Abstract
Chemokines (chemotactic cytokines) comprise a large family of proteins that recruit and activate leukocytes, giving chemokines a major role in both immune response and inflammation-related diseases. The poxvirus-encoded viral CC chemokine inhibitor (vCCI) binds to many CC chemokines with high affinity, acting as a potent inhibitor of chemokine action. We have used heteronuclear multidimensional NMR to determine the structure of an orthopoxvirus vCCI in complex with a human CC chemokine, MIP-1β (macrophage inflammatory protein 1β). vCCI binds to the chemokine with 1:1 stoichiometry, forming a complex of 311 aa. vCCI uses residues from its β-sheet II to interact with a surface of MIP-1β that includes residues adjacent to its N terminus, as well as residues in the 20′s region and the 40′s loop. This structure reveals the strategy used by vCCI to tightly bind numerous chemokines while retaining selectivity for the CC chemokine subfamily.
Keywords: inflammation, protein:protein complex, NMR, chemokine-binding protein
Chemokines play critical roles in the immune system, causing chemotaxis of a variety of cells to sites of infection and inflammation, as well as mediating cell homing and immune system development (1). To date, ≈50 chemokines have been identified, and these small proteins (7–14 kDa) are believed to function by binding with endothelial or matrix glycosaminoglycans (GAGs) to form a concentration gradient that is then sensed by high-affinity, seven-transmembrane domain, G protein-coupled chemokine receptors on the surface of immune cells, leading to activation and chemotaxis. The chemokine system is critical for host defense in healthy individuals and can lead to diseases such as asthma, arthritis, and atherosclerosis in the case of malfunction, often due to inappropriate inflammation and subsequent tissue damage (2). There are four subfamilies of chemokines, CC, CXC, C, and CX3C, named for the position of conserved N-terminal cysteine residues. Members of the same subfamily often have overlapping receptor- binding and cell activation abilities, whereas different subfamilies tend to function on different cell subsets (1).
Structures of chemokines from different subfamilies have been elucidated by NMR and x-ray crystallography (3–6). Despite the differences in amino acid composition and functionalities, most chemokines share a remarkably conserved tertiary structure, with an extended N terminus followed by three β-strands in a Greek key arrangement and a C-terminal α-helix. The solution structure of the human CC chemokine MIP-1β (macrophage inflammatory protein 1β) revealed a homodimer (7), and, subsequently, the dimer dissociation constant was determined to be 0.73 μM (8). Dimerization of other chemokines has been observed under in vitro conditions, and the ability to dimerize is necessary for some chemokines to function in vivo (9).
Given the importance of understanding the interaction between chemokines and their binding partners, considerable effort has been made to study the binding of chemokines to both GAGs and chemokine receptors by a range of techniques (10, 11). However, structure determinations of chemokines in complex with any biological molecule are rare and include only a chemokine:GAG complex (12), a chemokine:protein complex (13), and a chemokine:receptor fragment peptide complex (14).
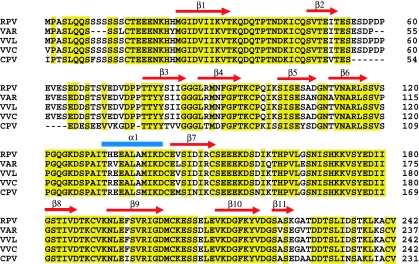
All known pox and herpes viruses encode proteins that interfere with the host chemokine network, likely as part of a strategy to manipulate and subvert the immune system (15). Such virally encoded proteins include chemokine mimics, chemokine receptor analogs, and a group of secreted, soluble chemokine-binding proteins (CKBPs) that exhibit little similarity to any mammalian protein (16). CKBPs competitively bind to chemokines and disrupt chemokine interactions with the host cell surface receptors or GAGs. Although some CKBPs interact with a very broad spectrum of chemokines across several chemokine subfamilies, the viral CC chemokine inhibitor (vCCI) proteins (previously called T1/35 kDa, also classified as type II CKBPs) produced by leporipoxviruses and orthopoxviruses bind selectively to members of the CC subfamily (16, 17). vCCI proteins have been shown to be potent inhibitors of chemokine action in vitro (17) and effective anti-inflammatory agents in vivo (18). Sequence alignment of vCCI proteins from five orthopoxvirus members [rabbitpox, cowpox, vaccinia-Lister, vaccinia-Copenhagen, and variola poxvirus (the causative agent of human smallpox)] shows at least 80% identity (Fig. 1), with some pairwise comparisons showing as much as 99% sequence identity, indicating that they have essentially identical structures and very likely share characteristics of chemokine binding. The x-ray crystal structure of unliganded cowpox vCCI by Carfi et al. (19) revealed a unique β-sandwich structure with no topological resemblance to any mammalian receptors. Although β-sheet I in this sandwich has two extended loops that effectively shield it from solvent, β-sheet II is exposed to solvent and was hypothesized by these researchers to include a chemokine-binding surface.
Fig. 1.
Sequence alignment of five members of the orthopox vCCI family. Alignments were made by using ClustalW. RPV, rabbitpox virus; VAR, variola virus; VVL, vaccinia virus Lister strain; VVC, vaccinia virus Copenhagen strain; CPV, cowpox virus. Conserved residues are highlighted in yellow.
Here, we report the solution structure of a complex between rabbitpox-encoded vCCI and a nonaggregating variant of the human CC chemokine MIP-1β. The structure reveals that the vCCI and MIP-1β form a complex in a 1:1 stoichiometry and that vCCI occludes the regions in the chemokine that are important for chemokine homodimerization, receptor binding, and GAG interaction. The structure also defines key interactions that form the basis for the affinity and selectivity of vCCI toward certain CC chemokines.
Results
Solution Structure Determination of the vCCI:MIP-1β Complex.
Although vCCI is a highly soluble and well folded protein at physiological pH, many CC chemokines prefer acidic conditions in vitro and aggregate near pH 7 at concentrations required for NMR experiments. Therefore, a nonaggregating human MIP-1β variant in which three positively charged amino acid residues in the so-called 40′s loop (45KRSK48) were replaced by Ala was chosen for the current study. The resulting protein, MIP-1β-45AASA48, has been found to be soluble at neutral pH while maintaining most spectral and functional characteristics of the WT protein with the exception of loss of GAG-binding ability (20). (For simplicity, when discussing the complex, MIP-1β-45AASA48 will hereafter be referred to as MIP-1β.)
Because of the relatively large size of vCCI (26 kDa), side-chain deuteration was used to reduce linewidths for better spectral resolution. Resonance assignments of both components in the complex were carried out with a series of standard 2D and 3D experiments by using 15N-labeled, 13C,15N-labeled, and 2H,13C,15N-labeled proteins. Essentially complete backbone and side-chain 1H, 13C, and 15N chemical-shift assignments for bound MIP-1β and 1HN, 13Cα, 13Cβ, 13C′, 15N chemical-shift assignments for bound and free vCCI were obtained and are reported elsewhere (21, 22). Deviation of chemical shift from random coil values allowed predictions of secondary structure for both protein components in the complex (23) and suggests that, in the complex, both proteins preserve the secondary structure elements that are present in their unliganded forms (7, 19).
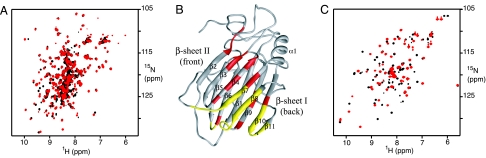
Upon titration of 15N-labeled vCCI with 14N MIP-1β, two sets of vCCI peaks in the 15N heteronuclear single quantum correlation spectra (HSQC) were evident when <1 equivalent of MIP-1β subunits was added, indicating slow exchange between two conformations on an NMR timescale. The spectrum resolved into only one peak per vCCI 15N-1H resonance after addition of 1 full equivalent of MIP-1β subunits, with additional amounts of MIP-1β causing no further spectral change, consistent with a 1:1 ratio of proteins in the complex (Fig. 2A). 15N relaxation experiments were carried out on the complex by using both 15N vCCI:14N MIP-1β and 14N vCCI:15N MIP-1β. The rotational correlation time was calculated to be 13.2 ± 0.7 ns for 15N vCCI in complex and 13.1 ± 0.6 ns for 15N MIP-1β in complex (data not shown). These values are both consistent with a molecular mass of ≈34 kDa, confirming a complex with 1:1 stoichiometry.
Fig. 2.
Observation of vCCI:MIP-1β interaction. (A) Overlay of the 15N heteronuclear single quantum correlation (HSQC) spectra of free (black) and liganded (red) vCCI. (B) Chemical shift perturbation mapping of vCCI upon addition of MIP-1β. Residues with weighted average HN and N chemical shift perturbation >1 standard deviation (SD) from the mean value are shown in red. Residues that are missing from the HSQC because of chemical exchange line broadening are shown in yellow. (C) Overlay of the 15N HSQC spectra of free (black) and liganded (red) MIP-1β.
Protein–protein complex formation is often accompanied by changes in 15N-1H chemical shifts of residues in regions that are affected by the binding interaction. Chemical-shift perturbation mapping reveals the likely binding sites on both proteins. For vCCI, the vast majority of chemical shift perturbations upon association with MIP-1β occur on β-sheet II and the nearby loops, with a few additional changes directly across the β-sandwich in β-sheet I, likely due to propagation of changes from β-sheet II (Fig. 2B). The MIP-1β spectra show significant change upon binding vCCI (Fig. 2C), particularly in the regions associated with the chemokine dimer interface, indicating alteration in this part of the protein caused by the breaking of the MIP-1β homodimer and binding with vCCI.
Residual dipolar coupling (RDC) constants were measured in the presence of alignment medium to obtain structural restraints as well as the relative orientation of the two binding partners (see the supporting information, which is published on the PNAS web site). NOESY data were also obtained by using complexes composed of proteins with various isotopic labeling schemes to facilitate resolution of the spectra. Three-dimensional 15N-NOESY of 1HN,15N,13C-labeled vCCI in complex with unlabeled MIP-1β and 3D 13C-NOESY and 15N-NOESY of 13C,15N-labeled MIP-1β in the presence of unlabeled vCCI were used for verification of chemical- shift assignments and to obtain intramolecular NOE constraints. These data also confirm that both proteins in the complex retain the same overall tertiary fold and secondary structures as in the unbound form.
Direct structural information of the binding interface was obtained by using a 4D 15N,13C-edited NOESY experiment (24) that was measured on a 2H,1HN,15N vCCI:13C,14N MIP-1β sample (see the supporting information). Because of the differential isotopic labeling of the proteins in the complex, this experiment shows resonances only for intermolecular contacts, providing distance information between the amide protons of vCCI and any nearby side-chain and backbone HC protons of MIP-1β. A total of 32 distinct intermolecular NOE restraints were obtained from this spectrum and from the 3D NOESY experiments mentioned above. The structure of the complex was calculated to be consistent with a total of 917 constraints, including RDC, dihedral angle, and NOE distance restraints (Table 1). The final ensemble of 15 structures shows a 0.78-Å rmsd for backbone atoms. The atomic coordinates for the ensemble of 15 NMR structures (Protein Data Bank ID code 2FIN) and the averaged energy-minimized structure (PDB ID code 2FFK) have been deposited in the Protein Data Bank.
Table 1.
NMR and refinement statistics for the vCCI:MIP-1β complex
| NMR distance and dihedral constraints | |
|---|---|
| Distance restraints | |
| Total NOE | 599 |
| Intraresidue | 81 |
| Interresidue | |
| Sequential (∣i − j∣ = 1) | 264 |
| Nonsequential (∣i − j∣ > 1) | 222 |
| Protein–protein intermolecular | 32 |
| Total dihedral angle restraints | |
| φ | 134 |
| ψ | 134 |
| Total RDCs | 184 |
| Q factor for average energy-minimized structure, % | 10.1 |
| Structure statistics* | |
| Violations (mean ± SD) | |
| Distance constraints, Å | 0.020 ± 0.003 |
| Dihedral angle constraints, ° | 0.54 ± 0.04 |
| Max. distance constraint violation, Å | 0.217 ± 0.068 |
| Max. dihedral angle violation, ° | 3.83 ± 0.53 |
| Deviations from idealized geometry | |
| Bond lengths, Å | 0.00457 ± 0.00014 |
| Bond angles, ° | 0.587 ± 0.015 |
| Improper angles, ° | 0.427 ± 0.026 |
| Average pairwise rmsd, Å | |
| vCCI | |
| Heavy | 1.64 ± 0.09 |
| Backbone | 0.42 ± 0.08 |
| MIP-1β | |
| Heavy | 1.61 ± 0.18 |
| Backbone | 0.38 ± 0.05 |
| Complex | |
| Heavy | 1.78 ± 0.12 |
| Backbone | 0.78 ± 0.25 |
*Fifteen structures from the final ensemble were included in the analysis. Residues 1–7 and 56–76 in vCCI were excluded from pairwise rmsd calculation. The force constants used for the structure calculation are as follows: 1,000 kcal·mol−1·Å−2 for bond lengths, 500 kcal·mol−1·rad−2 for angles and improper torsions, 4 kcal·mol−1·Å−4 for quartic van der Waals repulsions, 80 kcal·mol−1·Å−2 for experimental interproton distances, 200 kcal·mol−1·rad−2 for the TALOS-predicted torsion angle restraints, and 0.1–0.5 kcal·mol−1·Hz−2 for residual dipolar coupling.
Description of the vCCI:MIP-1β Structure.
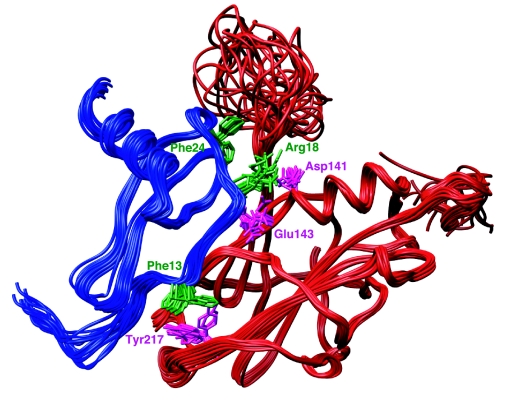
The vCCI:MIP-1β complex is formed by one monomer of vCCI interacting with one monomer of MIP-1β (Fig. 3). Each component of the complex is largely similar in conformation to the proteins in their unliganded forms. MIP-1β spans the entire width of vCCI, making contacts across the vCCI β-sheet II starting with MIP-1β residue 8, through the 20′s region, and including the 40′s loop and the third β-strand, constituting a protein:protein interface with a 2,200-Å2 buried solvent-accessible surface area (1.4-Å probe). The extended MIP-1β N-terminal fragment from Pro-8 to Ser-14 is positioned almost parallel to the vCCI β-strand 8, making numerous contacts with residues from Ser-182 to Thr-187. The residue Phe-13 of MIP-1β is proximal to a hydrophobic cluster formed by highly conserved vCCI residues, including Val-185 and Tyr-217. The strand of MIP-1β continues across the β-sheet II face of vCCI, with the Arg-18 side chain protruding toward the vCCI residues Asp-141 and Glu-143, consistent with involvement in electrostatic interactions. The 20′s region and 40′s loop of MIP-1β fit into a binding surface formed by vCCI strands β2 and β3 and the loop connecting these two strands. The third β-strand of MIP-1β is oriented roughly orthogonal to the vCCI β-strands and interacts with residues from vCCI β-strands 4, 7, and 8.
Fig. 3.
Solution structure of the vCCI:MIP-1β complex. Superposition of 15 NMR structures is shown, with the vCCI backbone colored red and selected side chains colored pink. The MIP-1β backbone is blue, and selected side chains are shown in green.
Binding of MIP-1β to vCCI by Using ELISA.
Quantitative ELISAs were carried out to determine the relative affinity for vCCI of WT MIP-1β and the variant used in the present structure determination. These studies reveal an EC50 for WT MIP-1β of 0.66 ± 0.4 nM and an EC50 for MIP-1β-45AASA48 of 0.64 ± 0.6 nM (see the supporting information).
Discussion
vCCIs comprise a group of poxvirus-encoded proteins with highly conserved sequence and unique structural topology. These proteins tightly bind and inhibit the action of CC chemokines but have low or no affinity for chemokines of other subfamilies, making them potent and specific anti-inflammatory agents with demonstrated effectiveness in vivo (18, 25). An extensive binding study with vaccinia vCCI using >80 chemokines from several organisms revealed 26 CC chemokines that bind with high affinity, including 13 human chemokines, such as MCP-1 (monocyte chemoattractant protein 1), MIP-1α, MIP-1β, and RANTES (26). To probe the chemokine residues that are important in vCCI binding, two previous studies using vCCI and variants of human CC chemokine MCP-1 suggested that the MCP-1 monomeric subunit binds vCCI through an interface that is partially overlapping with the site that MCP-1 uses to bind its natural receptors, with residues Tyr-13, Arg-18, and Arg-24 found to be particularly important (27, 28). The present work displays the structural underpinning of the high affinity between poxvirus vCCI and CC chemokines.
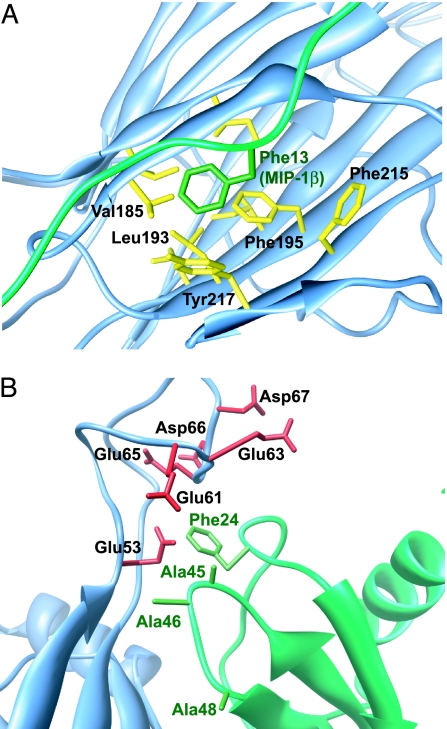
In the vCCI:MIP-1β structure, vCCI makes no contact with the first seven chemokine residues, which are involved in receptor activation (8). However, the highly conserved vCCI residues Ser-182 to Thr-187 make extensive contacts with the MIP-1β residues Pro-8 to Ser-14. This region of MIP-1β, and in particular Phe-13, has been shown to be critical to the chemokine both for receptor binding and for dimer formation (8), providing a clear rationale for the inhibitory capability of vCCI. The positioning of the MIP-1β N-terminal region allows its Phe-13 to make close contact with hydrophobic vCCI residues, from β8 at the edge of β-sheet II and β10 from the opposing β-sheet I (Fig. 4A). These residues are at the edge of a large hydrophobic network between the two sheets of the vCCI β-sandwich so that Phe-13 of MIP-1β makes a “cap” on the network. An amino acid with a shorter or a polar side chain would not be able to make these contacts and would therefore lack this component of binding affinity. Indeed, detailed mutagenesis studies of the CC chemokine MCP-1 show an ≈10-fold reduction in affinity when analogous residue Tyr-13 is replaced by Ala (27, 28). In all human chemokines that bind tightly to vCCI, this position is an aromatic or a large hydrophobic amino acid (26), again demonstrating the importance of this residue in both the function and inhibition of CC chemokines.
Fig. 4.
Detailed views of the VCCI:MIP–1β interaction. (A) MIP-1β Phe-13 (green) and the surrounding hydrophobic residues (yellow) from vCCI. (B) View of the 20′s region and the 40′s loop of MIP-1β (green) in proximity to vCCI acidic residues (red). In the present structure, MIP-1β residues 45, 46, and 48 are changed to Ala to enhance solubility.
Another important interaction in the complex involves a positively charged Arg-18 of MIP-1β. Our structure reveals that this residue is in close proximity to the conserved, negatively charged vCCI residues Asp-141 and Glu-143. In 12 of the final ensemble of 15 structures, the MIP-1β Arg-18 guanidino group is within 3.9 Å or less of the γ carboxyl group of Asp-141 of vCCI so that Asp-141 is well positioned for electrostatic interaction (Fig. 3). Although this interaction is not shielded from water, mutagenesis experiments on MCP-1 indicate its significance, showing a >20-fold reduction in affinity when Arg-18 is substituted with Ala (27, 28). Additionally, sequence analysis of human chemokines that bind vCCI shows a nearly universal positive charge at this position (Fig. 5).
Fig. 5.
Sequence alignment of human CC chemokines with high affinity to vCCI. Conserved cysteine residues are highlighted in yellow. Positions highlighted with red indicate residues that likely confer high-affinity binding to vCCI. The numbering is according to the MIP-1β sequence.
A particularly intriguing interaction occurs between the chemokine 20′s region and the large, flexible vCCI loop that connects the β2 and β3 strands. Among the CC chemokines having high affinity with vCCI, some have a large hydrophobic residue at this position (such as MIP-1β with Phe-24), whereas others have a positively charged residue (such as MCP-1 with Arg-24). Both types of residues at this position are apparently allowed, which raises the question of whether this amino acid takes part in the binding interaction, and, if so, how two completely different amino acid types are tolerated here. The answer to this question is quite important from a structural standpoint, because it provides insight into the ability of vCCI to maintain selectivity yet bind such a large number of chemokines. The structure of the vCCI:MIP-1β complex shows that the 24th position in MIP-1β faces the loop in vCCI between the β2 and β3 strands (Fig. 4B). In all orthopox vCCI proteins, this loop region is highly acidic, with ≈50% of the residues being Asp or Glu. 15N relaxation studies reveal that this region is quite flexible regardless of the presence of the bound MIP-1β (data not shown). The arrangement of negative charges and the conformational plasticity in this region of vCCI allow favorable interactions to positively charged residues in the chemokine and have minimal hindrance to large hydrophobic residues. A negatively charged chemokine residue at this position would be expected to be disfavored by the highly acidic loop, which explains the observation that an Arg-24-Glu MCP-1 mutant has no detectable binding to vCCI by surface plasmon resonance (28). Regarding vCCI interactions with those chemokines having a neutral rather than a basic residue at this 24th position, the high flexibility of the acidic vCCI loop region surrounding MIP-1β residue Phe-24 makes it difficult to pinpoint specific contacts, so it is possible that Phe-24 itself does not contribute favorably to binding vCCI. However, the chemokine 40′s loop is close in space to the 24th position and often contains a basic amino acid(s) at the turn, such as Lys-45 in WT MIP-1β (Figs. 4B and 5), so productive electrostatic-binding interactions are expected with the acidic flexible loop of vCCI. This interaction compensates for the absence of basic residues at the 24th position. In MCP-1 studies, the loss of Arg-24 by mutation to Ala was not compensated by a nearby positive charge (WT MCP-1 has Ile and Val at the 45th and 46th positions, respectively, using MIP-1β numbering), causing a >10-fold drop in affinity. An analysis of the sequences of the human chemokines that bind tightly to vCCI shows that they all have at least one positive charge in the 24th or the 45th position (with one chemokine having a positive charge at position 46 instead; Fig. 5).
The MIP-1β variant used here has mutations K45A/R46A/K48A, which, overall, lead to an affinity for vCCI approximately the same as that of the WT chemokine, as determined by ELISA (see the supporting information). Although neutralization of the Lys at position 45 would be expected to decrease the affinity of the mutant by the above argument, replacing the large basic Lys residue with Ala at position 48 apparently compensates for this change. In the current structure, the 48th position is approached closely by the side chains of vCCI Tyr-80 and Arg-89, so substitution of Lys-48 by Ala in MIP-1β could alleviate steric crowding and poor electrostatic effects of the WT protein. In support of this, the analogous mutation in MCP-1 Lys-49–Ala actually showed an increase in affinity for vCCI (27, 28).
In the present structure, the GAG-binding region of MIP-1β, composed of residues from the 40′s loop as well as Arg-18 (11, 29), interacts with vCCI. Therefore, we expect vCCI to interfere with GAG binding by MIP-1β and by chemokines that share a similar GAG-binding site. Inhibition of chemokine GAG binding is a strategy that has been demonstrated for other viral proteins and may disrupt the chemokine concentration gradient that is necessary for directed chemotaxis of leukocytes (30, 31).
The only other reported protein:protein complex involving a chemokine is the x-ray crystal structure of the γ-herpesvirus protein M3 in complex with a monomeric variant of the CC chemokine MCP-1 (13). M3 is a large protein that is able to bind chemokines from all four chemokine subfamilies. Although both M3 and vCCI share some properties of chemokine binding, their strategies are distinct. Most obvious, the M3:MCP-1 binding interface is unrelated to the vCCI:MIP-1β interface reported here. M3 primarily uses the loops at the edge of two different domains to contact each MCP-1 subunit (binding in a 2:2 complex); in contrast, vCCI uses much of its β-sheet II to contact MIP-1β and forms a 1:1 complex. M3 makes close contacts to residues involved in the receptor- binding surface of MCP-1, such as Tyr-13 and Arg-18, but only peripherally contacts the residues of the 40′s loop of the chemokine, which appear to be largely solvent-exposed.
Overall, then, the ability of vCCI to inhibit many CC chemokines hinges on several key interactions, ranging from occlusion of the receptor-binding residues to a likely blockade of the chemokine GAG-binding site. Sequence analysis of human chemokines having high affinity to vCCI exhibits a conserved pattern (Fig. 5): Hydrophobic residue 13, as well as positively charged residue 18, is a near-absolute requirement. Residue 24 and members of the 40′s loop are close in space, and there is a requirement for at least one positive charge in the 24/45 position; the lack of a positive charge at one of these sites (e.g., 24) is always compensated by at least one positive charge at the other (e.g., 45, or, in one case, 46). CC chemokines that have high affinity to vCCI (such as MIP-1β and MCP-1) show adherence to these principles, whereas chemokines having lower affinity to vCCI (such as TARC/CCL17) do not. CXC chemokines almost universally lack several elements of this pattern, particularly at positions 18 and 24, and so have low or no affinity to vCCI. In summary, this work elucidates the structure of a vCCI:chemokine complex and reveals a general strategy used by vCCI for selective chemokine binding, which could provide a guideline for future therapeutic design.
Methods
Sample Preparation.
The gene encoding the human MIP-1β nonaggregating mutant K45A/R46A/K48A (MIP-1β45AASA48) was subcloned into a modified pET-32 Xa/LIC vector as described in ref. 8. The resulting plasmid was then transformed into BL21-(DE3) Escherichia coli cells. Unlabeled, 15N-labeled, 13C,14N-labeled, and 13C,15N-labeled MIP-1β 45AASA48 were produced and purified by following the protocol described in ref. 8.
The gene encoding rabbit poxvirus vCCI was cloned into pPIC9K plasmid and then transformed into Pichia pastoris strain SMD1168 (Invitrogen, Carlsbad, CA). Unlabeled, 15N-labeled, 2H,15N-labeled, and 2H,13C,15N-labeled protein samples were prepared by growing cells in buffered minimal glucose medium for initial biomass accumulation and then shifting cells into buffered minimal methanol media with 15N (or 14N for unlabeled samples) ammonium sulfate and 13C (or 12C for unlabeled samples) methanol as the sole nitrogen and carbon sources for induction (32). For deuterated samples, the medium was prepared with 95% (final concentration) D2O. The recombinant protein was purified by anion exchange chromatography followed by size-exclusion chromatography. To fully exchange amide protons in the deuterated samples, the vCCI protein was partially unfolded in H2O buffer with 2 M urea, followed by a 10-fold quick dilution. After allowing the protein to fully refold, the protein was exchanged into the final NMR buffer.
The final NMR samples contained 0.5–2.0 mM protein(s) (with concentration determined in monomeric subunits) in 93% H2O/7% D2O containing 100 mM NaCl, 20 mM sodium phosphate (pH 7.0), 0.1 mM DSS, 0.01% sodium azide, and Complete Protease Inhibitor (Roche Applied Science, Indianapolis, IN).
NMR Spectroscopy.
Spectra were recorded at 37°C on Inova 600, 750, and 800 spectrometers (Varian, Palo Alto, CA). NMR data were processed by using NMRPipe (33). Bound and free 2H,15N,13C-labeled vCCI were used to obtain backbone and Cβ assignments (34). Bound 13C,15N-labeled MIP-1β 45AASA48 was used to obtain backbone and side-chain assignments. The observed chemical shift change (Δδobs) for each backbone amide between bound and free vCCI was measured as the weighted average of the proton and nitrogen chemical shift changes by using the equation Δδobs = [(Δδ2HN + Δδ2N/25)/2]1/2 (35).
3D 15N-NOESY of 2H,15N,13C-labeled vCCI in complex with unlabeled MIP-1β and 3D 13C-NOESY and 15N-NOESY of 13C,15N-labeled MIP-1β in the presence of unlabeled vCCI were carried out as described in ref. 36. To detect intermolecular restraints, a 4D 13C,15N-edited NOESY (24) was recorded on a complex of 2H,1HN,15N vCCI with 13C,14N MIP-1β. Details of NMR experiments can be found in the supporting information.
15N T1, T2, and 15N-HN NOE values were measured for both components of the complex (37). Data were excluded from the correlation time calculation if the corresponding 15N-HN NOE was <0.65 or if either T1 or T2 differed from the average value by >1 SD.
RDC.
Polyacrylamide gel (4.8%) was used to align the sample. The samples were prepared by using the apparatus developed by Chou et al. (38) and purchased from New Era Enterprise (Vineland, NJ), following a similar protocol to that described by Mohana-Borges et al. (39). The RDCs of resolved 1H-15N were measured by using the IPAP scheme (40) for both components of the complex (41).
Structure Calculation.
Torsion angles φ and ψ were derived from the TALOS database (42). Structures were calculated by using Dynamo (45). For MIP-1β-45AASA48, the high-resolution NMR structure of the WT MIP-1β (PDB ID code 1HUM) was used to prepare a starting input for the calculation. For rabbitpox vCCI, the sequence was first aligned with the unliganded cowpox vCCI (PDB ID code 1CQ3) and then threaded by Modeller 8v1 (43) to obtain a starting set of coordinates for calculation. As the first step, separate structural refinement of each protein was carried out. The high temperature in the annealing steps was kept at 500 K so that large changes in conformation were not likely unless heavy violation of the NMR data occurred. Ten structures were chosen from a calculation of 50 structures based on adherence to experimental restraints and low overall energy. These 10 structures were then used to obtain an averaged energy-minimized structure, which was further refined with RDC restraints. The resulting refined structure of vCCI showed 0.25-Å backbone rmsd from the 1CQ3 structure for 213 aligned residues, showing the same overall fold. Similarly, the resulting refined structure of MIP-1β 45AASA48 showed 0.16-Å backbone rmsd from a monomeric unit of the WT structure. These two initially refined protein structures were then used as inputs to calculate the complex structures. At this stage, the backbone atoms of each protein were constrained within 0.2 Å of the input coordinates without penalty, and the side chains were allowed to move freely to satisfy other restraints. The final ensemble of 15 structures was selected from an ensemble of 100 structures based on lowest energy and no violation of NOE restraints by >0.4 Å or dihedral restraints by >5°. PROCHECK analysis shows that, for the ensemble of 15 structures, 70.2% of the residues are in the most favored region, 26.5% are in the additionally allowed region, 2.7% are in the generously allowed region, and 0.6% are in the disallowed region.
ELISAs.
Ninety six-well OptiPlate-HB plates (PerkinElmer, Wellesley, MA) were coated with 100 μl of 65 nM vCCI overnight at 4°C. The plate was then washed with 0.1% Tween 20 in PBS (PBS-Tween), and excess binding sites were blocked with 300 μl of blocking buffer (0.5% BSA in PBS-Tween) for 2 h at room temperature with constant agitation. The wells were again washed with PBS-Tween, after which serial dilutions of chemokines in blocking buffer were added and incubated for 2 h at room temperature with constant agitation. After washing, 100 μl of 400 ng/ml polyclonal antibody to human MIP-1β (R & D Systems, Minneapolis, MN) was added, incubated for 1 h, and then washed. One hundred microliters of 400 ng/ml mouse anti-goat IgG conjugated with horseradish peroxidase (Pierce, Rockford, IL) was then incubated for 1 h and washed. One hundred microliters of Amersham ECL reagent (GE Healthcare, Buckinghamshire, U.K.) was added and incubated for 10 min, followed by detection with an Orion microplate luminometer (Berthold, Oak Ridge, TN). The EC50 was determined by using three separate experiments, each performed in triplicate.
Figure Preparation.
Structure figures were prepared by using UCSF Chimera (44).
Supplementary Material
Acknowledgments
We thank Drs. Frank Delaglio, Xiangming Kong, and Kirsten Frank for helpful discussions and the William R. Wiley Environmental Molecular Sciences Laboratory for use of high-field spectrometers. The NMR instrumentation at Texas A&M University is funded by National Science Foundation Grant DBI-9970232 and the Texas Agricultural Experiment Station. This work was supported by National Institutes of Health Grants AI47832 and AI070993 and Robert Welch Foundation Grant A1472.
Abbreviations
- vCCI
viral CC chemokine inhibitor
- MIP-1β
macrophage inflammatory protein 1β
- GAG
glycosaminoglycan
- RDC
residual dipolar coupling
- MCP-1
monocyte chemoattractant protein 1.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2FIN and 2FFK).
References
- 1.Baggiolini M. J Int Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 2.Gerard C, Rollins BJ. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 3.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Biochemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 4.Mayer KL, Stone MJ. Biochemistry. 2000;39:8382–8395. doi: 10.1021/bi000523j. [DOI] [PubMed] [Google Scholar]
- 5.Crump MP, Gong J-H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J-L, Baggiolini M, Sykes B, Clark-Lewis I. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez EJ, Lolis E. Annu Rev Pharmacol Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 7.Lodi PJ, Garrett DS, Kuszewski J, Tsang ML-S, Weatherbee JA, Leonard WJ, Gronenborn AM, Clore GM. Science. 1994;263:1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 8.Laurence JS, Blanpain C, Parmentier M, Burgner JW, LiWang PJ. Biochemistry. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- 9.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Proc Natl Acad Sci USA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau EK, Allen S, Hsu AR, Handel TM. Adv Protein Chem. 2004;68:351–391. doi: 10.1016/S0065-3233(04)68010-7. [DOI] [PubMed] [Google Scholar]
- 11.McCornack MA, Cassidy CK, LiWang PJ. J Biol Chem. 2003;278:1946–1956. doi: 10.1074/jbc.M207440200. [DOI] [PubMed] [Google Scholar]
- 12.Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, Harrenga A, Wells TN, Proudfoot AE. Structure (London) 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Alexander JM, Nelson CA, van Berkel V, Lau EK, Studts JM, Brett TJ, Speck SH, Handel TM, Virgin HW, Fremont DH. Cell. 2002;111:343–356. doi: 10.1016/s0092-8674(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 14.Skelton NJ, Quan C, Reilly D, Lowman H. Structure (London) 1999;7:157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 15.Boomker JM, de Leij LF, The TH, Harmsen MC. Cytokine Growth Factor Rev. 2005;16:91–103. doi: 10.1016/j.cytogfr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Seet BT, McFadden G. J Leukoc Biol. 2002;72:24–34. [PubMed] [Google Scholar]
- 17.Lalani AS, Ness TL, Singh JK, Harrison JK, Seet BT, Kelvin DJ, McFadden G, Moyer RW. Virology. 1998;250:173–184. doi: 10.1006/viro.1998.9340. [DOI] [PubMed] [Google Scholar]
- 18.Dabbagh K, Xiao Y, Smith C, Stepick-Biek P, Kim SG, Lamm WJ, Liggitt DH, Lewis DB. J Immunol. 2000;165:3418–3422. doi: 10.4049/jimmunol.165.6.3418. [DOI] [PubMed] [Google Scholar]
- 19.Carfi A, Smith CA, Smolak PJ, McGrew J, Wiley DC. Proc Natl Acad Sci USA. 1999;96:12379–12383. doi: 10.1073/pnas.96.22.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurence JS, Blanpain C, De Leener A, Parmentier M, LiWang PJ. Biochemistry. 2001;40:4990–4999. doi: 10.1021/bi002593w. [DOI] [PubMed] [Google Scholar]
- 21.DeRider ML, Zhang L, LiWang PJ. J Biomol NMR. 2006;1:0. doi: 10.1007/s10858-005-6276-x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, LiWang P. J Biomol NMR. 2006;1:0. doi: 10.1007/s10858-006-9021-1. [DOI] [PubMed] [Google Scholar]
- 23.Wishart DS, Sykes BD. Methods Enzymol. 1994;239:363–393. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 24.Muhandiram DR, Xu GY, Kay LE. J Biomol NMR. 1993;3:463–470. [Google Scholar]
- 25.Alcami A, Symons JA, Collins PD, Williams TJ, Smith GL. J Immunol. 1998;160:624–633. [PubMed] [Google Scholar]
- 26.Burns JM, Dairaghi DJ, Deitz M, Tsang M, Schall TJ. J Biol Chem. 2002;277:2785–2789. doi: 10.1074/jbc.M109884200. [DOI] [PubMed] [Google Scholar]
- 27.Beck CG, Studer C, Zuber JF, Demange BJ, Manning U, Urfer R. J Biol Chem. 2001;276:43270–43276. doi: 10.1074/jbc.M106305200. [DOI] [PubMed] [Google Scholar]
- 28.Seet BT, Singh R, Paavola C, Lau EK, Handel TM, McFadden G. Proc Natl Acad Sci USA. 2001;98:9008–9013. doi: 10.1073/pnas.171069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koopmann W, Ediriwickrema C, Krangel MS. J Immunol. 1999;163:2120–2127. [PubMed] [Google Scholar]
- 30.Lalani AS, Graham K, Mossman K, Rajarathnam K, Clark-Lewis I, Kelvin D, McFadden G. J Virol. 1997;71:4356–4363. doi: 10.1128/jvi.71.6.4356-4363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb LM, Smith VP, Alcami A. FASEB J. 2004;18:571–573. doi: 10.1096/fj.03-0485fje. [DOI] [PubMed] [Google Scholar]
- 32.Morgan WD, Kragt A, Feeney J. J Biomol NMR. 2000;17:337–347. doi: 10.1023/a:1008313530207. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Hu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. J Am Chem Soc. 1994;116:11655–11666. [Google Scholar]
- 35.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. Nat Struct Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 36.Zhang O, Forman-Kay JD, Shortle D, Kay LE. J Biomol NMR. 1997;9:181–200. doi: 10.1023/a:1018658305040. [DOI] [PubMed] [Google Scholar]
- 37.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 38.Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. J Biomol NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 39.Mohana-Borges R, Goto NK, Kroon GJA, Dyson HJ, Wright PE. J Mol Biol. 2004;340:1131–1142. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Hall JB, Dayie KT, Fushman D. J Biomol NMR. 2003;26:181–186. doi: 10.1023/a:1023546107553. [DOI] [PubMed] [Google Scholar]
- 41.Bax A. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 43.Fiser A, Do RK, Sali A. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F, Kuszewski J. DYNAMO: A Structure Calculation and Analysis Program. Bethesda: National Institutes of Health; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.