Abstract
Affinity of integrin lymphocyte function-associated antigen 1 (LFA-1) is enhanced by conformational changes from the low-affinity closed form to the high-affinity (HA) open form of the ligand-binding inserted (I) domain as shown by work with purified I domains. However, affinity up-regulation of LFA-1 on the cell surface by physiological agonists such as chemokines has yet to be demonstrated by monovalent reagents. We characterize a mAb, AL-57 (activated LFA-1 clone 57), that has been developed by phage display that selectively targets the HA open conformation of the LFA-1 I domain. AL-57 discriminates among low-affinity, intermediate-affinity, and HA states of LFA-1. Furthermore, AL-57 functions as a ligand mimetic that binds only upon activation and requires Mg2+ for binding. Compared with the natural ligand intercellular adhesion molecule-1, AL-57 shows a tighter binding to the open I domain and a 250-fold slower off rate. Monovalent Fab AL-57 demonstrates affinity increases on a subset (≈10%) of lymphocyte cell surface LFA-1 molecules upon stimulation with CXCL-12 (CXC chemokine ligand 12). Affinity up-regulation correlates with global conformational changes of LFA-1 to the extended form. Affinity increase stimulated by CXCL-12 is transient and peaks 2 to 5 min after stimulation.
Keywords: conformational changes, activation, cell adhesion, phage display
The integrin LFA-1 (lymphocyte function-associated antigen 1) plays a critical role in lymphocyte recirculation, diapedesis, and regulation of immune responses (1, 2). A remarkable property of LFA-1 is the large dynamic range of adhesion strength that is observed commensurate with the physiological context. LFA-1 participates in slowing rolling interactions (3–5), which requires rapid bond formation and rupture. LFA-1, however, is most important in mediating fluid shear-resistant firm adhesion and cell migration (1, 6, 7) and in maintaining the integrity of the immunological synapse, which requires stable, less labile bonds (8).
The adhesion of LFA-1 to ligands on opposed cell surfaces is controlled by the combination of two distinct mechanisms: an increase in the affinity of individual receptors to bind ligand (affinity modulation) and an increase in the total number of bonds formed (valency modulation) (9–16). Affinity of LFA-1 is regulated by conformational changes of individual monovalent receptors, whereas valency is regulated by the lateral mobility and clustering of receptors that change the number of bonds engaged at the contact area.
In the complex environment of the cell, affinity and valency modulation are potentially cooperative, and it is difficult to dissect the roles of the two mechanisms in mediating stable adhesion and migration. For example, whereas stimulation by chemoattractants or T cell receptor cross-linking is known to enhance adhesion of LFA-1-expressing cells to intercellular adhesion molecule 1 (ICAM-1) substrates (11), which enables rapid arrest of flowing leukocytes on endothelial cells (16, 17), direct affinity up-regulation stimulated by receptor tyrosine kinases or G protein-coupled receptors has not been demonstrated. This problem stems from a lack of reagents that are suitable for detecting affinity modulation of LFA-1 on the cell surface, where affinity up-regulation must be determined by binding of soluble monovalent ligands (9, 10, 12). Even in the high-affinity (HA) conformation, the binding of soluble monovalent ICAM-1, the strongest LFA-1 ligand identified (18), is weak and barely detectable (13, 15, 19). In lieu of direct detection of binding of monovalent ICAM-1, surrogate approaches have been adopted for detecting distinct activation states of LFA-1. Lollo et al. (15) used an indirect measurement, the displacement by soluble ICAM-1 of 125I-labeled Fab fragment, to estimate the affinity increase upon activation with phorbol myristoyl acetate (PMA). The binding of bivalent and multivalent ICAM-1 also allows one to assess increased affinity after activation (13, 16); however, with use of these reagents, complete separation of the affinity and valency states is not obtained.
The use of mAbs that bind selectively to the active conformation of LFA-1 represents an alternative, if indirect, approach for studying affinity up-regulation on the cell surface. Activation-dependent LFA-1 mAbs reported thus far map outside of the ligand-binding inserted (I) domain: NKI-L16 (20, 21) maps to the genu of the αL subunit, m24 (22–24) and 327C (25) bind to the β2 I-like domain, and KIM127 (26, 27) maps to I-EGF domain 2 in the β2 leg. Integrins that lack I domains have been shown to exist in at least three conformational states: bent (low affinity), extended with a closed headpiece putative intermediate affinity, and extended with an open headpiece (high affinity) (28, 29). Thus, extension, which is sufficient for exposure of the leg epitopes, coexists with two distinct conformations of the ligand-binding headpiece. Because affinity of LFA-1 is enhanced by conformational changes of the I domain (18, 30, 31), activation-dependent mAbs that map outside of the ligand-binding I domain cannot be used to ascertain LFA-1 affinity. Here, we functionally characterize a mAb, AL-57 (activated LFA-1 clone 57), that was selected by phage display for reactivity with the isolated αL I domain mutationally stabilized in the open, HA conformation. We demonstrate that binding of AL-57 is extremely sensitive to LFA-1 activation and can discriminate between low-affinity, intermediate-affinity (IA), and HA states of the integrin. We use this ligand-mimetic property of AL-57 to conclusively demonstrate affinity up-regulation of LFA-1 in lymphocytes stimulated by chemokine.
Results
Surface Plasmon Resonance (SPR) Analysis.
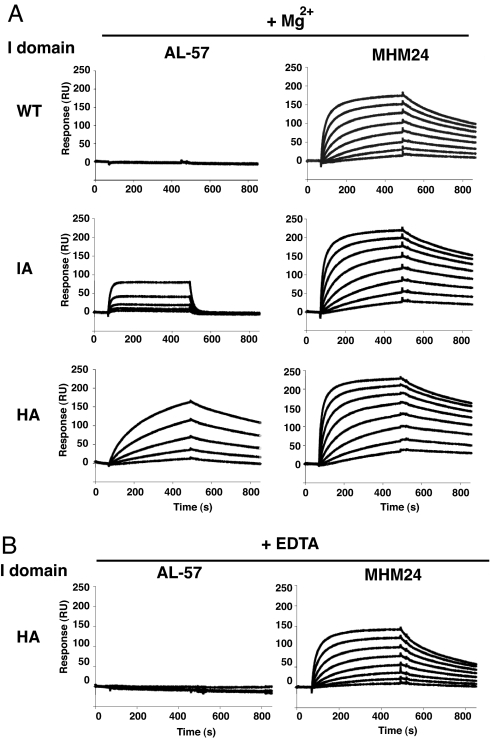
Selection of a human Fab library by phage display and identification of AL-57 is described in detail in ref. 32. We studied binding kinetics and selectivity of AL-57 for alternative conformations of the αL I domain by using SPR. For comparison, we used MHM24, a conventional activation-insensitive mAb to the αL I domain. MHM24 is a mouse mAb to human LFA-1 (33) that was humanized as efalizumab (Raptiva) and is used for the treatment of psoriasis patients (34–36). To exclude bivalent interactions, mAbs were immobilized on CM-5 sensor chips, and monovalent I domains were injected as analytes. MHM24 showed comparable binding and dissociation constants (KD) to WT disulfide-locked, IA L161C/F299C and HA K287C/K297C mutant I domains (31) (Fig. 1A and Table 1). By contrast, AL-57 showed no binding to the WT I domain, intermediate binding to the IA I domain, and good binding to the HA I domain (Fig. 1A and Table 1). Furthermore, unlike MHM24, binding of AL-57 depended on the presence of Mg2+, because EDTA abolished the binding (Fig. 1B and Table 1). The higher affinity to the HA mutant I domain than to the IA mutant I domain, the lack of detectable binding to the WT low-affinity I domain, and the absolute requirement for Mg2+ are key features shared with ICAM-1, showing that AL-57 is ligand mimetic.
Fig. 1.
SPR analysis of binding by the αL I domains to Abs AL-57 and MHM24. The HA (K287C/K294C), IA (L161C/F299C), or low-affinity WT I domain was perfused onto immobilized Abs in the presence of 1 mM MgCl2 (A) or 10 mM EDTA (B). The concentration series for MHM24 was 3.91, 7.81, 15.63, 31.25, 62.5, 125, 250, and 500 nM. For AL-57, the concentration series was 15.6, 31.3, 62.5, 125, and 250 nM for the HA I domain and 31.25, 62.5, 125, 250, and 500 nM for WT and IA I domains. In all cases, higher concentrations gave higher responses (except that differences are not visible for WT with AL-57 in Mg2+ and for HA with AL-57 in EDTA).
Table 1.
Dissociation constant of antibodies to alternative conformations of the αL I domain
| I domain |
KD, nM |
|||
|---|---|---|---|---|
| Mg2+ |
EDTA |
|||
| AL-57 | MHM24 | AL-57 | MHM24 | |
| WT | ND | 1.9 ± 0.4 | ND | 7.8 ± 0.5 |
| IA | 4,700 ± 3,200* | 2.0 ± 0.0 | ND | 4.6 ± 0.1 |
| HA | 23 ± 16 | 6.3 ± 0.2 | ND | 22 ± 0.0 |
SPR analysis was performed by using a BIAcore 3000 instrument. The I domains (IA, L161C/F299C mutant; HA, K287C/K294C mutant) were perfused onto antibodies immobilized on a CM-5 chip in buffer containing 1 mM Mg2+or 10 mM EDTA at a flow rate of 20 μl/min at room temperature. The data were fit by using a parallel reactions model plus drift to account for baseline offset, and KD was calculated as koff/kon. Data are means ± SEM of at least three independent experiments. ND, binding not detected.
*Determined by steady-state analysis.
An Acidic Residue in AL-57 Is Required for Binding to the HA I Domain.
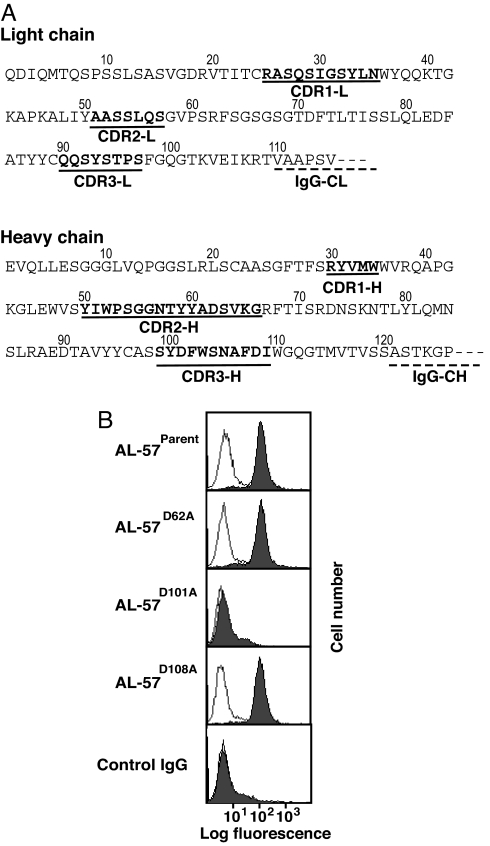
Most ligands to integrins, including ICAM-1, use an acidic residue to directly coordinate a magnesium ion at the metal ion-dependent adhesion site of the integrin I domain (31, 37, 38). The ligand-mimetic properties of the AL-57/LFA-1 interaction, especially the Mg2+ dependence of binding, led us to hypothesize that an interaction with an acidic residue in AL-57 is also required for I domain binding. The sequence of the complementarity-determining regions (CDRs) of the light and heavy chains revealed only three acidic residues: one Asp residue in CDR2-H and two Asp residues in CDR3-H (Fig. 2A), which we mutated individually to alanine.
Fig. 2.
Acidic residue D101 of AL-57 is critical for binding. (A) Primary sequence of variable regions of AL-57. The CDRs are shown in bold and are underlined. The beginning of a constant region is underlined with a dashed line. (B) Binding of parental and mutant AL-57 IgG (3 μg/ml) to the isolated, locked HA I domain expressed on K562 cells was examined by immunofluorescent flow cytometry in the presence of 1 mM Mg2+ (filled histograms) or 5 mM EDTA (open histograms). Binding of AL-57D101A in Mg2+ and EDTA is hardly distinguishable from background binding of control IgG.
Binding of parental and mutant AL-57 IgG to an isolated HA mutant I domain expressed on the surface of K562 cells (39) was tested by using immunofluorescent flow cytometry (Fig. 2B). Parental AL-57 bound well to the HA I domain in the presence of Mg2+. Binding of parental AL-57 was abolished by EDTA, confirming Mg2+-dependent binding (Fig. 2B, open histograms). Although the mutations D62A and D108A had no effect, the mutation D101A in heavy chain CDR3 completely abolished binding to the HA I domain (Fig. 2B). This result further supports the ligand-mimetic nature of recognition by AL-57 of the αL I domain.
Binding to WT αLβ2 Activated by Agonists.
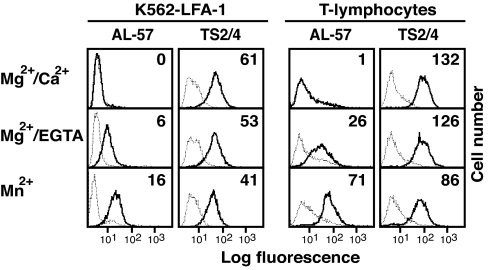
Binding of AL-57 to WT αLβ2 was examined by using WT LFA-1 on K562 transfectants as well as endogenously expressed LFA-1 on human T lymphocytes. In either context, LFA-1 activation, either with Mg2+/EGTA or Mn2+, was required to observe significant AL-57 binding (Fig. 3).
Fig. 3.
Binding of AL-57 IgG to WT LFA-1 activated by agonists as determined by immunofluorescent flow cytometry. K562 transfectants expressing WT LFA-1 and IL-15-cultured human primary T lymphocytes were stained for 20 min at 37°C with 20 μg/ml AL-57, TS2/4, or isotype-matched human or mouse control IgG in Hepes-buffered saline containing 1 mM MgCl2/1 mM CaCl2, 5 mM MgCl2/1 mM EGTA, or 1 mM MnCl2. Cells were washed and stained with FITC-conjugated goat anti-human or anti-mouse Abs. Staining with AL-57 and TS2/4 is shown as solid lines, and background staining with control IgGs is shown as dashed lines. Numbers within the panels show the specific mean fluorescence intensity of AL-57 and TS2/4.
Chemokine Activation.
AL-57 was used to observe LFA-1 activation after treatment with CXCL-12 (SDF-1) or the protein kinase C activator PMA. To exclude avidity (valency) effects, we used monovalent AL-57 Fab as the detection reagent. To obtain a large dynamic range of AL-57-detected activation, we used IL-15-cultured primary T cells. In mice (40) and humans (M.K., M.S., and T.A.S., unpublished data), stimulation of T lymphocytes followed by culture in IL-15 primarily induces memory cells that express latent LFA-1 with low basal ICAM-1-binding activity, whereas culture in IL-2 induces effector cells that express activated LFA-1 with increased basal ICAM-1 binding. The conformation-sensitive Ab KIM127 that maps to the leg of the β2 subunit (27, 41) was used to correlate global conformational rearrangement of LFA-1 with stimulation; the activation-insensitive Ab TS2/4 was used to quantify the total expression of LFA-1 on the cell surface.
We measured Ab epitope site density by using beads containing known numbers of mouse IgG-binding sites as described in Methods. In the resting condition, primary T cells cultured in IL-15 had a total of 27,000 LFA-1 sites per cell as determined with the conformation-insensitive mAb TS2/4. Resting T cells showed few AL-57- and KIM127-binding sites (Table 2). Upon stimulation with PMA or the chemokine CXCL-12 (SDF-1), the total number of LFA-1 sites increased slightly to 34,000 sites per cell (Table 2). By contrast, the number of AL-57 and KIM127 sites increased 10-fold or more, from 200–300 sites on resting cells to 2,200–3,900 sites on PMA- and CXCL-12-stimulated cells (Table 2). These results demonstrated that AL-57 could detect affinity up-regulation of LFA-1 by stimulation of both protein kinase C and a G protein-coupled receptor. In addition, the results showed that AL-57 and KIM127 activation-dependent neoepitopes are expressed on subpopulations of ≈10% of the LFA-1 molecules on the surface of agonist-stimulated T cells. Furthermore, AL-57 site density and KIM127 site density are roughly the same on agonist-stimulated T cells.
Table 2.
Antibody-binding sites on LFA-1 on lymphocytes
| Stimulation | Binding sites per cell |
||
|---|---|---|---|
| AL-57 | KIM127 | TS2/4 | |
| None | 200 ± 100 | 300 ± 100 | 27,000 ± 3,300 |
| PMA | 3,900 ± 1,300* | 2,800 ± 700* | 34,000 ± 3,800 |
| CXCL-12 | 2,200 ± 800* | 3,300 ± 500* | 34,000 ± 3,800 |
Human T lymphocytes were stimulated with PMA (100 nM) or CXCL-12 (SDF-1, 100 ng/ml) for 20 min at 37°C in the continued presence of antibody. Binding sites were determined with FITC anti-κ light chain and AL-57 Fab or FITC-KIM127 and FITC-TS2/4 IgG, and standardization was performed with beads containing known numbers of mouse IgG-binding sites as described in Methods. Data are the average of at least three independent experiments ± SEM.
*, P < 0.05 vs. no stimulation.
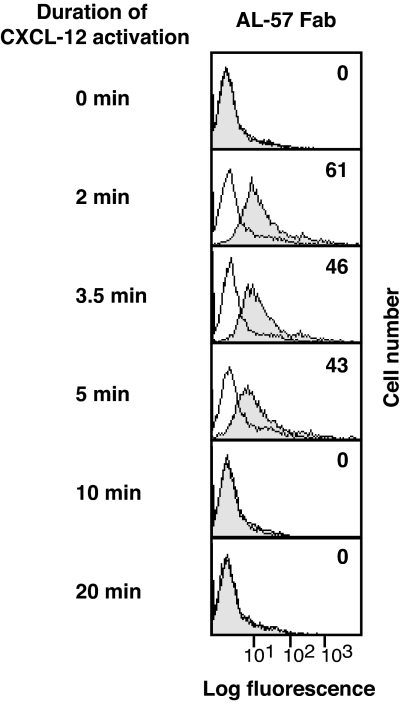
The adhesiveness of LFA-1 stimulated by the T cell receptor and G protein-coupled receptors has been shown to be transient (11, 16, 42). We therefore used AL-57 Fab to examine the kinetics of the increase in LFA-1 affinity after stimulation with CXCL-12. By the 2-min time point (the earliest time observable in our assay), AL-57 binding was significantly increased (Fig. 4). Binding peaked within 2–5 min after stimulation and was unobservable after 10 min.
Fig. 4.
The kinetics of LFA-1 activation after stimulation with CXCL-12. Representative histograms for a time course of AL-57 epitope expression are shown. T lymphocytes were stimulated with 50 ng/ml CXCL-12 for the indicated time period. Cells were stained with Fab AL-57 or control Fab labeled with Alexa 488 for 2 min by adding Abs 2 min before the end of stimulation. Cells were then fixed and subjected to flow cytometry. Background staining with control Fab is shown in the open histograms. Numbers within the panels show specific mean fluorescence intensity values for Fab AL-57. At 0, 10, and 20 min, differences between AL-57 Fab and control Fab binding are hardly visible.
Discussion
We demonstrate that the human mAb AL-57 specifically recognizes the active conformation of the LFA-1 I domain and use AL-57 to characterize the affinity state of LFA-1 on activated lymphocytes. Several lines of evidence demonstrate that AL-57 mimics ligands of LFA-1, such as ICAM-1. First, AL-57 binds to LFA-1 in an activation-dependent manner: This mAb binds to LFA-1 on the cell surface only when LFA-1 is activated; the affinity of AL-57 progressively increases as the affinity of the LFA-1 I domain for ICAM-1 rises, and binding of AL-57 to the low-affinity I domain is undetectable. Second, formation of the AL-57/I domain complex is Mg2+-dependent. Third, an aspartic acid residue in CDR3 of the heavy chain is required for binding. This residue is hypothesized to be responsible for the Mg2+-dependent binding through a direct coordination to the metal ion-dependent adhesion site metal. The interaction between an acidic residue in the ligand and the I domain-bound metal is a characteristic feature of authentic integrin-ligand architecture (31, 38, 43). Finally, we show in ref. 32 that AL-57 inhibits binding of ICAM-1 to WT activated LFA-1 as well as to LFA-1 containing the disulfide-locked HA I domain. Because ligand binding by the disulfide-locked HA I domain is blocked by competitive inhibition but not by allosteric inhibition (44), these results show that AL-57 directly competes with ICAM-1 for the I domain.
Compared with ICAM-1, AL-57 has comparable affinity for the mutant IA I domain and seven times higher affinity for the mutant HA I domain (Table 3). Furthermore, compared with ICAM-1, the koff of AL-57 is 8 times slower for the IA I domain and 250 times slower for the HA I domain (Table 3). The greater stability of the Ab–receptor complex compared with the ligand–receptor complex makes AL-57 an excellent reagent for investigating affinity up-regulation of LFA-1, as demonstrated here on chemokine and PMA-stimulated lymphocytes. The slow koff of AL-57 enhances its ability to probe affinity up-regulation when used for immunostaining of cells, in which dissociation of probes during washing hampers sensitive detection.
Table 3.
Comparison of binding kinetics of AL-57 and ICAM-1
| I domain |
kon, M−1·s−1 × 10−3 |
koff, s−1 × 102 |
KD, nM |
|||
|---|---|---|---|---|---|---|
| AL-57* | ICAM-1† | AL-57* | ICAM-1† | AL-57* | ICAM-1† | |
| WT | ND | 3.1 ± 0.1 | ND | 460 ± 36 | ND | 1,500,000 ± 200,000 |
| IA | 16.6 ± 13.6 | 133 ± 10 | 5.4 ± 5.2 | 43 ± 7 | 4,700 ± 3,200 | 3,000 ± 440 |
| HA | 2.1 ± 0.7 | 115 ± 7 | 0.0055 ± 0.0057 | 1.4 ± 0.1 | 23 ± 16 | 150 ± 16 |
Binding kinetics to the alternative conformations of the αL I domain (IA, L161C/F299C mutant; HA, K287C/K294C mutant) was measured by SPR with a BIAcore instrument in the presence of 1 mM MgCl2.
*For binding to the IA I domain, kon and koff were obtained by curve-fitting using a 1:1 binding model. KD was calculated by a Scatchard plot using binding at steady state. For binding to the HA I domain, kon and koff were obtained by using a parallel reactions model plus drift to account for baseline offset, and KD was calculated by koff/kon. Data are means ± SEM of at least three independent experiments. ND, binding not detected.
†Data are from ref. 31.
The ligand-mimetic feature of AL-57 is unique among several activation-dependent mAbs to integrin I domains that have been described. A function-blocking and activation-dependent mAb to the Mac-1 I domain, CBRM1/5 (45), maps adjacent to, rather than on, the ligand-binding site and does not require divalent cations (46). An α2 I domain mAb, IAC-1, binds preferentially to the active form but does not compete with ligand (47). A function-blocking mAb 107 binds to the Mac-1 I domain in a divalent cation-dependent manner but favors the closed conformation (48). An α1 I domain mAb, AQC2, is not activation-dependent yet is revealed by a crystal structure to coordinate to the metal ion-dependent adhesion site (MIDAS) through an Asp residue in CDR3 of the heavy chain (49). In contrast to AL-57, binding of AQC2 is decreased but not abolished in the absence of a metal or by Ala substitution of the MIDAS metal-coordinating Asp residue in CDR3 (49).
Activation-dependent Abs have been used to establish the structural mechanism of LFA-1 activation (20–22, 25, 27, 41, 44, 50). For example, the epitope of mAb KIM127 used in this study maps to a β2 leg epitope that is shielded in the bent, resting integrin conformation and becomes exposed in the extended, activated state (27, 28, 41, 51). The extended LFA-1 can contain a low-affinity, IA, or HA I domain, depending on the strength of activation and the density of ligand (9, 52). Therefore, the ability of AL-57 to directly report I domain conformation allows us to define LFA-1 conformations more precisely. The specificity of AL-57 for ligand-unoccupied LFA-1 further distinguishes it from previously available activation-dependent mAbs to LFA-1 described above, which bind whether or not LFA-1 is bound to ligand. Because affinity of AL-57 to the disulfide-locked HA I domain is 200 times higher than affinity to the disulfide-locked IA I domain, AL-57 would detect HA LFA-1 more efficiently than IA LFA-1 on the cell surface. However, one needs to consider the possibility that, with WT LFA-1 on live cells, AL-57 binds to both IA and HA conformations of LFA-1.
We have demonstrated by using Fab AL-57 that LFA-1 on lymphocytes increases its affinity upon activation of protein kinase C with PMA and physiological stimulation of G protein-coupled receptor CXCR4 with the chemokine CXCL-12. We have also shown that only a subset of ≈10% of the total number of LFA-1 molecules on T cells shows affinity up-regulation after stimulation with PMA and CXCL-12. These results with AL-57 Fab to LFA-1 on T cells correlate well with previous results with CBRM1/5 Fab to Mac-1 on neutrophils (45). Whereas neutrophils barely express CBRM1/5-binding sites in the resting condition, stimulation with PMA and the chemoattractant f-Met-Leu-Phe induced expression of the CBRM1/5 epitope on 10–30% of cell surface Mac-1 molecules. Both CBRM1/5 and AL-57 completely block ligand binding. Thus, after physiologic activation, a small subset of Mac-1 molecules on neutrophils and LFA-1 molecules on lymphocytes becomes competent to mediate adhesion.
The active, extended conformation of LFA-1 probed by KIM127 also represents a subset of ≈10% of the total LFA-1 molecules on activated T cells. Interestingly, the number of AL-57-binding sites is approximately equal to the number of KIM127-binding sites. This result strongly suggests that affinity up-regulation by chemokine as measured with AL-57 coincides with receptor extension as measured by KIM127. These results are consistent with electron microscopy and crystal structure studies on integrins that show that extension is essential for hybrid domain swing-out, which is allosterically linked to conversion of the ligand-binding site to the HA state (28, 29).
We used AL-57 Fab to study the kinetics and mechanism of LFA-1 activation after stimulation with CXCL-12. A transient increase in LFA-1 activity has been shown to be induced by chemokines through G protein-coupled receptor signals (16, 42, 53) and T cell receptor cross-linking (11). Although a rapid and reversible increase in affinity by dynamic conformational change of LFA-1 has been proposed, it has not been possible previously to measure monovalent affinity separately from multivalent avidity. Previous studies have measured adhesion to ICAM-1 immobilized on substrates (11, 42, 53), binding of multivalent protein micelles of full-length ICAM-1 (16), or competition with Fab binding (15) to overcome the inherently weak binding of ligand. Previous studies have reported that adhesiveness or multivalent ligand binding peaked at 2 min (16) or 10 min (53) after G protein-coupled receptor stimulation or 10 min after T cell receptor stimulation (11). Using ligand-mimetic AL-57 Fab, we unambiguously measured an increase in monomeric affinity of LFA-1 that was transient and peaked at 2–5 min after chemokine stimulation. The similarities in the kinetics of up- and down-regulation of monomeric affinity and adhesiveness suggests that affinity up-regulation is the dominant factor in T cell receptor-induced and G protein-coupled receptor-induced, transient, LFA-1-mediated adhesiveness.
Methods
Abs.
Details of selection of a human Fab library by phage display are described elsewhere (32). Briefly, the library was positively selected with the disulfide-locked HA K287C/K294C mutant I domain and negatively selected with the WT low-affinity I domain (31). Phage clones expressing Fab fragments were examined for preferential binding to the locked HA I domain over the disulfide-locked, low-affinity L289C/K294C mutant I domain (31). We found that clone 57 selectively recognized the HA I domain, and we designated it “AL-57” for activated LFA-1 clone 57. For further characterization, a Fab format of clone 57 was converted to an intact human IgG1 format. AL-57 IgG1 was expressed in CHO cells and purified with a protein L affinity column, followed by gel filtration on a Superdex 200 column (Amersham Pharmacia, Piscataway, NJ). AL-57 Fab and control Fab (from human IgG1; Calbiochem, San Diego, CA) were generated by papain digestion with a Fab preparation kit (Pierce, Rockford, IL) and further purified on a MonoQ HR 5/5 ion exchange column (Amersham Pharmacia). For studying interactions of acidic residues of AL-57 with the I domain, three Asp residues in CDRs were individually mutated to alanine by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Parental and mutant AL-57 IgGs were expressed in 293T cells.
SPR.
SPR analysis was performed by using a BIAcore 3000 instrument (BIAcore, Uppsala, Sweden). Abs were immobilized on a CM-5 sensor chip as described in ref. 54. WT locked intermediate and open I domains were as described in ref. 44. SPR measurements were performed in 10 mM Tris·HCl (pH 7.4)/140 mM NaCl/1 mM MgCl2/0.005% Tween 20 at a flow rate of 20 μl/min. The data were fit by using a parallel reactions model plus drift to account for baseline offset. The second binding constant was at least one order of magnitude weaker.
Cells.
K562 transfectants that express WT LFA-1 are described in refs. 23 and 39. Peripheral blood mononuclear cells were generated by centrifuging fresh blood through Histopaque (Sigma, St. Louis, MO) and taking the buffy coat. Lymphocytes were prepared by adsorbing the monocytes to gelatin-coated plates and culturing nonadherent lymphocytes in RPMI medium 1640 supplemented with 10% FBS and phytohemagglutinin (1 μg/ml) for 3 d, followed by culture in IL-15 (20 ng/ml) for 3–6 d. Flow cytometric analysis demonstrated that these cells were 97% CD3 positive and CD56 negative.
Site-Number Determination by Immunofluorescent Cytometry.
Epitope expression in terms of sites per cell was determined by comparing the mean fluorescence intensity (MFI) of bound mAb to Quantum Simply Cellular beads (Bangs Laboratories, Fishers, IN), which contain five bead sets with increasing numbers of goat anti-mouse-binding sites on their surface. From this analysis, a linear relation between MFI and binding site number was determined for FITC-conjugated KIM127 and TS2/4 IgG. For quantifying AL-57 Fab-binding sites, we compared binding of FITC-mouse monoclonal anti-human κ light chain (KPL, Gaithersburg, MD) to beads and to AL-57 Fab bound to cells. IL-15-cultured T lymphocytes were stained for 20 min at 37°C with AL-57 (20 μg/ml), KIM127 (10 μg/ml), or TS2/4 (10 μg/ml) in L15 medium/2.5% FCS in the presence or absence of PMA (100 nM) or CXCL-12 (100 ng/ml). Cells were fixed with cold 3.7% formaldehyde in PBS for 30 min on ice. Cells stained with AL-57 Fab were then incubated with FITC-mouse monoclonal anti-human κ light chain Ab. Cells and beads were subjected to immunofluorescent cytometry.
Time Course of Epitope Expression of AL-57 Fab.
To determine the expression of AL-57 epitope as a function of the time after CXCL-12 stimulation, IL-15-cultured human primary T lymphocytes (106 cells per point) in Hanks' balanced salt solution (HBSS) containing 2% glucose, 2% BSA, 1 mM MgCl2, and 1 mM CaCl2 were stimulated at time 0 with 50 ng/ml CXCL-12 for the indicated duration. Fab AL-57-Alexa Fluor 488 (20 μg/ml) or control Fab-Alexa Fluor 488 (20 μg/ml) was added 2 min before the end of stimulation. At the end of stimulation, cells were immediately fixed in cold 2% formaldehyde in HBSS. After fixation, cells were washed three times with HBSS containing 2% glucose and 2% BSA, resuspended in HBSS, and immediately subjected to immunofluorescent flow cytometry.
Acknowledgments
We thank Simone Piva for flow cytometry, Mary Mohrin and Ronnie Yoo for technical assistance, Illa Roy (Dyax) for DNA constructs, and Lili Huang for sharing unpublished data. This work was supported by National Institutes of Health Grants AI063421 (to M.S.) and CA31798 (to T.A.S.), the American Society of Hematology (M.S.), and the American Heart Association (M.K.).
Abbreviations
- LFA-1
lymphocyte function-associated antigen 1
- HA
high-affinity
- IA
intermediate-affinity
- I
inserted
- AL-57
activated LFA-1 clone 57
- ICAM-1
intercellular adhesion molecule 1
- CXCL-12
CXC chemokine ligand 12
- PMA
phorbol myristoyl acetate
- CDR
complementarity-determining region
- SPR
surface plasmon resonance.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Springer TA. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 2.Dustin ML, Springer TA. In: Guidebook to the Extracellular Matrix and Adhesion Proteins. Kreis T, Vale R, editors. New York: Sambrook & Tooze; 1999. pp. 228–232. [Google Scholar]
- 3.Perry MA, Granger DN. J Clin Invest. 1991;87:1798–1804. doi: 10.1172/JCI115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber C, Springer TA. J Clin Invest. 1997;100:2085–2093. doi: 10.1172/JCI119742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson RB, Lim LH, Tessier PA, Gavins FN, Mathies M, Perretti M, Hogg N. J Exp Med. 2001;194:219–226. doi: 10.1084/jem.194.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luscinskas FW, Kansas GS, Ding H, Pizcueta P, Schleiffenbaum BE, Tedder TF, Gimbrone MA., Jr J Cell Biol. 1994;125:1417–1427. doi: 10.1083/jcb.125.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carman CV, Springer TA. J Cell Biol. 2004;167:377–388. doi: 10.1083/jcb.200404129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dustin ML, Bivona TG, Philips MR. Nat Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- 9.Carman CV, Springer TA. Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Kim M, Carman CV, Yang W, Salas A, Springer TA. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dustin ML, Springer TA. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 12.Bazzoni G, Hemler ME. Trends Biochem Sci. 1998;23:30–34. doi: 10.1016/s0968-0004(97)01141-9. [DOI] [PubMed] [Google Scholar]
- 13.Stewart MP, Cabanas C, Hogg N. J Immunol. 1996;156:1810–1817. [PubMed] [Google Scholar]
- 14.van Kooyk Y, Figdor CG. Curr Opin Cell Biol. 2000;12:542–547. doi: 10.1016/s0955-0674(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 15.Lollo BA, Chan KW, Hanson EM, Moy VT, Brian AA. J Biol Chem. 1993;268:21693–21700. [PubMed] [Google Scholar]
- 16.Constantin G, Majeed M, Giagulli C, Piccib L, Kim JY, Butcher EC, Laudanna C. Immunity. 2000;13:759–769. doi: 10.1016/s1074-7613(00)00074-1. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence JB, Villnave CA, Singer RH. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 18.Shimaoka M, Lu C, Palframan R, von Andrian UH, Takagi J, Springer TA. Proc Natl Acad Sci USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganpule G, Knorr R, Miller JM, Carron CP, Dustin ML. J Immunol. 1997;159:2685–2692. [PubMed] [Google Scholar]
- 20.Keizer GD, Visser W, Vliem M, Figdor CG. J Immunol. 1988;140:1393–1400. [PubMed] [Google Scholar]
- 21.Xie C, Shimaoka M, Xiao T, Schwab P, Klickstein LB, Springer TA. Proc Natl Acad Sci USA. 2004;101:15422–15427. doi: 10.1073/pnas.0406680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dransfield I, Cabañas C, Barrett J, Hogg N. J Cell Biol. 1992;116:1527–1535. doi: 10.1083/jcb.116.6.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Shimaoka M, Zang Q, Takagi J, Springer TA. Proc Natl Acad Sci USA. 2001;98:2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamata T, Tieu KK, Tarui T, Puzon-McLaughlin W, Hogg N, Takada Y. J Immunol. 2002;168:2296–2301. doi: 10.4049/jimmunol.168.5.2296. [DOI] [PubMed] [Google Scholar]
- 25.Beals CR, Edwards AC, Gottschalk RJ, Kuijpers TW, Staunton DE. J Immunol. 2001;167:6113–6122. doi: 10.4049/jimmunol.167.11.6113. [DOI] [PubMed] [Google Scholar]
- 26.Robinson MK, Andrew D, Rosen H, Brown D, Ortlepp S, Stephens P, Butcher EC. J Immunol. 1992;148:1080–1085. [PubMed] [Google Scholar]
- 27.Lu C, Ferzly M, Takagi J, Springer TA. J Immunol. 2001;166:5629–5637. doi: 10.4049/jimmunol.166.9.5629. [DOI] [PubMed] [Google Scholar]
- 28.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 29.Xiao T, Takagi J, Wang J-H, Coller BS, Springer TA. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huth JR, Olejniczak ET, Mendoza R, Liang H, Harris EA, Lupher ML, Jr, Wilson AE, Fesik SW, Staunton DE. Proc Natl Acad Sci USA. 2000;97:5231–5236. doi: 10.1073/pnas.97.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimaoka M, Xiao T, Liu J-H, Yang Y, Dong Y, Jun C-D, McCormack A, Zhang R, Joachimiak A, Takagi J, et al. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L, Shimaoka M, Rondon I, Roy I, Chang Q, Po M, Dransfield DT, Ladner RC, Edge AS, Salas A, et al. J Leukocyte Biol. 2006 doi: 10.1189/jlb.1105649.. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hildreth JEK, Gotch FM, Hildreth PDK, McMichael AJ. Eur J Immunol. 1983;13:202–208. doi: 10.1002/eji.1830130305. [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb AB, Krueger JG, Wittkowski K, Dedrick R, Walicke PA, Garovoy M. Arch Dermatol. 2002;138:591–600. doi: 10.1001/archderm.138.5.591. [DOI] [PubMed] [Google Scholar]
- 35.Gordon KB, Papp KA, Hamilton TK, Walicke PA, Dummer W, Li N, Bresnahan BW, Menter A. J Am Med Assoc. 2003;290:3073–3080. doi: 10.1001/jama.290.23.3073. [DOI] [PubMed] [Google Scholar]
- 36.Leonardi CL, Papp KA, Gordon KB, Menter A, Feldman SR, Caro I, Walicke PA, Compton PG, Gottlieb AB. J Am Acad Dermatol. 2005;52:425–433. doi: 10.1016/j.jaad.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 37.Song G, Yang Y, Liu J-H, Casasnovas J, Shimaoka M, Springer TA, Wang J-H. Proc Natl Acad Sci USA. 2005;102:3366–3371. doi: 10.1073/pnas.0500200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 39.Lu C, Shimaoka M, Ferzly M, Oxvig C, Takagi J, Springer TA. Proc Natl Acad Sci USA. 2001;98:2387–2392. doi: 10.1073/pnas.041606398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. J Clin Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beglova N, Blacklow SC, Takagi J, Springer TA. Nat Struct Biol. 2002;9:282–287. doi: 10.1038/nsb779. [DOI] [PubMed] [Google Scholar]
- 42.Campbell JJ, Hedrick J, Zlotnik A, Siani MA, Thompson DA, Butcher EC. Science. 1998;279:381–384. doi: 10.1126/science.279.5349.381. [DOI] [PubMed] [Google Scholar]
- 43.Shimaoka M, Takagi J, Springer TA. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 44.Shimaoka M, Salas A, Yang W, Weitz-Schmidt G, Springer TA. Immunity. 2003;19:391–402. doi: 10.1016/s1074-7613(03)00238-3. [DOI] [PubMed] [Google Scholar]
- 45.Diamond MS, Springer TA. J Cell Biol. 1993;120:545–556. doi: 10.1083/jcb.120.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oxvig C, Lu C, Springer TA. Proc Natl Acad Sci USA. 1999;96:2215–2220. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoolmeester A, Vanhoorelbeke K, Katsutani S, Depraetere H, Feys HB, Heemskerk JM, Hoylaerts MF, Deckmyn H. Blood. 2004;104:390–396. doi: 10.1182/blood-2003-12-4224. [DOI] [PubMed] [Google Scholar]
- 48.Li R, Haruta I, Rieu P, Sugimori T, Xiong JP, Arnaout MA. J Immunol. 2002;168:1219–1225. doi: 10.4049/jimmunol.168.3.1219. [DOI] [PubMed] [Google Scholar]
- 49.Karpusas M, Ferrant J, Weinreb PH, Carmillo A, Taylor FR, Garber EA. J Mol Biol. 2003;327:1031–1041. doi: 10.1016/s0022-2836(03)00203-1. [DOI] [PubMed] [Google Scholar]
- 50.Woska JR, Jr, Shih D, Taqueti VR, Hogg N, Kelly TA, Kishimoto TK. J Leukocyte Biol. 2001;70:329–334. [PubMed] [Google Scholar]
- 51.Kim M, Carman CV, Springer TA. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 52.Salas A, Shimaoka M, Kogan AN, Harwood C, von Andrian UH, Springer TA. Immunity. 2004;20:393–406. doi: 10.1016/s1074-7613(04)00082-2. [DOI] [PubMed] [Google Scholar]
- 53.Shimonaka M, Katagiri K, Kakayama T, Fujita N, Tsuruo T, Yoshie O, Kinashi T. J Cell Biol. 2003;161:417–427. doi: 10.1083/jcb.200301133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vorup-Jensen T, Ostermeier C, Shimaoka M, Hommel U, Springer TA. Proc Natl Acad Sci USA. 2003;100:1873–1878. doi: 10.1073/pnas.0237387100. [DOI] [PMC free article] [PubMed] [Google Scholar]