Abstract
In the uterus, progesterone (P4) acts early in G1 as a physiological inhibitor of estradiol-17β (E2)-induced epithelial cell proliferation. Gene expression profiling of uterine epithelial cell RNA isolated 3 h after hormonal treatment of ovariectomized mice revealed the co-coordinate down-regulation by P4 of >20 genes whose functions are associated with DNA replication. This group included all of the minichromosome maintenance (MCM) proteins that are required for DNA replication licensing. E2 regulated loading of these MCM proteins onto chromatin in parallel with its induction of DNA synthesis. E2 caused this chromatin loading by retention of MCM proteins in the nucleus and through the induction of the loading factor Cdt1, which is necessary for the MCM heterohexamer to bind to the origin of DNA replication. P4 dramatically reduced the binding of the MCMs to chromatin by a number of mechanisms. First, MCM mRNA and protein abundance was down-regulated. Second, P4 inhibited the E2 induction of Cdt1. Third, P4 treatment sequestered the normally nuclear MCM proteins into the cytoplasm. This reduced MCM binding resulted in the complete inhibition of E2-induced DNA synthesis by P4. These data reveal mechanisms not only for female sex steroid hormone action but also in the regulation of DNA replication licensing.
Keywords: DNA replication, minichromosome maintenance, uterus, cell cycle, microarray
In uteri of mice and women, the female sex steroid hormones estradiol-17β (E2) and progesterone (P4) interact to regulate cell proliferation (1). In mice, E2 synthesized at proestrus stimulates uterine luminal and glandular epithelial cell proliferation (2). In contrast, P4 synthesized by corporea lutea formed after mating blocks the E2-induced epithelial cell proliferation but permits E2 to induce a single wave of stromal cell division (3, 4). P4 also induces differentiation of these epithelial cells so that they are receptive to the hatched blastocysts in order that implantation can proceed. These uterine cellular dynamics can be mimicked by administration of exogenous E2 and P4 to ovariectomized mice in regimens that parallel their physiological secretion (2, 5, 6). This hormone treatment provides a controllable model in which to study the mechanism of E2 induction of cell proliferation in target epithelial tissues and also the action of a physiological inhibitor of cell division, in this case P4. These studies are highly relevant because exposure to the mitogenic effects of E2 is thought to be the major risk factor for the development of endometrial and breast cancer (7).
DNA replication needs to be a highly regulated process, and most cells seek to ensure one (and only one) round of replication per cell cycle. DNA synthesis is initiated by the formation of the prereplicative complex (pre-RC) at origins of replication during early G1, a process known as replication licensing (8, 9). Pre-RC formation involves the sequential assembly of >20 replication factors in a process that is largely conserved from yeast to human. The origin of DNA replication is first marked by the origin recognition complex, a heterohexameric complex, which serves as a scaffold for the loading of additional proteins. The binding of Cdc6 and Cdt1 to the origin recognition complex facilitates the loading of the minichromosome maintenance (MCM) proteins (MCM2–7) onto the replication origins in stoichiometric amounts to form the pre-RC. Once loaded, origin firing can be activated by two kinases, Cdc7-Dbf4 kinase and cyclin-dependent kinase (CDK), to commence DNA synthesis. This origin firing involves the orderly recruitment of additional replication factors including Cdc45, proliferating cell nuclear antigen (PCNA), and DNA polymerase α, the latter of whose primase activity initiates DNA replication. Pre-RC formation confers competence on the origins to replicate only once in S phase. Studies in different organisms suggest that this process of pre-RC formation is a key mechanism that coordinates DNA replication with cell-cycle division.
Our previous studies in the adult mouse uterus have identified the nuclear localization of the cell-cycle regulatory molecule cyclin D1 as a major event in steroid hormone regulation of epithelial cell proliferation (10). E2 causes the nuclear accumulation of cyclin D1 together with its CDK partners, CDK4 and CDK6, to phosphorylate the retinoblastoma family of proteins. Thereafter, cyclin E/CDK2 is activated, cyclin A is induced, and the cells are propelled into DNA synthesis. P4 pretreatment completely blocks this cyclin D1 nuclear localization, and, as a consequence, retinoblastoma phosphorylation and cell-cycle progression are inhibited. Our recent studies have identified the mechanism that links the antagonistic actions of these two hormones and the subcellular localization of cyclin D1 (11). In the uterine epithelial cells E2 causes an inhibitory phosphorylation of GSK-3β at Ser9 through phosphatidylinositol 3-kinase activation of AKT. The inhibition of phosphatidylinositol 3-kinase activity by P4 results in active GSK-3β activity that phosphorylates cyclin D1 at Thr286 and its export from the nucleus (12). These data were confirmed by direct inhibition of GSK-3β that reversed the cyclin D1 accumulation into the nucleus in response to E2 even in the presence of P4. This cyclin D1 nuclear accumulation caused progression of the cells toward S phase as shown by the induction of the S phase markers PCNA and Ki67. However, despite the induction of these markers, no true DNA synthesis could be detected by the incorporation of BrdU. These data suggest that a second pathway (independent of the retinoblastoma one) needs to be activated by E2 for DNA synthesis to be initiated and that this pathway is also inhibited by P4.
In this study, we used analysis of gene expression patterns of the uterine luminal epithelial cells after E2 and P4E2 treatment to attempt to identify pathways that might be regulated by E2 and inhibited by P4. Remarkably, >20 genes associated with DNA replication were rapidly down-regulated by P4. Noticeably among this group were all six MCM proteins, suggesting that replication licensing is a key regulatory point in sex steroid hormone regulation of cell proliferation. This article shows that this is the case, with P4 regulating the abundance, activity, and cellular localization of MCM proteins indicating mechanisms controlling replication licensing in the uterine epithelium by sex steroid hormones in vivo.
Results
Microarray Analysis Revealed That Genes Associated with DNA Replication Are Down-Regulated by P4.
To study the molecular mechanism of P4 inhibition of the E2-induced proliferation in the uterine epithelium, a cDNA microarray comparison was performed between mice treated with E2 and P4E2. Ovariectomized adult mice were pretreated with P4 for 3 days before a final combined injection of P4 and E2 on the fourth day in regimens that parallel the physiological concentrations (13). The RNA samples obtained were compared with mice given E2 treatment that mimicked the preovulatory surge of the estrus cycle (14). Given these different responses in different cell types of uteri and because P4 exerts its inhibitory action on the uterine epithelium within the first 3 h (15), we isolated luminal epithelial cells at >95% purity (16, 17) over a time window of 3–4 h after P4E2 or E2 treatment and prepared RNA samples from groups of three mice. These samples were cross-compared by using cDNA microarrays as described in Supporting Methods, which is published as supporting information on the PNAS web site.
After LOWESS normalization, data analysis, and data cutoffs by the filter criteria described in Materials and Methods, our analysis revealed that of the 27,396 gene array transcripts, 222 (0.810%) and 208 (0.759%) gene sequences were up-regulated and down-regulated, respectively, in the uterine epithelia after the treatment of P4E2 compared with E2. The complete list of transcripts whose abundance was decreased is shown in Table 2, which is published as supporting information on the PNAS web site, and those 222 gene transcripts that were up-regulated have been discussed in another publication (17). Our analysis in the down-regulated group focused on the genes with known function, and this resulted in removal of 82 unknown genes such as EST and RIKEN cDNA gene sequences. To gain insight into the biological significance of the down-regulation in gene expression, classification of the remaining down-regulated gene sequences by Gene Ontology annotations showed that 32 of 126 known sequences were involved in cell-cycle control, DNA replication, and modification processes (Table 1). This percentage is a significant enrichment of this function category (25.3%). Well defined cell-cycle-related genes, such as Mki-67, E2f1, Ccnd1, Ccne1, and Tk1, are present. Striking is the coordinate down-regulation of all MCM-deficient (Mcm2–7) genes. These data were further validated because Mcm2, Mcm4, and Mcm5 are repeated twice on the chips by different cDNA sequences. Additionally, there was a concomitant reduction of transcripts of DNA elongation genes including Fen1, Pcna, and Lig1. Finally, transcripts of a number of genes involved in chromatin assembly and modification were also down-regulated. Examples included Chaf1a, Chaf1b, Hells, and Ppp1r1b (Table 1). Collectively, our data indicate that the entire DNA replication process in the uterine luminal epithelium, including DNA prereplication assembly, replication elongation, and nucleosome assembly and modification, is a major target of P4 in early G1 phase.
Table 1.
List of genes associated with cell cycle and DNA replication whose transcripts are down-regulated by P4
| Name | Symbol | ID* | Average M† |
|---|---|---|---|
| DNA pre-RC licensing genes | |||
| MCM-deficient 4 homolog | Mcm4 | AA259788 | −1.444577042 |
| AI325074 | −1.371608965 | ||
| MCM-deficient 5 | Mcm5 | AA031056 | −1.286345349 |
| AW536273 | −1.263153464 | ||
| MCM-deficient 3 | Mcm3 | AW536712 | −1.236508743 |
| MCM-deficient 2 mitotin | Mcm2 | AA011839 | −1.02171739 |
| AW553939 | −1.004775339 | ||
| MCM-deficient 7 | Mcm7 | AA064230 | −0.9295692 |
| MCM-deficient 6 | Mcm6 | AA016759 | −0.883079258 |
| DNA replication genes | |||
| Flap structure-specific endonuclease 1 | Fen1 | AW538437 | −1.114619134 |
| PCNA | Pcna | AW545318 | −0.990405638 |
| Ligase I, DNA, ATP-dependent | Lig1 | C77364 | −0.91547759 |
| Chromatin assembly and modification genes | |||
| Chromatin assembly factor 1, subunit B (p60) | Chaf1b | AW547084 | −1.468701553 |
| AA387585 | −1.070805707 | ||
| W64706 | −0.91775875 | ||
| Helicase, lymphoid specific | Hells | AW555541 | −1.397495102 |
| Chromatin assembly factor 1, subunit A (p150) | Chaf1a | AU016029 | −1.196072702 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 1B | Ppp1r1b | AU040756 | −1.194267115 |
| Other cell-cycle-related genes | |||
| Stratifin | Sfn | AA009229 | −1.556398182 |
| AU043198 | −0.915968886 | ||
| AW536416 | −1.423732936 | ||
| Antigen identified by monoclonal antibody Ki 67 | Mki67 | AW536416 | −1.423732936 |
| E2F transcription factor 1 | E2f1 | AA396123 | −1.206181967 |
| CDK-like 2 (CDC2-related kinase) | Cdkl2 | AA414632 | −1.072730785 |
| Cyclin D1 | Ccnd1 | AU015041 | −1.01337925 |
| AI894115 | −0.940600249 | ||
| MAD2 (mitotic arrest deficient, homolog)-like | Mad2l1 | AA174630 | −0.993491411 |
| 1 (yeast) | C87726 | −0.913837172 | |
| Thymidine kinase 1 | Tk1 | AW544533 | −0.959189527 |
| Myeloblastosis oncogene | Myb | AA267899 | −0.977284174 |
| Myeloblastosis oncogene-like 2 | Mybl2 | AW555561 | −0.916437637 |
| Cyclin E1 | Ccne1 | AA465987 | −0.893640476 |
*NCBI nonredundant BLAST.
†log2 P4E2/E2.
Down-Regulation of MCM Protein Requires the Synergistic Action of P4 and E2.
MCM family members are highly conserved proteins found in all eukaryotes and play an important role in DNA replication licensing (8, 9). We focused on this family of proteins and the assembly of the pre-RC in the uterine epithelium. First, we performed quantitative real-time PCR (QRT-PCR) analysis for all Mcm genes to confirm the validity of the microarray result. As shown in Fig. 1A, there was similar inhibition pattern by P4 of 2- to 4-fold on Mcm transcript expression as determined by the microarrays and QRT-PCR, although the down-regulation of Mcm5 and Mcm6 was estimated to be greater by QRT-PCR than by the microarrays. In each case, this down-regulation was statistically significant. However, Mcm7 was an exception because QRT-PCR did not confirm the microarray result. To determine which of the two was correct, two other pairs of primers to Mcm7 were designed for QRT-PCR, and both of these indicated that there was no change in Mcm7 expression.
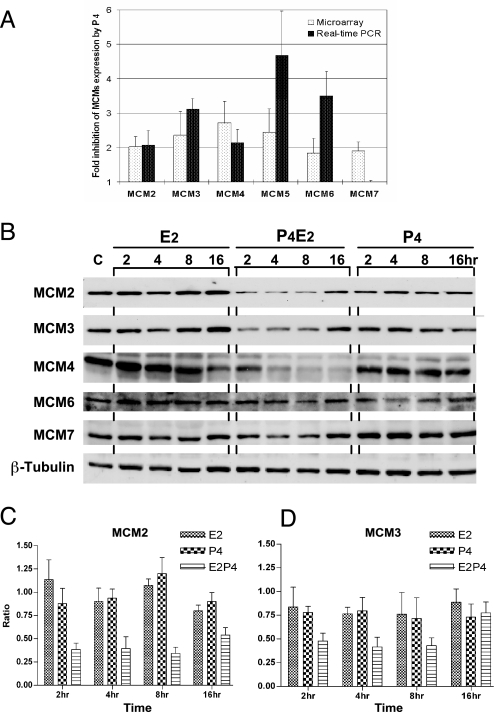
Fig. 1.
Hormonal regulation of Mcm expression. (A) Validation of cDNA microarray data by QRT-PCR by using the gene-specific primers shown in Table 3, which is published as supporting information on the PNAS web site. The y axis shows the amplitude of down-regulation determined by Microarray or QRT-PCR of the genes shown on the x axis. Data shown are the mean ± SD of three experiments and show significant down-regulation by P4 compared with E2 treatment. (B) Synergistic down-regulation of MCM2, MCM3, and MCM4 protein concentration by E2 and P4. Ovariectomized mice were killed at the times shown after the different hormone treatments as indicated. Equivalent amounts (60 μg) of protein isolated from the total epithelial cell lysates were separated by SDS/PAGE, blotted onto Nylon membranes, and probed with the antibodies against the proteins listed on the side. Detection of β-tubulin with an anti-β-tubulin antibody was used as a protein loading control. The Western blots shown are representative of those obtained from three independent experiments. (C and D) Densitometric analysis of the expression of MCM2 (C) and MCM3 (D) after the various treatments shown. Data shown are the mean ± SD of three experiments.
To examine the hormonal regulation of the MCM proteins in the uterine luminal epithelium, we treated three independent cohorts of mice with the appropriate hormonal regimens followed by the preparation of epithelial cell lysates over a 16-h period after treatment that were subjected to Western blotting. Compared with the hormone-primed but otherwise untreated control mice (referred to as controls), the concentration of MCM2, MCM3, MCM4, MCM6, and MCM7 proteins barely showed any change after E2 or P4 treatment alone (Fig. 1B). In contrast, P4 pretreatment in combination with E2 caused an ≈40% down-regulation of MCM2 (Fig. 1 B and C) and MCM3 (Fig. 1 B and D) between 2 and 4 h after treatment. A similar down-regulation of MCM4 was also seen (Fig. 1B) However, the protein concentrations of MCM6 and MCM7 did not show a significant reduction after P4E2 treatment. There was a similar kinetic in the regulation of MCM2 and MCM3, with a drop 2 h after treatment to reach a nadir at ≈8–9 h, followed by a progressive return until 16 h. However, P4E2 induced a gradual decrease of MCM4 from 2 h until 16 h. These data show that E2 and P4 synergize to down-regulate MCM2, MCM3, and MCM4 protein abundance, although with different kinetics.
P4 and E2 Regulate MCM Cellular Distribution.
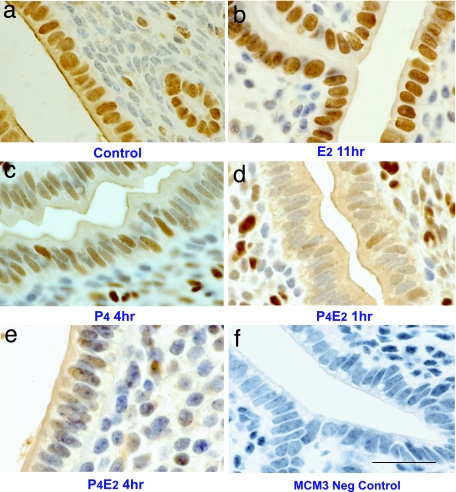
In metazoans, MCM are nuclear proteins organized in heterohexameric complexes that are primarily retained in the nucleus in a cell-cycle-independent manner (9). Unexpectedly, when we analyzed the cellular distributions of the MCMs by immunohistochemistry (IHC) of transverse sections of uteri, we found that the different hormone treatments altered their cellular localization. MCM3 had a predominant nuclear localization in the uterine epithelium of control mice, although there was some weak cytoplasmic staining (Fig. 2a). E2 treatment caused, as determined by the intensity of the IHC stain, a recruitment of this minor cytoplasmic fraction into the nucleus, and by 11 h after E2 treatment there was prominent nuclear staining with very little detected in the cytoplasm (Fig. 2b). In contrast, P4 treatment alone compared with the control reduced the nuclear MCM3 signal and concomitantly increased it in the cytoplasm (Fig. 2c). In the P4E2-treated uteri more MCM3 signal was detected in the cytoplasm within 1–2 h together with a concordant reduction in the nuclear signal (Fig. 2d). Consistent with the decrease in total protein caused by P4E2 treatment, the intensity of MCM3 staining in both the nucleus and cytoplasm was decreased by 4 h and reached its lowest level ≈8 h after hormone treatment (Fig. 2e), followed by a gradual increase in total MCM3 staining intensity at 11 h with some nuclei staining positive (data not shown). As a control for antibody specificity, we incubated the anti-MCM3 antibody with the cognate peptide before IHC, and this treatment completely inhibited the signal, confirming that MCM3 was being detected (Fig. 2f).
Fig. 2.
MCM3 localization is controlled by E2 and P4 in the mouse uterine epithelium. Shown is immunostaining for MCM3 of uterine transverse sections isolated at the indicated times after the different hormonal treatments. (a) Control. (b) Fifty nanograms of E2 at 11 h. (c) One milligram of P4 for 4 days. (d and e) One milligram of P4 for 4 days and killed at 1 h (d) and 4 h (e) after 50 ng of E2 on the fourth day. (f) Inhibition of the IHC signal by a competitive MCM3 peptide. Brown indicates positive staining, and the columnar cells are the luminal epithelium. (Scale bar: 50 μm.)
Examination of MCM2 by IHC demonstrated that the cellular distribution of MCM2 in the uterine epithelium was also reversely regulated by E2 and P4 treatments (Fig. 6, which is published as supporting information on the PNAS web site). The kinetics of redistribution of MCM2 in the P4E2-treated uteri was similar to that of MCM3. However, unlike MCM3, MCM2 was solely retained in the uterine epithelial cell nuclei in both the untreated control and E2-treated uteri. Similar to MCM3, preincubation with cognate peptide reduced the signal to background (Fig. 6g).
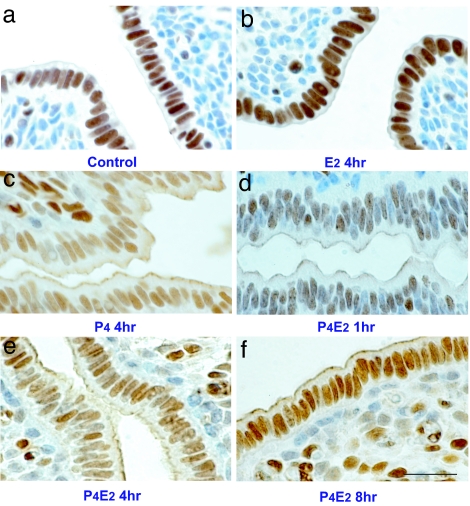
We further analyzed the distribution of MCM6 and MCM7 in the uterine epithelium after the different hormone treatments by IHC. Because MCM6 and MCM7 showed exactly the same pattern, only the distribution of MCM6 in the uterine epithelium is reported here (Fig. 3). MCM6 was found only in nuclei of control and E2-treated uterine epithelial cells (Fig. 3 a and b). P4 treatment caused a redistribution of MCM6 from the nucleus to the cytoplasm of the epithelial cells (Fig. 3c). In contrast to MCM2 and MCM3, after P4E2 treatment, although some of MCM6 stayed in the cytoplasm at 1 h (Fig. 3d), MCM6 shifted back to the nucleus from the cytoplasm from 2 h after hormone administration, and the majority of MCM6 had a nuclear localization by 8 h (Fig. 3 e and f). No competitive peptide was available for either MCM6 or MCM7, although omission of primary antibody reduced the IHC signal to background (Fig. 6h). The localization of MCM4 and MCM5 in the uterine epithelium could not be ascertained because of the lack of appropriate antibodies for IHC.
Fig. 3.
P4 transiently excludes MCM6 from the nucleus. Shown is immunostaining for MCM6 of uterine transverse sections isolated at the indicated times after the different hormonal treatments. (a) Control. (b) Fifty nanograms of E2 at 4 h. (c) One milligram of P4 for 4 days. (d–f) One milligram of P4 for 4 days and killed 1 h (d), 4 h (e), and 8 h (f) after 50 ng of E2 on the fourth day. (Scale bar: 50 μm.)
Thus, in contrast to the observations in almost all species that MCM are solely nuclear proteins, in the mouse uterine epithelium MCMs shift between the nucleus and cytoplasm under the influence of the different hormone regimens.
MCM Complex Loading onto the Chromatin Is Induced by E2 and Inhibited by P4.
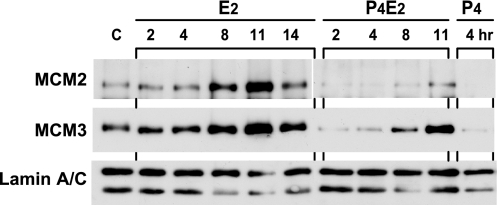
The stepwise recruitment of origin recognition complex, Cdc6 and Cdt1, followed by MCM complex onto the chromatin for pre-RC assembly appears to be conserved in eukaryotes and is required for the initiation of DNA replication (18, 19). Therefore, MCM are present in the nucleus in two different forms: one is soluble and extractable by nonionic detergents, and the other is tightly associated with nuclear structure and resistant to this extraction (20, 21). The redistribution of MCMs from nucleus to cytoplasm combined with the reduction of the total MCM protein led us to examine the chromatin association of MCMs regulated by female steroid hormones. Using lamin A/C as a loading control, some chromatin-associated MCM2 and MCM3 protein could be detected by Western blotting in control samples (Fig. 4). This finding is consistent with the low rate of epithelial cell proliferation in these control mice (≈5%). After E2 treatment, additional MCM2 and MCM3 chromatin binding occurred cumulatively throughout G1 phase, starting from 4 h and peaking between 8 and 11 h. After this time there was a gradual dissociation from the chromatin with a return to almost the basal level at 14 h (Fig. 4). This loading of MCMs onto the chromatin is restricted to the G1 phase of the cell cycle, consistent with the role of MCM in replication licensing. P4 treatment of the mice alone resulted in the dissociation of MCM2 and MCM3 from the epithelial chromatin to an almost undetectable level (Fig. 4). This pretreatment with P4 almost completely abolished the E2-induced MCMs loading onto chromatin through the first 8 h of E2 treatment. However, after 8 h of P4E2 treatment some MCM2 and MCM3 was detected in the chromatin fraction, although the level of chromatin-bound MCMs remained significantly lower when compared with that bound in the samples treated with E2 alone (Fig. 4). Thus, P4 not only dissociated the existing MCMs from chromatin during the first 3 days of pretreatment but also significantly delayed and lowered the E2-induced MCMs binding onto chromatin (Fig. 4).
Fig. 4.
P4 inhibits the E2-induced chromatin binding of MCM complexes in the mouse uterine epithelium. Uterine luminal epithelial cells were purified at indicated times after treatment with vehicle alone (C), E2, P4, or P4E2 as described. Chromatin-bound insoluble proteins were separated from total epithelial lysates as described in Materials and Methods. Equal amounts (30 μg) of protein were separated by SDS/PAGE and blotted onto nylon membranes. These blots were probed as indicated with antibodies to MCM2, MCM3, and lamin A/C. Lamin A/C served as the loading control for the chromatin-bound MCM2 and MCM3 proteins. Shown are representative Western blots from three independent experiments.
E2 and P4 Exert Their Regulation Through Cdt1.
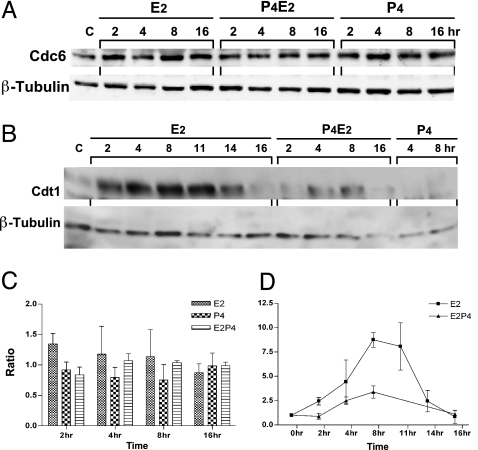
At the replication origin, the binding of the members of the pre-RC to chromatin is strictly ordered. Origin recognition complex must first bind to chromatin to allow Cdc6 and Cdt1 binding, and this is a prerequisite for MCM binding (18, 19). To investigate the E2 regulation of MCM recruitment onto the chromatin and its inhibition by P4 pretreatment, we explored the hormonal regulation of Cdc6 and Cdt1 in the uterine epithelial cells. Cdc6 protein level in the total cell lysates of the uterine epithelial cells was found to remain approximately constant over the entire 16-h time course after the different hormone treatments (Fig. 5A and C). No significant differences in the subcellular localization of Cdc6 in the uterine epithelium were observed after the different hormone treatments (data not shown). We further investigated Cdt1 protein levels in the total cell lysates of the epithelial cells after hormonal treatment. Interestingly, the Cdt1 level in the epithelial cells was very low in control mice, and similar levels were found after P4 treatment (Fig. 5 B and D). However, E2 treatment increased Cdt1 protein abundance within 2 h followed by an ≈10-fold accumulation that reached a plateau at the G1/S phase transition between 8 h and 11 h, followed by a rapid loss to basal levels at 16 h (Fig. 5 B and D). The tight control of Cdt1 protein level by E2 stimulation is precisely coincident with the loading of MCM onto chromatin during the cell-cycle progression of these epithelial cells (Fig. 4). P4 pretreatment significantly attenuated the E2-induced elevation of Cdt1, although there was still a rise that began from 4 h and reached a maximum by 8 h before declining to the basal level at 16 h (Fig. 5B). This induction parallels the kinetics of MCM loading onto chromatin in the P4E2-treated sample (Fig. 4). Taken together, P4 and E2 differentially regulate the two pre-RC loading factors, Cdc6 and Cdt1. Cdt1 rather than Cdc6 is the limiting factor for either the association of MCM with chromatin in the cell-cycle epithelial cells or the displacement of MCM complexes from chromatin in the P4E2-induced differentiation cells.
Fig. 5.
P4 and E2 differentially regulate the two DNA replication initiation factors Cdc6 and Cdt1 in the uterine epithelium. Shown are representative Western blots from three independent experiments of total uterine luminal epithelial cell extracts prepared at various time points after hormonal treatments as indicated and probed with anti-Cdc6 antibody (A) and anti-Cdt1 antibody (B). Detection of β-tubulin was used as the loading control. (C and D) Densitometric analysis of the expression of Cdc6 (C) and Cdt1 (D) at different time points after the hormonal treatments as indicated. Data shown are the mean ± SD of three experiments.
Discussion
P4 blocks the mitogenic activity of E2 in the uterine luminal and glandular epithelium of mice and women. In mice, if given 2 days before the E2, mimicking the physiological condition, this suppression of cell division is complete (2). Studies altering the timing of P4 administrations indicate that its inhibitory actions occur early in G1 and acts within the first 3 h (4).
Both E2 and P4 require their transcription factor receptors for their actions on cell proliferation, suggesting that they work via transcriptional mechanisms (22, 23). Thus, there have been several studies that have profiled gene expression patterns in the uterus after sex steroid hormone treatment of ovariectomized mice or of endometrial samples isolated at different stages of the menstrual cycle in humans (24–27). In general these studies have taken RNA extracted from whole uterine tissue to profile. However, the uterus is a complex multicellular tissue, and it also shows dynamic changes in cell type under different hormonal conditions. Thus, the approach of taking whole-organ lysates for analysis obfuscates the cell-type-specific responses, particularly those in the uterine epithelium, which comprises only 5% of the uterine cell mass. In the current studies we isolated the uterine luminal epithelium before RNA purification and expression profiling on cDNA microarrays. We chose the 3-h time point after E2 and P4E2 treatments because of the aforementioned fact that P4 acts early in G1.
This study revealed ≈200 expressed gene sequences whose transcripts were down-regulated 3 h after P4E2 treatment. Of these, Gene Ontology analysis showed that >20 genes (32 sequences) were involved in the DNA replicative process, including replication elongation and nucleosome assembly (Table 1). Remarkably, transcripts for five Mcm genes (Mcm2–6) involved in the prereplication licensing were down-regulated at this time, suggesting that this pathway is a major target of P4 action.
Interestingly, unlike other cell types in which reduction of DNA synthesis is always accompanied by a concomitant down-regulation of RNA splicing and transport and protein synthesis during the cell differentiation process (28), these processes are unaffected by P4. Indeed, the hypertrophy induced by E2 is comparable between the epithelial cells regardless of whether the mice have been treated with P4 (29). E2 also stimulates the induction of immediate early gene expression, including the protooncogenes c-fos, c-myc, and a little later c-rasHa, and this expression is also unaffected by P4 (1). This result was confirmed by analysis of the present cDNA microarray expression data (Table 2).
In this study we concentrated on the MCM proteins because of their importance in the regulation of DNA replication licensing. MCM proteins were originally identified in genetic screens for mutants defective in MCM in Saccharomyces cerevisiae and then characterized in a variety of eukaryotes, including mouse and human (8, 9). All six MCM members are closely related in sequence and structure with several highly conserved regions. Most of the MCM protein members cosediment on glycerol gradients and coelute after gel filtration as a complex with a molecular mass of ≈600 kDa, and which is composed of each MCM protein in an equimolar stoichiometry (8, 30, 31). MCM complex assembly occurs in the cytoplasm followed by entry into the nucleus. Because only MCM2 and MCM3 possess identifiable nuclear localization sequences (32, 33), these two MCM members probably provide the nuclear targeting signal for the entire complex. Although MCM proteins function as a hexameric complex, each MCM member is uniquely required for DNA replication because mutation in any single MCM gene in budding, and fission yeast has an equal effect on inhibiting DNA replication and cell viability (34, 35).
Our study shows that preinitiation licensing is tightly controlled during the cell cycle in the E2-induced uterine epithelium. Pre-RC assembly is regulated mostly at the level of loading of the MCM proteins onto the chromatin without change in overall MCM protein level and distribution in the luminal epithelium. This finding is consistent with the previous finding in most organisms that MCM proteins do not fluctuate during the cell cycle and are constitutively retained in the nucleus (8, 9). E2 stimulates MCM complex chromatin binding early in G1 phase, starting from 2 h and peaking at 8–11 h at the G1-to-S phase transition.
The data presented in this study show that P4 completely inhibits this E2-induced uterine epithelial cell proliferation by targeting replication licensing in early G1 phase through several mechanisms. First, measurements with both cDNA microarray and QRT-PCR showed that P4 reduced the transcript abundance of Mcm2–6. Furthermore, Western blotting indicated that MCM2, MCM3, and MCM4 protein concentration was also reduced by P4. This down-modulation required the E2 to be in combination with P4, because treatment with P4 alone did not have a significant effect on the concentrations of these proteins compared with the hormone-untreated control mice. Because a modest reduction in any one of MCM levels in other systems results in a dramatic inhibition of DNA synthesis (34, 35), it seems probable that this P4-induced down-regulation of MCM is part of the cause of the profound block of DNA synthesis by P4.
P4 pretreatment significantly lowered the E2-induced chromatin binding of MCM proteins. This reduction in chromatin binding paralleled the reduction of Cdt1 but not Cdc6 protein. Several lines of evidence indicate that Cdc6 and Cdt1 physically interact with each other and functionally cooperate in the recruitment of MCM complex onto chromatin (36, 37). However, our data indicate that female steroid hormone differentially regulates these two licensing factors in the uterine epithelial cells during both proliferation and differentiation. This finding strongly suggests that P4 regulates DNA replication licensing by inhibiting MCM complex loading through reducing the abundance of Cdt1 in exact opposition to E2 action that enhances Cdt1 concentration and consequently MCM loading.
A third level of regulation by P4 was achieved by alterations in cellular localization of MCM proteins. In all other cases documented, MCM are predominantly nuclear proteins (9). Such a nuclear localization for MCM2, MCM3, MCM6, and MCM7 was observed in uterine epithelial cells in control and E2-treated mice. However, as assessed by IHC, P4 pretreatment resulted in a redistribution of these proteins to the cytoplasm, with the effect being greatest on MCM2 and MCM3. The subsequent E2 treatment in the face of P4 resulted in a further depletion from the nucleus over the first 2 h of treatment, although there was some rebound at 8 h after E2. The exact mechanism of this exclusion remains to be determined, but these results show a mechanism for the control of replication licensing through changes in MCM cellular localization.
Our previous studies indicated that P4 inhibited the canonical cell-cycle regulatory machinery by blocking cyclin D1/CDK4 nuclear localization (10, 11). Here we demonstrate that P4 inhibits the replication licensing machinery at many levels. Thus, P4 exerts its complete block of cell proliferation by inhibiting two E2-induced pathways in early G1 phase that apparently run in parallel but both of which are required for the E2 induction of DNA synthesis.
Materials and Methods
Animal Treatment and Purification of Uterine Epithelial Cells.
Adult female CD1 mice (Charles River Laboratories, Wilmington, MA) were ovariectomized at 8–10 weeks of age under anesthesia and rested for 2–3 weeks before any hormonal treatment. E2 and P4 were purchased from Sigma Chemical (St. Louis, MO) and given s.c. in peanut oil as described before (10). All experiments and all time points were independently repeated at least three times with similar results.
After hormone treatment, uteri were removed, split longitudinally, and vortexed with Teflon beads (Small Parts, Miami, FL) in 1 ml of extraction buffer for 2.5 min as described to obtain an epithelial cell preparation that is >95% pure (16). This preparation was used for either protein or total RNA isolation as described (10, 38).
Supporting Information.
For further details, see Supporting Methods.
Supplementary Material
Acknowledgments
We thank Dr. F. Hanaoka (RIKEN, Wako, Japan) for the anti-mouse Cdt1 antibody; Aldo Massimi and Shufen Chen (Albert Einstein College of Medicine Cancer Center Microarray Facility) for support and assistant with the microarray experiments; Cheng Fan (Albert Einstein College of Medicine Cancer Center Biotechnology Facility) for help with microarray analysis; and Dr. Liyin Zhu and Jim Lee for help with the mouse experiments. This research was supported by National Institutes of Health Grants R01 CA 89617 and P30 CA13330 (to the Albert Einstein Cancer Center). J.W.P. is the Sheldon and Betty E. Feinberg Senior Faculty Scholar in Cancer Research.
Abbreviations
- MCM
minichromosome maintenance
- CDK
cyclin-dependent kinase
- QRT-PCR
quantitative real-time PCR
- pre-RC
prereplicative complex
- E2
estradiol-17β
- P4
progesterone
- IHC
immunohistochemistry
- PCNA
proliferating cell nuclear antigen.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tong W, Pollard JW. In: The Endometrium. Glasser SR, Aplin JD, Giudice LC, Tabibzadeh S, editors. London: Taylor & Francis; 2002. pp. 94–109. [Google Scholar]
- 2.Martin L, Finn CA, Trinder G. Endocrinol. 1973;56:303–307. doi: 10.1677/joe.0.0560303. [DOI] [PubMed] [Google Scholar]
- 3.Finn CA, Martin L. J Endocrinol. 1969;45:57–65. doi: 10.1677/joe.0.0450057. [DOI] [PubMed] [Google Scholar]
- 4.Martin L, Das RM, Finn CA. J Endocrinol. 1973;57:549–554. doi: 10.1677/joe.0.0570549. [DOI] [PubMed] [Google Scholar]
- 5.Martin L, Finn CA, Trinder G. J Endocrinol. 1973;56:133–144. doi: 10.1677/joe.0.0560133. [DOI] [PubMed] [Google Scholar]
- 6.Martin L, Pollard JW, Fagg B. J Endocrinol. 1976;69:103–115. doi: 10.1677/joe.0.0690103. [DOI] [PubMed] [Google Scholar]
- 7.Travis RC, Key TJ. Breast Cancer Res. 2003;5:239–247. doi: 10.1186/bcr628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tye BK. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 9.Forsburg SL. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong W, Pollard JW. Mol Cell Biol. 1999;19:2251–2264. doi: 10.1128/mcb.19.3.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B, Pan H, Zhu L, Deng Y, Pollard JW. Mol Endocrinol. 2005;19:1978–1990. doi: 10.1210/me.2004-0274. [DOI] [PubMed] [Google Scholar]
- 12.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin L, Finn CA. J Endocrinol. 1970;48:109–115. [PubMed] [Google Scholar]
- 14.Finn CA, Martin L. J Endocrinol. 1970;47:431–438. doi: 10.1677/joe.0.0470431. [DOI] [PubMed] [Google Scholar]
- 15.Das RM, Martin L. J Endocrinol. 1973;59:205–206. doi: 10.1677/joe.0.0590205. [DOI] [PubMed] [Google Scholar]
- 16.Fagg B, Martin L, Rogers L, Clark B, Quarmby VE. J Reprod Fertil. 1979;57:335–339. doi: 10.1530/jrf.0.0570335. [DOI] [PubMed] [Google Scholar]
- 17.Pan H, Zhu L, Deng Y, Pollard JW. Endocrinology. 2006 doi: 10.1210/en.2006-0140. in press. [DOI] [PubMed] [Google Scholar]
- 18.Coleman TR, Carpenter PB, Dunphy WG. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 19.Maiorano D, Moreau J, Mechali M. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 20.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. J Biol Chem. 1996;271:4349–4354. doi: 10.1074/jbc.271.8.4349. [DOI] [PubMed] [Google Scholar]
- 21.Todorov IT, Attaran A, Kearsey SE. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korach KS. Science. 1994;266:1524–1527. doi: 10.1126/science.7985022. [DOI] [PubMed] [Google Scholar]
- 23.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 24.Cheon YP, Li Q, Xu X, DeMayo FJ, Bagchi IC, Bagchi MK. Mol Endocrinol. 2002;16:2853–2871. doi: 10.1210/me.2002-0270. [DOI] [PubMed] [Google Scholar]
- 25.Reese J, Das SK, Paria BC, Lim H, Song H, Matsumoto H, Knudtson KL, DuBois RN, Dey SK. J Biol Chem. 2001;276:44137–44145. doi: 10.1074/jbc.M107563200. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe H, Suzuki A, Kobayashi M, Takahashi E, Itamoto M, Lubahn DB, Handa H, Iguchi T. J Mol Endocrinol. 2003;30:347–358. doi: 10.1677/jme.0.0300347. [DOI] [PubMed] [Google Scholar]
- 27.Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC. Endocrinology. 2002;143:2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- 28.Mariadason JM, Arango D, Corner GA, Aranes MJ, Hotchkiss KA, Yang W, Augenlicht LH. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- 29.Cheng SV, MacDonald BS, Clark BF, Pollard JW. Exp Cell Res. 1985;160:459–470. doi: 10.1016/0014-4827(85)90193-4. [DOI] [PubMed] [Google Scholar]
- 30.Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- 31.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasion SG, Forsburg SL. Mol Biol Cell. 1999;10:4043–4057. doi: 10.1091/mbc.10.12.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young MR, Suzuki K, Yan H, Gibson S, Tye BK. Genes Cells. 1997;2:631–643. doi: 10.1046/j.1365-2443.1997.1510349.x. [DOI] [PubMed] [Google Scholar]
- 34.Labib K, Tercero JA, Diffley JF. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 35.Liang DT, Hodson JA, Forsburg SL. J Cell Sci. 1999;112:559–567. doi: 10.1242/jcs.112.4.559. [DOI] [PubMed] [Google Scholar]
- 36.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 37.Cook JG, Chasse DA, Nevins JR. Biol Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 38.Cheng SVY, Pollard JW. FEBS Lett. 1986;196:309–314. doi: 10.1016/0014-5793(86)80269-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.