Abstract
The identification of genes and deduced pathways from the mature human oocyte can help us better understand oogenesis, folliculogenesis, fertilization, and embryonic development. Human metaphase II oocytes were used within minutes after removal from the ovary, and its transcriptome was compared with a reference sample consisting of a mixture of total RNA from 10 different normal human tissues not including the ovary. RNA amplification was performed by using a unique protocol. Affymetrix Human Genome U133 Plus 2.0 GeneChip arrays were used for hybridizations. Compared with reference samples, there were 5,331 transcripts significantly up-regulated and 7,074 transcripts significantly down-regulated in the oocyte. Of the oocyte up-regulated probe sets, 1,430 have unknown function. A core group of 66 transcripts was identified by intersecting significantly up-regulated genes of the human oocyte with those from the mouse oocyte and from human and mouse embryonic stem cells. GeneChip array results were validated using RT-PCR in a selected set of oocyte-specific genes. Within the up-regulated probe sets, the top overrepresented categories were related to RNA and protein metabolism, followed by DNA metabolism and chromatin modification. This report provides a comprehensive expression baseline of genes expressed in in vivo matured human oocytes. Further understanding of the biological role of these genes may expand our knowledge on meiotic cell cycle, fertilization, chromatin remodeling, lineage commitment, pluripotency, tissue regeneration, and morphogenesis.
Keywords: genechip, microarray, RT-PCR, pluripotency, RNA amplification
The mammalian oocyte is responsible for a number of extraordinary biological processes. It has the ability to haploidize its DNA, to reprogram sperm chromatin into a functional pronucleus, to drive early embryonic development, and to give rise to pluripotent embryonic stem cells (ESCs). Identifying the genes in the oocyte that are essential for oogenesis, folliculogenesis, fertilization, and early embryonic development will provide a valuable genomic resource in reproductive and developmental biology. However, the oocyte transcriptome and its functional significance in the human are relatively unknown because of ethical and technical limitations.
Although extensive genomic studies of oocytes and preimplantation embryos have been conducted in mouse oocytes (1–6), in human the accessibility of mature oocytes i.e., metaphase II (MII) oocytes, has been a major barrier to studying oocyte genomics using microarrays. Attempts have been made to address this problem by using candidate gene approaches employing RT-PCR and differential display (7–22). In addition, serial analysis of gene expression (SAGE) and cDNA libraries was generated from human oocytes, and SAGE tags and expressed sequence tags were sequenced for rapid gene discovery and expression profiling in the oocytes (see reviews in refs. 23 and 24). However, these molecular approaches resulted in a small number of genes analyzed in each sample. Recently, four reports described initial transcriptome analyses of human oocytes using microarrays (25–28). Although they provided valuable information, these studies did not present a comprehensive picture of the human oocyte transcriptome because of a number of biological and technical constraints. Among the biological impediments are the use of discarded human oocytes that have failed to fertilize (25, 28), limited coverage of the microarrays (25, 27), in some cases lack of sufficient biological replications (26, 28), and technical issues (27). Among the technical shortcomings, the most important is the use of a potentially unfaithful RNA amplification protocol. Li et al. (27) seem to have synthesized the first-strand cDNA using only a simple oligo(dT) primer, which makes target amplification unfeasible (27); however, the actual procedure used for RNA amplification is unclear. Although we recognize this issue could have been a mistake on their described-published protocol, the actual procedure used for RNA amplification remains elusive. Our study addresses these concerns. First, we used young oocytes, as opposed to aged and fertilized ones, which could have quite significantly different expression profiles; second, we used the most comprehensive human microarray platform; and third we took advantage of publicly available gene expression databases to interpret our own data in a more meaningful way. Thus, the present study was conducted to identify the gene transcripts present in young, untreated MII oocytes within minutes after isolation from the ovary in three independent replicates and to compare these genes with a reference RNA (a mixture of total RNA from 10 different normal human tissues not including the ovary) by using Affymetrix GeneChip technology. To achieve this goal, a protocol that combined template-switching PCR and T7-based amplification methods was developed for the analysis of gene expression in samples of small quantity. We amplified RNA from the oocyte and reference samples. Results were later compared with available transcriptome databases of mouse oocytes, and human ESCs (hESCs) and mouse ESCs (mESCs). Here, we report the transcript profile of in vivo matured human MII oocytes using the most recent Affymetrix human GeneChip array, interrogating >47,000 transcripts including 38,500 well characterized human genes.
Results and Discussion
Validation of Amplification Fidelity (Amplified vs. Nonamplified RNA).
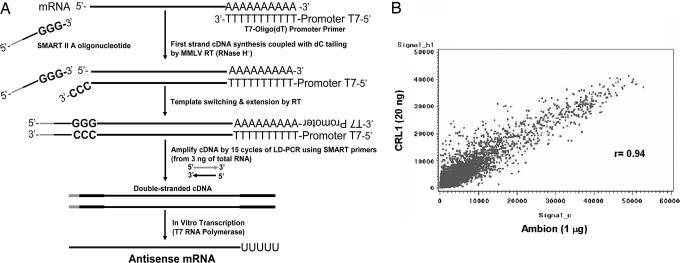
A critical step in the analysis of gene expression on small samples is the faithful amplification of mRNA molecules present in the sample. We have designed a PCR-based amplification system using the combination of SMART II A oligonucleotide (Clontech, Mountain View, CA) and T7-Oligo(dT) promoter primers (CRL RNA amplification protocol) (Fig. 1A). We isolated total RNA from a human cell line and 20, 3, and 1.5 ng input total RNA was amplified using the CRL amplification protocol. For each experiment, 15 μg of fragmented amplified RNA (aRNA) was hybridized to a single Affymetrix Human Genome U133 Plus 2.0 array. Nonamplified RNA from the same original sample (1 μg) was run in parallel by using the MessageAmp II aRNA Kit (Ambion, Austin, TX). Gene expression results from both amplified vs. nonamplified RNA samples were compared, and the correlation coefficients were found to be 0.94 (Fig. 1B), 0.93, and 0.91 for 20 ng, 3 ng, and 1.5 ng of total input RNA, respectively. The CRL Amplification protocol was repeated two times with 20 ng of initial total RNA from the same cell type, and the correlation between the two experiments was 0.99. These results show that our RNA amplification strategy faithfully and consistently amplifies small amounts of RNA to quantities required to perform microarray experiments. The CRL amplification protocol provides a practical approach to facilitate the analysis of gene expression in samples of small quantity while maintaining the relative gene expression profile throughout reactions.
Fig. 1.
Summary of CRL RNA amplification protocol. (A) Flow Chart of the CRL amplification protocol. (B) Representative plot of gene intensities comparing the CRL and Ambion amplification methods using 20 ng and 1 μg of total RNA, respectively.
Validation of Microarray Data.
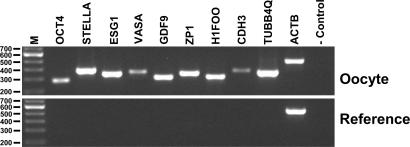
A selected list of genes was used to validate the microarray results by RT-PCR (Fig. 2). These genes were found to be present in the oocyte sample and absent in the reference RNA.
Fig. 2.
RT–PCR verification of the GeneChip array result. Loading orders of the gel were as following: M, 100 bp molecular weight standards with sizes as indicated on the left margin; OCT4, POU domain, class 5, transcription factor 1; STELLA, DPPA3, developmental pluripotency-associated 3; ESG1, embryonal stem cell-specific gene 1; VASA, DEAD box RNA helicase; GDF9, growth differentiation factor 9; ZP1, zona pellucida glycoprotein 1; H1FOO, H1 histone family, member O, oocyte-specific; CDH3, cadherin 3, type 1, P-cadherin (placental); TUBB4Q, β-tubulin; ACTB, β-actin; and negative control with no DNA template.
Differentially Up-Regulated Genes in the Human Oocyte.
We generated a database of the human oocyte transcriptome by comparing the transcripts in the oocyte with the reference samples, which contain mRNA from several somatic tissues. A complete list of up- and down-regulated genes and functional, comparative, and correlation analyses are available (see Data Sets 1 and 2 at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). Although the oocytes were thoroughly denuded manually from their surrounding cells, we were concerned over the risk for potential contamination of RNA belonging to cumulus cells. We then specifically checked for the levels of expression of cumulus cell-specific genes, such as GREM1, PTGS2 and PTX3, and found them absent or down-regulated in the oocyte RNA samples.
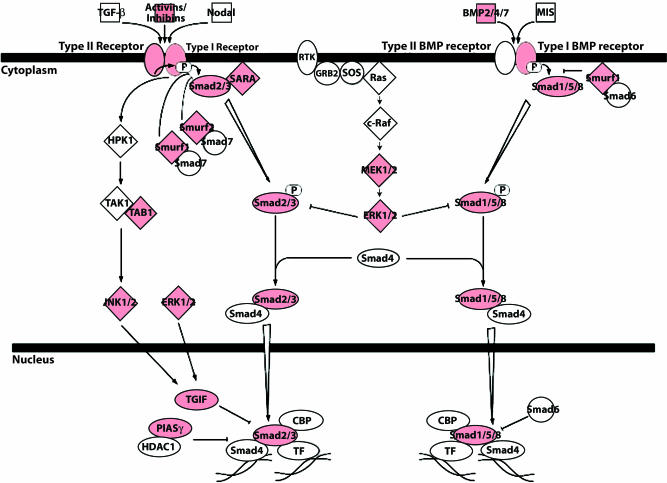
Compared with reference samples, there were 5,331 transcripts significantly up-regulated and 7,074 transcripts significantly down-regulated in the oocyte. Genes up-regulated in oocyte samples included most of the well-known germ cell-specific genes, such as FIGLA, STELLA, VASA, DAZL, GDF9, ZP1, ZP2, MOS, OCT4, NPM2, and H1FOO. Using Ingenuity Software Knowledge Base (Redwood City, CA), we confirmed the presence of pathways previously described in the mouse, in particular the TGF-β pathway (Fig. 3).
Fig. 3.
TGF-β signaling pathway. Genes shown in red are differentially up-regulated in human oocytes.
The number of genes expressed in young MII human oocytes was 7,560 in our study (based on Unigene build 189; see Data Set 3, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html) whereas the only other study employing the same microarray (28) reported the gene number as 5,633 in aged human oocytes. As the complete list of genes is not available, a direct comparison of these data sets is not feasible. Although there is overlap between the two data sets, the difference in the number of genes detected could be because the oocytes assayed in the two studies were not equivalent to each other (young MII oocytes vs. unfertilized and aged oocytes) or the effect of different RNA amplification protocols used.
Functional Annotation of Genes Overexpressed in the Human Oocyte.
To examine the biological processes performed by the oocyte, we implemented Expression Analysis Systematic Explorer (EASE) (29), contrasting the genes overexpressed in the oocyte with all of the genes present in the Affymetrix chip (Table 1, which is published as supporting information on the PNAS web site). One of the top overrepresented categories found in oocytes was related to RNA metabolism. This finding is in agreement with the fact that oocytes store RNA to support the events of fertilization and early embryonic development until the embryonic genome is activated (30, 31). DNA metabolism and chromatin modification were also overrepresented categories, in agreement with the need of the oocyte to remodel the sperm chromatin upon fertilization. Detection of gametogenesis and reproduction as overrepresented categories further suggests the accuracy of this transcriptional profiling. An important category highly represented in the oocyte was related to nucleic acid metabolism and regulation of transcription. Although transcriptionally silent at the MII stage, the oocyte is very active in transcription and translation throughout its growth phase and must be prepared to initiate transcription during embryonic genome activation at the four- to eight-cell stage in human (32). Many of the genes in this category represent Zinc-finger proteins that are not yet fully characterized, providing an opportunity to discover new transcriptional regulatory networks that operate during embryonic genome activation.
Chromatin remodeling genes are also represented in the human oocyte. Genes in this category included the following: DNA methyltransferases (DNMT1, DNMT3A and DNMT3B), histone acetyltransferases (NCOA1 and -3, SRCAP, GCN5L2, and TADA2L), histone deacetylases (HDAC3, HDAC9, and SIRT7), methyl-CpG-binding proteins (MBD2 and MBD4), histone methyltransferases (EHMT1 and SET8), ATP-dependent remodeling complexes (SMARCA1, SMARCA5, SMARCAD1, SMARCC2, and SMARCD1), and other chromatin-modifying genes (ESR1, NCOA6, HMGB3, HMGN1, and HMGA1). These Gene Ontology results not only validate our transcriptome analysis when compared with candidate gene analysis already reported in other species but more importantly, shed new light into a large number of biological processes that take place in the human oocyte.
Intersection Between Human Oocyte and Mouse Oocyte Transcriptome.
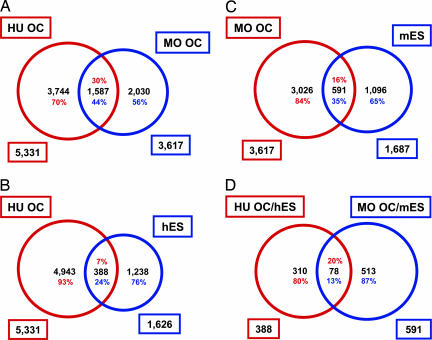
Mouse has been the best model for genetic studies, and several groups have already reported the transcriptome analysis of mouse oocytes (4, 6). In an effort to find differences and similarities between the human and mouse oocyte, we compared our human oocyte transcriptome results with that of mouse oocyte transcriptome derived from data of Su et al. (33). We intersected differentially up-regulated genes in the human and the mouse oocyte transcriptome and found a set of 1,587 genes to be in common, indicating genes of conserved function in mammalian oocytes (Fig. 4A and Data Set 4, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). The functional characterization of 15 of the top 100 intersected genes that have their functions described in mouse oocytes is summarized in Table 2, which is published as supporting information on the PNAS web site. Many of these genes relate to oocyte maturation, from the first meiotic division to MII arrest, encompassing various controls of cell cycle checkpoints and cellular machinery for DNA segregation and cell division. It was surprising to find at the intersection of these data sets the up-regulation of the estrogen receptor (ER) signaling pathway (Fig. 5, which is published as supporting information on the PNAS web site). Genes significantly up-regulated in this pathway were CTBP2, ESR1, GTF2H1, GTF2H2, MAP2K1, NCOA1, NCOA3, PCQAP, PHB2, POLR2C, POLR2J, RBM9, TAF3, TAF4 and 4B, TAF5, TAF6, TAF12, and TBP. Recent studies in knockout models for aromatase have shown that estrogen is not required for the generation of preimplantation embryos (34); our study, however, in agreement with previous reports (35, 36) suggests that some genes associated with the ER pathways are indeed transcribed in the oocyte, perhaps in response to hormonal stimulation during folliculogenesis and oocyte maturation. It remains to be determined whether the ER pathway has a role during preimplantation development in human embryos. Considering the high degree of similarity in early embryonic development between mouse and human, these 1,587 common genes deserve particular attention and must be considered for future candidate gene-approach studies related to fertility disorders, developmental defects, and assisted reproductive technologies. Furthermore, with the inherent ethical and technical difficulties of studying human oogenesis in the laboratory, the mouse model will continue to provide a platform for the functional characterization of other highly conserved genes that may bear significance in understanding human germ cell formation and maturation.
Fig. 4.
Venn diagrams showing the intersection between differentially up-regulated genes in the human (HU OC) and mouse oocytes (MO OC) (1,587 transcripts were found to be in common in both species) (A); HU OC and hESCs (388 transcripts were found to be common in both cell types) (B); MO OC and mESCs (591 transcripts were found to be common in both cell types) (C); and HU OC/hESC and MO OC/mESC (78 transcripts were found to be common in all four cell types (D).
Intersection Between Oocytes and ESC Transcriptomes.
The oocyte is derived from germ cell precursors believed to have segregated from pluripotent precursors before somatic tissue differentiation (37). Primordial germ cells (PGCs) undergo mitotic proliferation followed by meiosis. By the time the oocyte reaches the MII stage, it is already a highly specialized cell capable of remodeling the sperm nucleus and restoring totipotency to the diploid zygote. In addition, somatic cell nuclear transfer (SCNT) into enucleated oocytes has shown that, when challenged with a somatic nucleus, the oocyte cytosol will attempt to completely erase the somatic epigenetic phenotype and transform the nucleus to a totipotent state. Although failures in this epigenetic reprogramming have been reported elsewhere, there are reported cases in which animals produced by SCNT have developed normally (38). Reinforcing the notion that, when SCNT is performed under ideal circumstances (yet to be described), the oocyte cytosol can turn a somatic nucleus into a totipotent one. Recent somatic cell–ESC fusion experiments suggest that ESCs retain similar as yet undefined components that can initiate the reprogramming of introduced somatic nucleus to confer pluripotency to the somatic nuclei (as measured by phenotypic and by transcriptional analyses). In this respect, the cytoplasmic environment of both ESCs and oocytes shares the capacity to reprogram a somatic nucleus (39–41). Furthermore, recent work suggests that mESCs can give rise to PGCs that can differentiate into cells similar to oocytes and sperm in a period significantly shorter when compared with in vivo gametogenesis (42, 43). Taken together, this evidence indicates that there is a common set of genes between oocytes and ESCs responsible for reprogramming somatic cells. To identify these genes, differentially up-regulated transcripts in the oocyte were compared and intersected with recently published data for genes that are expressed preferentially in ESCs (44). Our analysis of the Sato et al. (44) data revealed 1,626 hESC differentially up-regulated genes. When these hESC genes were intersected with our human differentially up-regulated oocyte transcripts, we found an overlap of 388 transcripts (Fig. 4B and Data Set 5, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). Our final intersection was drawn between these 388 human transcripts and a list of 591 genes common to mouse oocyte and mESCs (Fig. 4C and Data Set 6, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). A final list of 66 unique genes (78 transcripts) common amongst mouse oocyte, mESCs, human oocyte, and hESCs was obtained; five of these genes have unknown functions (Fig. 4D; Table 3, which is published as supporting information on the PNAS web site; and Data Set 7, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). Among these 66 unique genes, there is a high abundance of chromatin and DNA modifying genes, suggesting that the genomes of both the ESCs and oocytes are maintained in a responsive or primitive-naive state, potentially primed for the activation of a whole repertoire of genes leading the generation of all tissue lineages. Whether any of these genes are involved in the ability to reprogram somatic nuclei should be further explored.
Conclusions
Our report provides a comprehensive expression baseline of gene transcripts present in in vivo matured human MII oocytes. Using the most recent Affymetrix Human GeneChip, we have identified 5,331 transcripts highly expressed in human oocytes, including well known genes such as FIGLA, STELLA, VASA, DAZL, GDF9, ZP1, ZP2, MOS, OCT4, NPM2, NALP5/MATER, ZAR1, and H1FOO. More importantly, 1,430 of these up-regulated genes have unknown functions, arguing for the need for further studies aimed to elucidate the functional role of these genes in the human oocyte.
We have also identified a significant number of genes common between hESCs and MII oocytes. Such genes may provide the missing link between ESCs and MII oocytes and may serve as genetic resources to identify ESCs that have full potential for differentiation into an oocyte.
As in the case of many microarray studies, profiling of the genes in the tissues of interest is the first step of a comprehensive experimental approach toward dissecting biological processes and their players at the molecular level. The data sets created in our experiments (which are available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html) will provide some of the means to address hypothesis-driven questions regarding unique functions of the oocyte.
Further understanding of the biological role of these genes may expand our knowledge of the meiotic cell cycle, fertilization, chromatin remodeling, lineage commitment, pluripotency, tissue regeneration, and morphogenesis. The practical implications of compiling gene expression information on human gametes and embryos would be enormous by bolstering efforts to solve problems from infertility to degenerative diseases.
Materials and Methods
Oocyte Collection, Total RNA Extraction, and Reference RNA.
Human oocytes were obtained from three patients undergoing an assisted reproductive treatment (ART) at the Unit of Reproductive Medicine of Clinica Las Condes, Santiago, Chile. It is important to emphasize that the routine in vitro fertilization protocol at Clinica Las Condes calls for fertilizing only those oocytes that will be transferred into the uterus of the patient. Therefore, there is always a surplus of oocytes. We then had the opportunity to use specific criteria to select donors as follows: (i) <35 years old, (ii) reproductively healthy with regular ovulatory cycles, (iii) male factor as the only cause of infertility, and (iv) considerable number of developing follicles that assured spared oocytes. The experimental protocol was reviewed and approved by a local independent Ethics Review Board. All donors signed informed consent. At the time this manuscript was submitted, all three donors had already conceived; two of them got pregnant during the ART cycle in which our samples were collected, and the third one got pregnant after a spontaneous cycle with artificial insemination using donated sperm. Ovarian stimulation, oocyte retrieval, and cell lysis were performed as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Three groups of 10 oocytes each were used. Total RNA was isolated following the guanidium thiocyanate method (45) by using the PicoPure RNA isolation kit (Arcturus, Sunnyvale, CA) following the manufacturer's instructions. However, only 6.5 μl of elution buffer (Arcturus) was used, and the elution was repeated at least three times by using the first eluate. All RNA samples within the purification column were treated with the RNase-Free DNase (Qiagen, Valencia, CA). Extracted RNA was stored at −80°C until used as template for cDNA synthesis. The quality and quantity of extracted total RNA from 8 matured oocytes (independent from the 30 oocytes used in this study) was evaluated on the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). Each mature oocyte was found to have ≈330 pg of total RNA when the Arcturus RNA isolation kit was used. The quality of RNA was intact as shown in Fig. 6, which is published as supporting information on the PNAS web site. Reference RNA (100 μg) was prepared by mixing 10 μg of total RNA from each of 10 different normal human tissues, including skeletal muscle, kidney, lung, colon, liver, spleen, breast, brain, heart, and stomach (Ambion).
First-Strand cDNA Synthesis and cDNA Purification.
The following reagents were added to each 0.5 ml of RNase-free tube: 5 μl total RNA (i.e., 3 ng for the reference and ≈3 ng for the oocyte samples) and 300 ng of an anchored T7-Oligo(dT)24V promoter primer (Ambion). The reaction tubes were incubated in a preheated PCR machine at 70°C for 2 min and transferred to ice. After denaturation, the following reagents were added to each reaction tube: 1.4 μl of SMART II A oligonucleotide (5′-AAGCAGTGGTATCAACGCAGAGTACGCGrGrGr-3′) (Clontech), 4 μl of 5× first-strand buffer, 2 μl of 20 mM DTT, 0.6 μl of 5 mg/ml T4 Gene 32 Protein (Roche, Indianapolis, IN), 2 μl of 10 mM dNTPs, 20 units of RNase inhibitor (Ambion), and 1 μl of PowerScript Reverse Transcriptase (Clontech). The final first-strand reaction volume was 20 μl for all experiments. After gently mixing, reaction tubes were incubated at 42°C for 60 min in a hot-lid thermal cycler. The reaction was terminated by heating at 70°C for 15 min and purified by NucleoSpin Extraction Kit (Clontech) following the manufacturer's instructions.
Double-Stranded cDNA Synthesis by Long-Distance PCR and cDNA Purification.
PCR Advantage 2 mix (9 μl) was prepared as follows: 5 μl of 10× PCR Advantage buffer (Clontech), 1 μl of 10 mM dNTPs, 100 ng of 5′ SMART upper primer (5′-AAGCAGTGGTATCAACGCAGAGTA-3′), 100 ng of 3′ SMART lower primer (5′-CGGTAATACGACTCACTATAGGGAGAA-3′), and 1 μl of Polymerase Mix Advantage 2 (Clontech). This mix was added to 41 μl of the first-strand cDNA synthesis reaction product, and thermal cycling was carried out in the following conditions: 95°C for 1 min, followed by 15 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 62°C for 30 s, and extension at 68°C for 10 min. The cDNA was purified by the NucleoSpin Extraction Kit (Clontech).
For in vitro transcription, biotin-labeled aRNA purification and aRNA fragmentation, hybridization, washing, staining and imaging, and RT-PCR analysis, see Supporting Materials and Methods.
Microarray Analysis.
Transcriptional profile of each sample was probed by using Affymetrix Human Genome U133 Plus 2.0 GeneChips. The raw data obtained after scanning the arrays were analyzed by dChip (46). A smoothing spline normalization method was applied before obtaining model-based gene expression indices, also known as signal values. There were no outliers identified by dChip so all samples were carried on for subsequent analysis.
When comparing two groups of samples to identify genes enriched in a given group, we used the lower confidence bound (LCB) of the fold change (FC) between the two groups as the cut-off criteria. If 90% LCB of FC between the two groups was >2, the corresponding gene was considered to be differentially expressed (DE). LCB is a stringent estimate of the FC and has been shown to be the better ranking statistic (46). Recently, dChip's LCB method for assessing DE genes has been shown to be superior to other commonly used approaches, such as MAS 5.0 and Robust Multiarray Average-based methods (47, 48).
Pathways analysis was performed by using Ingenuity Software Knowledge Base (Redwood City, CA), which is a manually created database of previously published findings on mammalian biology from the public literature. We used the network analysis, using the knowledge base to identify interactions of input genes within the context of known biological pathways.
Gene Ontology was performed by using the EASE software package (http://david.niaid.nih.gov/david/ease.htm). Given a list of genes, EASE forms subgroups based on the functional categories assigned to each gene. EASE assigns a significance level (EASE score) to the functional category based on the probability of seeing the number of subgroup genes within a category given the frequency of genes from that category appearing on the microarray (29).
Comparison with External Data Sets.
Mouse MII oocyte transcriptome data were obtained from Su et al. (33), who used custom designed Affymetrix chips to obtain gene expression profiles of oocytes and 60 other mouse tissue types. Using their expression database, we identified 3,617 differentially up-regulated transcripts in the mouse oocyte using the median expression value of the remaining 60 samples as the baseline (see Data Set 8, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). We selected transcripts with an expression value in oocyte samples that are 2-fold higher than the baseline.
Human ESC data were derived from the work of Sato et al. (44), who profiled human stem cells and their differentiated counterparts using Affymetrix HG-U133A representing ≈22,000 transcripts.
We analyzed the raw data using dChip and identified 1,626 hESC genes by selecting transcripts significantly up-regulated in human stem cells compared with their differentiated counterparts (see Data Set 9, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html).
Finally, for mESCs, we used a list of 1,687 differentially up-regulated mESC genes published by Fortunel et al. (49), which were identified by comparing mESCs to differentiated cells by using Affymetrix MG-U74Av2 chips representing ≈12,000 transcripts (see Data Set 10, which is available at www.canr.msu.edu/dept/ans/community/people/cibelli_jose.html). We used Affymetrix's NetAffx tool (https://www.affymetrix.com/analysis/netaffx/index.affx) for mapping genes across organisms and platforms used in the respective studies.
Supplementary Material
Acknowledgments
We thank the members of the Cellular Reprogramming Laboratory for their support during the course of this investigation and Drs. Annette Thelen and Jeff Landgraf at the Research Technology Support Facility, Michigan State University (MSU), for providing technical assistance with the Affymetrix array experiments. Funding was provided by the Office of the Vice President for Research and Graduate Studies at MSU, the Michigan Agricultural Experiment Station, and the MSU Foundation. This work was also partially supported by National Institutes of Health Grant DK047636 (to B.L. and H.H.O.) and by a grant from A-Star, Singapore (to B.L.).
Abbreviations
- ESC
embryonic stem cell
- hESC
human ESC
- mESC
mouse ESC
- MII
metaphase II
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brambrink T, Wabnitz P, Halter R, Klocke R, Carnwath J, Kues W, Wrenzycki C, Paul D, Niemann H. BioTechniques. 2002;33:376–378. 380, 382–385. doi: 10.2144/02332rr04. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka TS, Ko MS. Eur J Obstet Gynecol Reprod Biol. 2004;115(Suppl 1):S85–S91. doi: 10.1016/j.ejogrb.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. Dev Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- 4.Zeng F, Baldwin DA, Schultz RM. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Hamatani T, Carter MG, Sharov AA, Ko MS. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 7.Pitera JE, Milla PJ, Scambler P, Adjaye J. Mech Dev. 2001;109:377–381. doi: 10.1016/s0925-4773(01)00530-5. [DOI] [PubMed] [Google Scholar]
- 8.Verlinsky Y, Morozov G, Gindilis V, Strom CM, Freidin M, Rechitsky S, Verlinsky O, Ivakhnenko V, Zdanovsky V, Kuliev A. Mol Reprod Dev. 1995;41:127–132. doi: 10.1002/mrd.1080410202. [DOI] [PubMed] [Google Scholar]
- 9.Adjaye J, Daniels R, Monk M. J Assist Reprod Genet. 1998;15:344–348. doi: 10.1023/A:1022565115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adjaye J, Bolton V, Monk M. Gene. 1999;237:373–383. doi: 10.1016/s0378-1119(99)00329-7. [DOI] [PubMed] [Google Scholar]
- 11.Adjaye J, Monk M. Mol Hum Reprod. 2000;6:707–711. doi: 10.1093/molehr/6.8.707. [DOI] [PubMed] [Google Scholar]
- 12.Daniels R, Lowell S, Bolton V, Monk M. Hum Reprod. 1997;12:2251–2256. doi: 10.1093/humrep/12.10.2251. [DOI] [PubMed] [Google Scholar]
- 13.Daniels R, Adjaye J, Bolton V, Monk M. Mol Hum Reprod. 1998;4:785–789. doi: 10.1093/molehr/4.8.785. [DOI] [PubMed] [Google Scholar]
- 14.Hinkins M, Huntriss J, Miller D, Picton HM. Reproduction. 2005;130:883–888. doi: 10.1530/rep.1.00675. [DOI] [PubMed] [Google Scholar]
- 15.Liu HC, He ZY, Tang YX, Mele CA, Veeck LL, Davis O, Rosenwaks Z. Fertil Steril. 1997;67:733–741. doi: 10.1016/s0015-0282(97)81375-1. [DOI] [PubMed] [Google Scholar]
- 16.Salpekar A, Huntriss J, Bolton V, Monk M. Mol Hum Reprod. 2001;7:839–844. doi: 10.1093/molehr/7.9.839. [DOI] [PubMed] [Google Scholar]
- 17.Goto T, Adjaye J, Rodeck CH, Monk M. Mol Hum Reprod. 1999;5:851–860. doi: 10.1093/molehr/5.9.851. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh RH, Au HK, Yeh TS, Chang SJ, Cheng YF, Tzeng CR. Fertil Steril. 2004;81(Suppl 1):912–918. doi: 10.1016/j.fertnstert.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Huntriss J, Hinkins M, Oliver B, Harris SE, Beazley JC, Rutherford AJ, Gosden RG, Lanzendorf SE, Picton HM. Mol Reprod Dev. 2004;67:323–336. doi: 10.1002/mrd.20030. [DOI] [PubMed] [Google Scholar]
- 20.Huntriss J, Gosden R, Hinkins M, Oliver B, Miller D, Rutherford AJ, Picton HM. Mol Hum Reprod. 2002;8:1087–1095. doi: 10.1093/molehr/8.12.1087. [DOI] [PubMed] [Google Scholar]
- 21.Monk M, Holding C, Goto T. Reprod Fertil Dev. 2001;13:51–57. doi: 10.1071/rd00073. [DOI] [PubMed] [Google Scholar]
- 22.Goto T, Jones GM, Lolatgis N, Pera MF, Trounson AO, Monk M. Mol Reprod Dev. 2002;62:13–28. doi: 10.1002/mrd.10118. [DOI] [PubMed] [Google Scholar]
- 23.Ko MS. Trends Biotechnol. 2001;19:511–518. doi: 10.1016/s0167-7799(01)01806-6. [DOI] [PubMed] [Google Scholar]
- 24.Adjaye J. Reprod Fertil Dev. 2005;17:37–45. doi: 10.1071/rd04075. [DOI] [PubMed] [Google Scholar]
- 25.Bermudez MG, Wells D, Malter H, Munne S, Cohen J, Steuerwald NM. Reprod Biomed Online. 2004;8:325–337. doi: 10.1016/s1472-6483(10)60913-3. [DOI] [PubMed] [Google Scholar]
- 26.Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 27.Li SS, Liu YH, Tseng CN, Singh S. Biochem Biophys Res Commun. 2006;340:48–53. doi: 10.1016/j.bbrc.2005.11.149. [DOI] [PubMed] [Google Scholar]
- 28.Assou S, Anahory T, Pantesco V, Carrour TL, Pellestor F, Klein B, Reyftmann L, Dechaud H, Vos JD, Hamamah S. Hum Reprod. 2006;21:1705–1719. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachvarova R. Dev Biol. 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. [DOI] [PubMed] [Google Scholar]
- 31.Wassarman PM, Kinloch RA. Mutat Res. 1992;296:3–15. doi: 10.1016/0165-1110(92)90028-8. [DOI] [PubMed] [Google Scholar]
- 32.Braude P, Bolton V, Moore S. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 33.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh K, Jones G, Thouas G, Britt KL, Simpson ER, Jones ME. Biol Reprod. 2004;70:1263–1269. doi: 10.1095/biolreprod.103.022111. [DOI] [PubMed] [Google Scholar]
- 35.Sar M, Welsch F. Endocrinology. 1999;140:963–971. doi: 10.1210/endo.140.2.6533. [DOI] [PubMed] [Google Scholar]
- 36.Hiroi H, Momoeda M, Inoue S, Tsuchiya F, Matsumi H, Tsutsumi O, Muramatsu M, Taketani Y. Endocr J. 1999;46:153–158. doi: 10.1507/endocrj.46.153. [DOI] [PubMed] [Google Scholar]
- 37.Cauffman G, Van de Velde H, Liebaers I, Van Steirteghem A. Mol Hum Reprod. 2005;11:405–411. doi: 10.1093/molehr/gah167. [DOI] [PubMed] [Google Scholar]
- 38.Lanza RP, Cibelli JB, Faber D, Sweeney RW, Henderson B, Nevala W, West MD, Wettstein PJ. Science. 2001;294:1893–1894. doi: 10.1126/science.1063440. [DOI] [PubMed] [Google Scholar]
- 39.Byrne JA, Simonsson S, Western PS, Gurdon JB. Curr Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- 40.Cowan CA, Atienza J, Melton DA, Eggan K. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 41.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 42.Hubner K, Fuhrmann G, Christenson LK, Kehler J, Reinbold R, De La Fuente R, Wood J, Strauss JF, III, Boiani M, Scholer HR. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 43.Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, Daley GQ. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 44.Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 45.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 46.Li C, Hung Wong W. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shedden K, Chen W, Kuick R, Ghosh D, Macdonald J, Cho KR, Giordano TJ, Gruber SB, Fearon ER, Taylor JM, Hanash S. BMC Bioinformatics. 2005;6:26. doi: 10.1186/1471-2105-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fortunel NO, Otu HH, Ng HH, Chen J, Mu X, Chevassut T, Li X, Joseph M, Bailey C, Hatzfeld JA, et al. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.