proteasomal degradation (24, 25). We investigated the interaction of a His-tagged C-terminal fragment of CoREST (residues 293–482) with two complementary deletion mutants of LSD1, LSD1ΔTower, and LSD1-Tower (amino acids 420–520) employing a pull-down assay (Fig. 5 B and D). We were surprised to find that the Tower domain of LSD1 is, by itself, sufficient for a stable interaction with CoREST-C, forming complexes that can survive extensive washing (Fig. 5D). The fact that LSD1ΔTower does not interact with CoREST indicates that the SWIRM and AOL domains of LSD1 do not significantly contribute to the interaction with CoREST (Fig. 5D). Based on these results, we conclude that the Tower domain of LSD1 contains the binding site for CoREST.
Discussion
The Functions of the SWIRM Domain.
Although both structure and biochemical data presented here indicate that the SWIRM domain is important for the stability of LSD1, the function of the SWIRM domain in LSD1 has not been established yet. Both the Swi3 and Ada2α SWIRM domains are able to interact with double-stranded DNAs nonspecifically, and these interactions are important for the in vivo functions of Swi3 and Ada2α (29, 30). These findings led us to hypothesize that the LSD1 SWIRM domain might also be capable of binding nucleic acids. However, the gel-mobility shift assay clearly demonstrated that the SWIRM domain of LSD1 did not shift DNA even at a very high protein concentration, indicating that the LSD1 SWIRM domain is not a DNA-binding motif (data not shown). A recent study showed that the SWIRM domain of LSD1 may interact with the N-terminal tail of histone H3 (28–30). Particularly, an H3 peptide bearing two point mutations (R8E/T11K) weakened the interaction between the SWIRM domain and the peptide (30). However, close examination of our crystal structure of LSD1 revealed that the SWIRM domain is too far away from the catalytic center of LSD1 (>24 Å). Without large conformational change in LSD1, it is unlikely that a histone H3 tail binds to LSD1 with its K4 located at the catalytic center and R8/T11 interacting with the SWIRM domain simultaneously. More biochemical and structural studies are needed to resolve this discrepancy and to better understand the functions of the SWIRM domain in LSD1.
The Functions of the Tower Domain.
The Tower domain directly interacts with one of the LSD1-interacting proteins, CoREST (Fig. 5D). The two SANT domains of CoREST have been proposed to be a histone-tail-presenting module (38). Consistent with this model, the SANT2 domain of CoREST was shown to play a key role in endowing LSD1 with the ability to demethylate nucleosomal substrates (24). Thus, the Tower domain of LSD1, together with the CoREST SANT2 domain, functions as a molecular bridge that connects LSD1 to its nucleosomal substrates.
In addition to mediating the interaction with CoREST, we also found that the Tower domain is indispensable for the histone demethylase activity of LSD1 (Fig. 5C). But how does the Tower domain affect the activity of LSD1 without direct interactions with the catalytic center? One possible mechanism is that the Tower may play a role in substrate binding. Alternatively, the involvement of the Tower domain on the LSD1 enzymatic activity could be due to an allosteric effect. That is, in the presence of the methylated histone substrate, the Tower domain induces some conformational changes in LSD1 that transform the catalytic center into an active conformation.
Implications for the Substrate Binding Mechanism.
So far, the precise mechanism by which LSD1 recognizes the histone peptide substrates and catalyzes histone demethylation remains unknown. In this respect, the crystal structure of LSD1 reported here reveals several unique features of LSD1. First, structural comparison of flavoproteins that carry out dehydrogenation reactions has revealed that the substrate carbon atom that undergoes flavin-dependent oxidation binds in a highly conserved position in front of the flavin N5-C4a locus, indicating that the substrate carbon atom of a flavoenzyme could be accurately located in its structure (22). On this basis, the Cζ atom of a methylated lysine was modeled in the catalytic site of LSD1 (Fig. 4A). The distance between the Cζ atom and the protein surface is 21 Å. It is unlikely that a histone peptide adopting an extended conformation would bind to LSD1 with its methylated lysine residue positioned in the catalytic center. Thus, we conclude that the histone peptide must enter the catalytic cavity and fold into certain unique conformation to position the methylated lysine residue in the correct location for catalysis. Second, the opening of the catalytic cavity is characterized by several highly conserved solvent-accessible glutamate and aspartate residues (Fig. 4 A and D), which seem to be suited to fulfilling the role of interacting with the basic residues of the histone peptide. Third, the catalytic cavity of LSD1 can accommodate at most a 10-residue polypeptide, which is shorter than the minimum length (>16 and <21) of the histone H3 peptide required for the substrate recognition by LSD1 (27). These structural features lead us to suggest that the acidic surface formed by helices Sα1 and Sα3 (Fig. 4C) at the entrance of the catalytic center makes additional sequence-specific interactions with the H3 peptide. Based on these observations, we propose a model of substrate binding by LSD1 in which the methylated histone H3 tail adopts a loop-like structure so that the methylated K4 is positioned at the catalytic center and the negative charged residues (for instance, R8 and K9) of the H3 peptide make sequence-specific contacts with the acidic residues of LSD1 both inside and outside of the catalytic cavity (Fig. 4). Although our proposed substrate-binding mechanism is plausible based on the LSD1 crystal structure and current biochemical data, additional structural studies of LSD1 complexed with peptide substrates and other interacting proteins are essential to verify this model and to clarify the catalytic and regulatory mechanisms of histone demethylation by LSD1.
Materials and Methods
Cloning, Expression, and Purification.
An N-terminal deleted human LSD1 (residues 172–834) was expressed in E. coli BL21(DE3) by using a modified pET28b vector with a SUMO protein fused at the N terminus after the 6×His tag. The SUMO moiety of the vector helped the expressed fusion proteins in a soluble native form. The His-SUMO-LSD1 protein was purified by Ni-NTA affinity column following standard procedures. Then the ULP1 protease was added to remove the His-SUMO tag. The LSD1 protein was further purified by gel-filtration chromatography. The LSD1 mutants and the C-terminal fragment of CoREST (CoREST-C) were cloned, expressed, and purified following the same procedures as described above.
Crystallization and Data Collection.
Crystals were grown by hanging-drop vapor diffusion against 5% PEG 8000, 150 mM NaCl, 10 mM MgCl2, and 50 mM Na2HPO4/KH2PO4 (pH 6.33). They are in space group P6122 (a = b = 187.11 Å; c = 106.58 Å) with one LSD1 molecule per asymmetric unit. For preparation of a mercury derivative, crystals were transferred gradually into 25% PEG 8000, 150 mM NaCl, 10 mM MgCl2, 50 mM Mes (pH 6.3), and 0.5 mM FAD and soaked in this buffer with 0.1 mM added HgAc2. Both native and derivative crystals were transferred stepwise into a stabilizing solution containing 30% PEG 8000, 25% glycerol, 150 mM NaCl, 10 mM MgCl2, 50 mM Na2HPO4/KH2PO4 (pH 6.33), and 0.5 mM FAD. The crystals were flash-frozen by immersion in liquid nitrogen. Diffraction data were collected at Advanced Photon Source beamline 23-ID. Data were integrated and scaled by using the program HKL2000 (Table 1) (39).
Structure Determination and Refinement.
Multiwavelength anomalous dispersion data from the mercury derivative were used to obtain initial phases. Three mercury atoms were located and refined, and the multiwavelength anomalous dispersion phases were calculated by using SHARP (40); the initial multiwavelength anomalous dispersion map was significantly improved by solvent flattening, which allowed us to fit a model of the maize PAO structure (Protein Data Bank ID code 1B37) as a rigid body. From this initial model, another round of density modification led to a clear and completely interpretable map. A model of the relevant residues of human LSD1 was built into the density by using the program O (41); the model was then transferred into the native unit cell by rigid-body refinement and further refined by using simulated annealing and positional refinement in CNS (42) with manual rebuilding using O.
Histone Demethylase Assay.
The histone demethylase assay was performed essentially as previously described (5). Briefly, core histone substrates methylated by GST-SET7 in the presence of 3H-SAM were incubated with wild-type or mutant recombinant LSD1 in histone demethylation buffer (100 mM glycine, pH 9/1 mM PMSF/1 mM DTT/50 mM KCl) at 37°C for 1 h. For detection of the released 3H-formaldehyde, a modified NASH method (43) was used. After trichloroacetic acid precipitation, an equal volume of NASH reagent (3.89 M ammonium acetate, 0.1 M acetic acid, and 0.2% 2,4-pentanedione) was added into the supernatant, and the mixtures were incubated at 37°C for 50 min before extraction with equal volume of 1-pentanol. The extracted radioactivity was measured by scintillation counting.
His-Tag Pull-Down Assay.
A total of 10 μg of purified His-SUMO-CoREST-C (residues 293–482) protein was incubated with 20 μg of LSD1, 18 μg of LSD1ΔTower, or 4 μg of LSD1-Tower, respectively, at 4°C for 16 h in 40 μl of binding buffer (25 mM Tris·HCl, pH 8.0/300 mM NaCl/3 mM imidazole) containing 5 μl of Ni2+-NTA beads. The beads were then washed three times by 100 μl of binding buffer and finally resuspended in 20 μl of 2× SDS protein sample buffer. The samples of the input, beads, and flow-through were analyzed by SDS/PAGE. The His-SUMO tag itself did not interact with LSD1.
Illustrations.
Figures were prepared by using the programs PyMOL (http://pymol.sourceforge.net) and GRASP (44).
Supplementary Material
Acknowledgments
We thank Dr. Yang Shi (Harvard Medical School, Boston, MA) for the LSD1 cDNA, Dr. Ramin Shiekhattar (The Wistar Institute, Philadelphia, PA) for the CoREST cDNA, and Dr. Janet Smith and Derek Yoder of beamline 23-ID at the Advanced Photon Source for assistance with data collection. Work in the laboratory of M.L. is supported by the University of Michigan Medical School. M.L. is a Kimmel Scholar of the Sidney Kimmel Foundation. Work in the laboratory of Y.Z. is supported by the Howard Hughes Medical Institute and National Institutes of Health Grant GM68804. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- LSD1
lysine-specific demethylase 1
- AOL
amine oxidase-like
- PAO
polyamine oxidase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HKO).
References
- 1.Fischle W, Wang Y, Allis CD. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 2.Margueron R, Trojer P, Reinberg D. Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 6.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 9.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 10.Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trojer P, Reinberg D. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Kubicek S, Jenuwein T. Cell. 2004;119:903–906. doi: 10.1016/j.cell.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Bannister AJ, Kouzarides T. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 14.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 15.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. J Biol Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 19.You A, Tong JK, Grozinger CM, Schreiber SL. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holbert MA, Marmorstein R. Curr Opin Struct Biol. 2005;15:673–680. doi: 10.1016/j.sbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Fraaije MW, Van Berkel WJ, Benen JA, Visser J, Mattevi A. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- 22.Fraaije MW, Mattevi A. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Iyer LM. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0039. RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Lee MG, Wynder C, Cooch N, Shiekhattar R. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 26.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 27.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 28.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, et al. Structure (London) 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Proc Natl Acad Sci USA. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Nat Struct Mol Biol. 2005;12:1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- 31.Mattevi A. Biophys Chem. 1998;70:217–222. doi: 10.1016/s0301-4622(97)00126-9. [DOI] [PubMed] [Google Scholar]
- 32.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. Structure (London) 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 33.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Nat Struct Biol. 2002;9:22–26. [Google Scholar]
- 34.Wierenga RK, Drenth J, Schulz GE. J Mol Biol. 1983;167:725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- 35.Binda C, Angelini R, Federico R, Ascenzi P, Mattevi A. Biochemistry. 2001;40:2766–2776. doi: 10.1021/bi002751j. [DOI] [PubMed] [Google Scholar]
- 36.Polticelli F, Basran J, Faso C, Cona A, Minervini G, Angelini R, Federico R, Scrutton NS, Tavladoraki P. Biochemistry. 2005;44:16108–16120. doi: 10.1021/bi050983i. [DOI] [PubMed] [Google Scholar]
- 37.Alber T. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 38.Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Mol Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.La Fortelle Ed, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 41.Jones TA, Zou JY, Cowan SW, Kjeldgaaed M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Kleeberg U, Klinger W. J Pharmacol Methods. 1982;8:19–31. doi: 10.1016/0160-5402(82)90004-3. [DOI] [PubMed] [Google Scholar]
- 44.Nicholls A, Sharp KA, Honig B. J Comput Chem. 1991;270:26184–26191. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.