Abstract
Sequence polymorphisms affect gene expression by perturbing the complex network of regulatory interactions. We propose a probabilistic method, called Geronemo, which directly aims to identify the mechanism by which genetic changes perturb the regulatory network. Geronemo automatically constructs a set of coregulated genes (modules), whose regulation can involve both sequence variations and expression of regulators. By exploiting the modularity of genetic regulatory systems, Geronemo reveals regulatory relationships that are indiscernible when genes are considered in isolation, allowing the recovery of intricate combinatorial regulation. By incorporating both expression and genotype of regulators, Geronemo captures cases where the effect of sequence variation on its targets is indirect. We applied Geronemo to a data set from the progeny generated by a cross between laboratory BY4716 (BY) and wild RM11-1a (RM) isolates of Saccharomyces cerevisiae. Geronemo produced previously undescribed hypotheses regarding genetic perturbations in the yeast regulatory network, including transcriptional regulation, signal transduction, and chromatin modification. In particular, we find a large number of modules that have both chromosomal characteristics and are regulated by chromatin modification proteins. Indeed, a large fraction of the variance in the expression can be explained by a small number of markers associated with chromatin modifiers. Additional analysis reveals positive selection for sequence evolution of elements in the Swi/Snf chromatin remodeling complex. Overall, our results suggest that a significant part of individual expression variation in yeast arises from evolution of a small number of chromatin structure modifiers.
Keywords: expression phenotype, gene regulation, probabilistic model, regulatory network, association studies
Although >99% of the human genome is conserved across the population (1), variations in DNA sequence have a major impact on an individual's response to environmental factors, disease, and therapies. Quantitative trait loci mapping (2–8) tackles the important problem of identifying DNA (typically single-nucleotide) polymorphisms that are linked or associated with a phenotype. Expression quantitative trait loci (eQTL) mapping (4–10) relates genotype to individual expression phenotypes by treating the expression of each gene as a quantitative trait. Yet, often the mechanism by which these genetic changes exert their effect on phenotype is far from obvious (11). Furthermore, association of complex traits such as disease status to an individual single-nucleotide polymorphism (SNP) is typically occluded by the large magnitude of other effects. One approach to address these two difficulties is to focus on the intermediary between genotype and phenotype, the complex regulatory network that governs the cell's activity. In this paper, we study the mechanisms by which an organism's genotype can perturb this network (Fig. 6, which is published as supporting information on the PNAS web site). In trans-G (genotype) regulation, polymorphisms in a regulator's coding region can affect its function and, thereby, its effect on its targets. In trans-E (expression) regulation, a change in the abundance of a regulator, whether due to the regulator's own cis regulation or an upstream perturbation, also can affect the activity of its targets. Finally, in cis regulation, a SNP in a gene itself can affect its affinity to its regulatory factors and, therefore, its abundance. Many genes are affected by combinations of several such perturbations (4, 9, 10, 12).
We present a computational method, called Geronemo (genetic regulatory network of modules) that aims to decipher both the cell's regulatory network and perturbations to it resulting from sequence variability. Geronemo (Fig. 7, which is published as supporting information on the PNAS web site) takes, as input, data for a set of individuals in a population, measuring both their gene expression profiles and genetic markers (see Materials and Methods). Extending the module network approach (13), which has been shown to successfully reconstruct regulatory relationships in yeast from gene expression data alone, Geronemo automatically constructs a set of regulatory modules (e.g., Fig. 2a), sets of coregulated genes, each associated with a regulatory program that “explains” the expression of the module genes in terms of a set of regulatory contexts, defined by a combination of both expression regulators and genotype regulators (Fig. 2ai). By comparison, the eQTL approach explains the expression of individual genes by one or two linked genotypes. Briefly, Geronemo begins by partitioning genes into modules with similar expression profiles. It then iterates over two steps: learning a regulatory program for each module and reassigning each gene to the module whose regulation program provides the best prediction for the gene's expression profile. Each of these two steps attempts to heuristically optimize a principled, Bayesian scoring function.
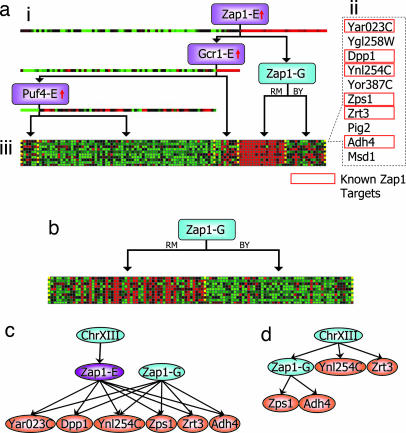
Fig. 2.
Zap1 module (module no. 79 in Table 2). (a) The regulatory module learned by Geronemo. (ai) In the regulation program, each node (rectangle) represents a query on the value of some particular regulator: a purple node (trans-E) corresponds to a regulator in our list and a particular split on the regulator's expression level; a blue node (trans-G) corresponds to a genetic marker and a split on its genotype. The expression/genotype of the regulators themselves is shown below their respective node. The key regulator (top of the tree) is Zap1 expression, and an additional node is the Zap1 genotype. (aii) List of genes in module; genes in boxes are known Zap1 targets. (aiii) Gene expression profiles, where the rows are genes, ordered as in aii, and the columns are arrays (segregants) arranged according to the regulation tree. For example, the rightmost context (group of arrays that follow the same branches down the tree) contains arrays in which Zap1 expression is up and Zap1 genotype is BY. (b) Module representation by using markers only; the Zap1 genotype alone does not give as coherent a split as the combination of both its expression and genotype. (c) Partial graph summarizing causal links from regulators to targets found by Geronemo, with ovals representing genes and arrows representing regulation: ChrXIII regulates Zap1 expression; Zap1 expression and genotype together regulate Zap1 target genes. (d) Graph as detected by Yvert et al. (19).
Geronemo offers several important benefits over eQTL. First, by using “expression regulators,” Geronemo can distinguish between associations induced by a direct effect of the SNP and those induced by an indirect relationship via the expression of a regulator. Second, Geronemo exploits the modularity of biological systems to robustly derive signal from limited, noisy data. Rather than treating each gene as a separate quantitative trait, Geronemo searches for regulatory programs that are predictive of entire groups of genes. Thus, it can discover a regulatory relationship between a regulator and a set of targets even when the signal in the expression data might be insufficient when genes are considered in isolation (14, 15). This property is particularly important when we wish to recover intricate combinatorial regulation, where the signal can rarely be detected robustly for individual genes.
Results
We applied Geronemo to the data set of Brem and Kruglyak (16), containing expression and genotype data for 112 Saccharomyces cerevisiae individuals, generated by crossing a lab strain (BY) with a wild vineyard strain (RM). We used a precompiled list of 304 putative regulators, spanning transcription factors, signal transduction proteins, chromatin modification factors, and mRNA processing factors (Table 1, which is published as supporting information on the PNAS web site).
We applied Geronemo to these data, resulting in a total of 165 regulatory modules. The model identified a total of 155 cis- regulated genes, genes whose expression is regulated by polymorphisms in their vicinity. Many of these cis genes reside in 82 modules that contain only 1–2 genes, which arise when a gene's expression profile is sufficiently unique that it does not fit well into any larger module. Cis-acting regulation can result from polymorphisms both in a gene's promoter and in its coding region. Indeed, these 155 cis genes show significantly more interstrain sequence variation (see Materials and Methods) in both regions than other genes in their vicinity (nonsynonymous coding: P < 1.84 × 10−6; promoter: P < 4.5 × 10−5). There were 79 modules involving trans-acting regulation and containing at least three genes, spanning both trans-E (71 of 79) and trans-G (45 of 79) regulation. In trans-G regulation, polymorphisms in a regulator's coding region affect the expression of its targets. Indeed, regulators in the vicinity of a locus identified by our model as a trans-G regulator show nonsynonymous variation in their coding regions (P < 1.52 × 10−2) but not in their promoter region (P > 0.2). Interestingly, although trans-E regulators are not necessarily associated with sequence variation, a nonsynonymous to synonymous substitution ratio dN/dS test (17) on the top expression regulators in the 14 largest modules (>50 genes) showed that these regulators are enriched for the genes under positive selection (P < 0.013), supporting their key role in the perturbation of this network (see Supporting Materials, which is published as supporting information on the PNAS web site).
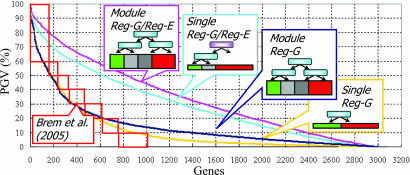
We evaluated our method statistically (Fig. 1) by estimating the proportion of the genetic variance (PGV) of expression values (see Materials and Methods) explained by the Geronemo model, compared with the results obtained by Brem and Kruglyak (16). The Geronemo model explains a significantly greater fraction of the variance: explaining >50% PGV for 828 genes, as compared with 238 in the analysis of Brem and Kruglyak (16) of the same data set. Our comparison to three simpler Geronemo models suggests that most of the improvement results from the incorporation of trans-E regulation, which captures indirect effects of sequence variation. Also significant is the association of regulatory programs with modules rather than individual genes, which helps in two ways: First, it allows us to ascribe linkages even when the signal is too faint to be detected by using a single gene-based statistic. Second, because of the larger number of data points in each module, we can robustly learn a much richer combinatorial regulation program. Indeed, Brem and Kruglyak (16) suggest that 50% of highly heritable transcripts can be explained only by using more than five loci. The use of modules allows the identification of complex combinatorial regulation, explaining more of the expression variation.
Fig. 1.
Explaining variance of gene expression. The PGV is explained by detected regulation programs for Geronemo (pink) and three simpler variants of Geronemo and for the eQTL analysis of Brem and Kruglyak (red boxes) applied to the same data set as reported in their paper (16). The graph shows the PGVg values (y axis) of 3,152 genes (x axis). The genes (x axis) are sorted by their PGVg, shown on the y axis. The simpler variants of Geronemo consist of: (i) allowing only markers as genetic regulators (ModuleReg-G; blue), (ii) forcing each gene to form a separate module (SingleReg-G/Reg-E; sky blue), and (iii) both of the constraints (SingleReg-G; yellow). A significant advantage is obtained by explicitly modeling regulatory effects by the expression values of regulators, and module-based models show higher PGV than the corresponding single gene-based models. Note that the module-based models identified more genetic regulators per gene than the single gene-based model (ModuleReg-G/Reg-E: 5.25; ModuleReg-G: 6.09; SingleReg-G/Reg-E: 1.36; SingleReg-G: 2.14). Importantly, because of the sharing of parameters between genes in a module, the Geronemo model actually has fewer estimated parameters than the method of Brem and Kruglyak despite allowing for rich combinatorial regulation programs. This result suggests that the module-based model achieves greater statistical power by using fewer parameters, both by correctly linking more genes and by recovering more intricate combinatorial interaction.
Overall, our analysis captured regulatory relationships spanning a wide range of mechanisms, including transcription factors, signaling molecules (kinases and phosphotases), chromatin modification factors, RNA processing, and other posttranscriptional regulation (Table 1). In a detailed analysis using a range of available resources, we found statistically significant experimental support for 13 of 79 of these modules and weaker support for an additional 41 (Table 2, which is published as supporting information on the PNAS web site).
The Zap1 module (Fig. 2a) demonstrates both the importance of the module-based analysis and Geronemo's ability to suggest fine-grained regulatory hypotheses. The module's key regulator is the zinc-regulated transcription factor Zap1 (10 nonsynonymous coding SNPs). The module contains 10 genes, of which six are known targets of Zap1. The regulatory program depends combinatorially on both Zap1 expression and the genotype of the region containing ZAP1; module genes are induced if Zap1 mRNA is present in abundance and is in its RM form. This program is poorly captured by genotype alone (Fig. 2b). Zap1 itself is in a module whose key regulator is a locus on chromosome XIII. The model obtained by standard eQTL mapping (19) correctly associates only two of the genes and confounds direct and indirect regulation (Fig. 2d). By contrast, Geronemo captures a more complete model of the regulatory influences, covering many more Zap1 targets, and correctly distinguishing direct and indirect interactions, illustrating the power of a module-based approach.
In some cases, Geronemo captured a module consisting of a coherent set of coregulated genes but identified only a proxy for its regulation program, an additional coregulated gene rather than the causal regulator. For example, the nucleosomal module (Fig. 8, which is published as supporting information on the PNAS web site) contains many histone genes and is enriched for cis-regulatory binding sites of the cell cycle regulator Fkh1 (P < 1.1 × 10−10), as well as for Mbp1 (P < 1.3 × 10−6) and Swi4 (P < 2.2 × 10−9), transcription factors that, in complex with Swi6, regulate cell cycle progression from G1 to S phase. The module's key regulator is Apg1, an unrelated signaling protein that appears to be a proxy for Fkh1 and Swi6, both of which regulate Apg1 expression (20) and were excluded from the analysis because of low expression variation. Promoter analysis reveals a systematic disruption, in RM, of many cell-cycle regulated binding sites in some module genes and their upstream regulators (Fig. 9, which is published as supporting information on the PNAS web site), suggesting a possible mechanism for the differential regulation of this module. Thus, even when the exact regulator is not correctly identified, a careful analysis of a module can reveal important information regarding regulatory events that vary between individuals.
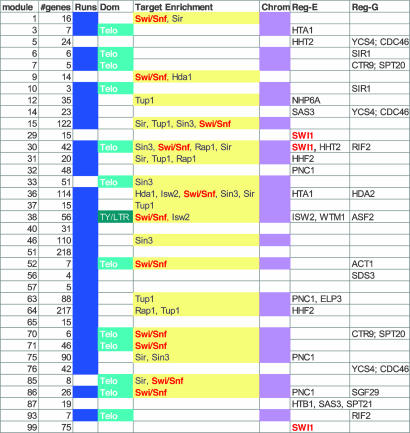
Particularly intriguing were the large number of regulators involved in chromatin modification and the prevalence of chromosomal features (e.g., enrichment for particular chromosomal regions) in many of our modules. To study this phenomenon systematically, we characterized each module in terms of three chromosomal characteristics (see Materials and Methods): modules that contain multiple and/or long runs of consecutive genes along the chromosome; modules enriched for proximity to particular chromosomal domains (e.g., telomeres or Ty/LTR elements); and modules enriched for chromatin modifying protein targets, defined by either differentially expressed genes (DEGs) (21–23) or ChIP assays (20, 24). There was significant overlap between these three sets of modules (Fig. 3), leading us to define a module as chromosomal if it had two of three of these characteristics. The 23 resulting chromosomal modules significantly overlap (10 of 16, P < 9.2 × 10−8) with the set of modules that have a chromatin modification factor as a trans-E regulator. Ten chromosomal modules have a trans-G regulator whose region contains a chromatin modification factor with nonsynonymous coding SNPs, and four modules are combinatorially regulated by both trans-G and trans-E chromatin modification factors. Altogether, 16 of 23 of these modules have a known chromatin modification factor in their regulation program. The targets of chromatin modification factors are generally not well characterized, making it difficult to verify these learned regulatory programs. However, in two cases, enough data were available to compare against our model's predictions.
Fig. 3.
Summary of chromosomal modules. Shown is a list of all modules containing chromosomal characteristics or that have chromatin modifiers as trans-E regulators. The columns in the table (in order) are as follows: module, module number; #genes, number of genes in the module; runs, whether the module contains multiple or long runs of genes along the chromosome (blue); dom, whether the module exhibits enrichment for some chromosomal domain (light cyan, telomeres; dark cyan, Ty/LTR); target enrichment, list of chromatin modification complexes such that the module is enriched for DEGs of some gene in the complex (sorted in order of P value); chrom, whether the module was characterized as chromosomal (purple); reg-E, chromatin modifiers that are trans-E module regulators; reg-G, chromatin modifiers with SNPs that are in the region of trans-G module regulators. The strong overlap between different chromosomal characteristics (runs, domains, and chromatin DEGs) supports our definition of a chromosomal module as one that contains two of three characteristics. There is significant overlap between chromosomal modules and modules predicted by our analysis to have a chromatin modifier as a regulator (see Results).
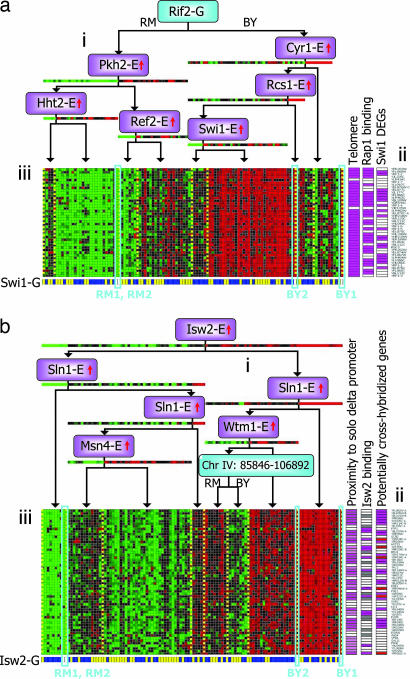
The telomere module (Fig. 4a) contains 42 genes, of which 40 are in the telomeres of multiple chromosomes. The module is repressed in the RM parent relative to BY, suggesting that telomeric silencing is enhanced in RM. The module's top regulator is a locus containing RIF2 (SNPs: six promoter and eight nonsynonymous coding), which controls telomere length and establishes telomeric silencing. Rif2 functions at telomeres with Rap1p, binding to its C terminus (26). Indeed, most of the module genes are bound by Rap1 (24). The module is combinatorially regulated by Swi1 (SNPs: 23 promoter and 51 non-syn coding), a component in the Swi/Snf chromatin remodeling complex. Indeed, many of the module genes are differentially expressed under Swi1 deletion (23). Swi1 itself is cis-regulated, but the effect of its genotype on the telomere module appears to be indirect, via changes to the Swi1 expression level.
Fig. 4.
Sample chromosomal modules. (a) Telomere module (no. 30). (ai) The module's top regulator is a region on chromosome XII containing RIF2, which controls telomere length and establishes telomeric silencing. At the fourth level in the tree, but with a distinct and statistically significant split (P < 6.5 × 10−77), we have trans-E regulation by Swi1, a component in the Swi/Snf chromatin remodeling complex. (aii) Relevant annotations for module genes: 40 of 42 genes are in the telomeric regions; genes that are ChIP-binding targets (24) of Rap1, which works in concert with Rif2; genes differentially expressed in Swi1 deletion mutants (23). (aiii) Expression data and Swi1 genotype data for the arrays. (b) Ty module (no. 38). (bi) The module's top regulator is the expression of Isw2, a member of the imitation-switch class of ATP-dependent chromatin remodeling complexes. (bii) Relevant annotations for module genes: (Left) The module contains 23 Ty elements (pink) and 16 genes that are in close proximity (within 7 genes) of the related LTR elements (P < 3.94 × 10−5, after accounting for cross-hybridization). (Center) Twenty-eight of 44 tested module genes (pink) are ChIP-binding targets of Isw2 (25) (6 of 23, P < 3.1 × 10−8, after accounting for cross-hybridization); 12 genes (gray) were not tested. (Right) Two groups of potentially cross-hybridized genes: 23 Ty elements (pink) and 6 YLR223C-class genes (red). (biii) Expression data and Isw2 genotype data.
The Ty module (Fig. 4b) contains 23 Ty elements, 6 paralogous genes similar to YLR223C, and 27 distinct genes. To limit the effect of cross-hybridization of the Ty and YLR223C sequences, we included only a single representative of these two groups in any enrichment analysis. Interestingly, 17 of the 29 remaining genes are within 5 genes of either Ty elements or the related LTR element (P < 3.9 × 10−5). The module's top regulator (trans-E) is Isw2 (SNPs: five promoter and zero coding), a member of the imitation-switch class of ATP-dependent chromatin remodeling complexes. Indeed, the module is significantly enriched (P < 3.1 × 10−8) for Isw2 ChIP targets (25). Transcription of Ty elements depends on several chromatin remodeling factors but has never been tested for the imitation-switch class. Note that, whereas Isw2 expression is strongly correlated with the expression of the module genes, its genotype is significantly less predictive (Fig. 4b).
Overall, modules that have trans-E chromatin regulators cover 971 genes, approximately one-third of the genes that are varying in the data set, suggesting that genetic variation in chromatin modification factors plays a significant role in explaining the gene expression variance in these progeny. To quantitatively test this hypothesis, we selected a subset of chromatin regulators, those whose DEGs are enriched in one of our modules and those that appeared as regulators in one of our modules and contain SNPs. The PGV explained by the 23 markers associated with these regulators is a significant fraction of the PGV explained by the entire set of markers, and much larger than 100 random subsets of 23 relevant markers (Fig. 5). The average PGV value (over the genes included in our analysis) in the chromatin model is 11.93, as compared with 14.44 in the marker-only Geronemo model. By contrast, the median model among the 100 random runs had an average PGV of 7.57. Overall, chromatin markers explain a larger fraction of the variance in these data than all 100 random groups of regulatory markers.
Fig. 5.
Chromosomal markers explain a significant fraction of the variance. Evaluation of the statistical power of different sets of markers through PGV. We selected a subset of 23 “chromatin markers” (see Results) and learned a Geronemo model by using only these as candidate genetic regulators. We compared the resulting PGV (pink) with that of our full genotype-only model. Also, we compared with 100 Geronemo runs by using 23 markers randomly chosen from linked, regulatory markers (see Results). The range of these runs is shown by the light-blue-shaded region, and the run corresponding to the curve with the median area is shown with a light-blue line.
A prominent feature of our chromosomal modules is the frequent appearance of the Swi/Snf chromatin remodeling complex, with 11 modules enriched for DEGs of swi1 or snf2 mutants (23) and 3 modules containing Swi1 as a trans-E regulator (Fig. 3). We noticed that SNF2 (SNPs: 9 promoter and 48 nonsynonmous coding) and SWI1 (SNPs: 23 promoter and 51 nonsynonymous coding) contained a large number of SNPs. In fact, the genes encoding components of the Swi/Snf genes are enriched (P < 4.2 × 10−4) for hypervariability of coding regions (see Methods). Moreover, a nonsynonymous to synonymous substitution ratio test (17) shows that the Swi/Snf complex is enriched (P < 1.2 × 10−3) for genes (ARP9, RTT102, SNF11, SNF12, SNF6, and TAF14) that are subject to positive evolutionary selection pressure. A similar analysis for other regulator families revealed no enrichment (see Supporting Materials). These findings suggest that there is evolutionary pressure to control the gene expression of multiple targets via sequence variation in these chromatin remodeling factors.
Discussion
Geronemo is capable of uncovering a broad range of regulatory interactions, including direct transcription, signaling, and chromatin modification. Importantly, it also provides significant insight into the mechanisms by which genotype perturbs the regulatory network, leading to expression changes. As in other forms of analysis, care must be taken when interpreting an inferred model, because neither genetic linkage nor correlation of gene expression necessarily imply causality; nevertheless, many of the interactions inferred by Geronemo are supported by additional data and literature.
We attribute the success of the analysis to two main factors. First, the use of expression regulators and the statistical robustness provided by the module-based approach allow us to uncover signals that are difficult to detect by using standard linkage methods. Second, the data itself, expression variation among individuals, appears particularly well suited to the detection of regulatory interactions. Unlike other types of data (e.g., individual gene deletions or environmental stimuli), these arrays represent small, natural perturbations to the system, allowing subtle changes to manifest. Moreover, each array represents a large set of such perturbations, providing a rich source of statistical variation that helps clarify the signal. Interestingly, many perturbations are revealed only in the offspring, with the parents showing no expression variation. We believe that the parent strains evolve so that perturbations in one part of the system are often “corrected” by perturbations in another, leading to similar responses. In their progeny, the effect of genetic perturbations is revealed clearly both in expression and in phenotype (8, 19).
One of our most interesting findings is the large role of chromatin remodeling in the expression variation of these individuals and the fact that the Swi/Snf complex specifically appears to be under positive selective pressure. This finding raises obvious questions: How do functional differences between this and other chromatin remodeling complexes influence this process? Are the effects the result of specific target genes whose expression depends on this activity or are there chromosomal structure constraints? Overall, our finding suggests that, at least in these yeast strains, it was advantageous to effect global changes in the regulatory network by evolving a small set of chromatin remodeling proteins. It would be of great interest to explore whether this phenomenon arises in other organisms.
The combination of genotype data and the expression perturbations across individuals was a powerful resource for uncovering regulatory mechanisms. Expanding the experimental data in two directions will enhance greatly the ability of our analysis to disentangle the regulatory network. First, in the data we used (16), gene expression was measured in rich media conditions, leaving parts of the network that are active only in other conditions unperturbed; probing these progeny under different environmental conditions and stimuli can help uncover these regions of the network. Second, the study of other strains can deconvolve additional mechanisms that remain unperturbed between the BY and RM strains.
An exciting extension of this work will be the application of Geronemo to mammalian data, using the recently published human and mouse HapMap data (1) and the increasingly available data on individual gene expression (5, 27, 28). Several features typical of mammalian systems will require significant extensions, including larger genomes, more regulators, the effect of lineage-specific gene regulation, and contributions of heterozygous alleles in diploid cells. Although mammalian systems are significantly more complex, we expect that the number of significant causal factors in the context of a single cell type to be similar to yeast. Therefore, given Geronemo's ability to learn a broad range of regulatory interactions, we believe that this application will allow us to uncover regulatory networks in higher-level organisms and to understand the mechanisms underlying complex phenotypes, including human disease.
Materials and Methods
Data Set.
We used gene expression data measured from 112 meiotic recombinant progeny of two yeast strains: BY4716 (BY; a laboratory strain) and RM11-1a (RM; a natural isolate). We selected the 3,152 genes for which >90% of the expression values are present, and that had SD >0.25 in expression level. We used the genotype values, measured in 2,957 genetic markers, merging adjacent, highly correlated markers, for a total of 581 markers (Table 3, which is published as supporting information on the PNAS web site). As candidate expression regulators, we compiled a large list of regulators that potentially might have transcriptional effect, including: transcription factors, signaling molecules, chromatin modification factors, and RNA factors (degradation and RNA processing). The list was derived by using Gene Ontology annotations in SGD (29) and further corrections through manual curation. We intersected this list with the 3,152 genes above, resulting in 304 candidate regulators (see Table 1).
Geronemo Learning Algorithm.
Our approach extends the module network approach of Segal et al. (13) to allow two types of regulatory factors: g regulators, the genotype of some chromosomal region defined by a marker that (in our data) has two possible split values for the two progenitor alleles; and e regulators, the expression level of some regulator R, whose set of possible splits is continuous. This modification required some substantial extensions to the module network algorithm, briefly summarized below (see Supporting Materials for full details).
Learning a Geronemo model involves two tasks: (i) assigning each gene into some regulatory module; and (ii) learning the regulation program for each module. We initialized the learning procedure with 500 modules obtained by k means clustering and then iterated over two phases: learning the regulatory program for the current modules and reassigning genes to modules. We use a Bayesian scoring approach, which roughly corresponds to the ability of each module's regulatory program to predict variation in the gene expression of the module genes.
Given a set of modules, we learned a regression tree (regulatory program) for each module by using the candidate e and g regulators as candidate queries for each decision node. We recursively learn the regulatory program by choosing, at each point, the regulator that best splits the gene expression of the module genes into two distinct behaviors. When considering a potential split, we evaluate all candidate regulators and split values and we picked that which achieves the highest improvement in score. No prior biological knowledge regarding the regulator is used in this procedure.
We added a number of important modifications to the original module network algorithm, which we briefly review here:
Rather than fixing the number of modules in advance, we allow this number to be selected automatically via steps that introduce modules (see below) and by deleting modules that become empty. Indeed, we found that the final number of modules learned was insensitive to the number used for initialization (see Fig. 10, which is published as supporting information on the PNAS web site).
To allow for cis-regulation effects, we introduced a step that allows genes to “break off” from their module and create a new, typically single gene, cis-linked module. This step also was performed only when it increased the model's overall score.
To improve the biological validity and statistical significance of our regulation program, we introduced an false discovery rate permutation test (computing δ scores for random permutations of regulators) when determining whether a split in the regulation tree is warranted. This test also helps to correct for the fact that the continuous-valued candidate e regulators have more possible split values than the discrete-valued ones and, therefore, are more likely to accidentally explain the data in the module.
To bias the model in favor of more biologically plausible regulation programs, we introduced a “power law” prior distribution on model structures that imposes sparsity both on the number of targets of each regulator and on the number of distinct split values that a regulator has.
We iterated the Geronemo procedure until convergence. This resulted in 198 regulatory modules. A small number of modules had low coherence, defined as the average Bayesian score per gene; these modules did not provide a good explanation of the data and, therefore, were less likely to represent true biological relationships. We filtered out the 33 least coherent modules, using this score, resulting in 165 modules that we then evaluated.
Enrichment Analysis for Number of Polymorphisms in a Gene Group.
We applied an enrichment analysis of polymorphisms for several groups of genes: cis genes, regulators in the vicinity of markers selected as genotype regulators, and the genes in the Swi/Snf complex. For coding sequences, we evaluated enrichment for nonsynonymous SNPs. We computed enrichment of the polymorphisms in a gene group of interest G by comparing the distribution of the number of polymorphisms in G and in a control group consisting of the neighbors of genes in G. We computed a P value by using a nonparametric permutation test described in detail in Supporting Materials.
Proportion of Genetic Variance Explained by Genetic Regulators.
We estimated the PGV explained by the identified genetic regulators (18) by following the procedure of Brem and Kruglyak (16); see Supporting Materials for details. We randomly divided the data of 112 segregants into a detection set and an estimation set. We used Geronemo on the detection set to learn a set of modules and regulation programs and used the estimation set to calculate the PGV for these regulation programs. The PGV formula uses a corrected single factor ANOVA, which automatically accounts for model complexity. We repeated this process 10 times with different random splits of data and estimated PGV of each gene by taking the average of its PGV over 10 runs.
Chromosomal Characteristics.
We defined three criteria for chromosomal features of modules: (i) enrichment for DEGs or ChIP targets of chromatin modifiers, as defined in the Supporting Materials; we used a very stringent cutoff for enrichment of P value <1.0 × 10−5 (Fig. 3, target enrichment). (ii) At least 20% of the modules genes appear consecutively along the chromosome in runs of length at least 2 (Fig. 3, runs). (iii) Tendency for a module's genes to be significantly close to chromosomal domains such as telomeres, Ty elements, or LTR elements, evaluated by a Kolmogorov–Smirnoff test (with cutoff P < 0.001) comparing the distribution of distances (measured in bps) between the module genes and the closest telomere/Ty/LTR element and the same distribution for the other genes (Fig. 3, dom). A chromosomal module was defined to be a module with at least two of the above three chromosomal features.
Supporting Information.
We briefly describe the key methods used in the analysis, deferring detailed explanation to Supporting Materials. Also, for more data, see Fig. 11 and Tables 4 and 5, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
S-I.L. and D.K. are supported by a grant from the National Science Foundation. D.P. is supported by a Burroughs Welcome Fund CASI award and a National Institute of General Medical Sciences Center of Excellence grant. A.M.D. and G.M.C. are supported by a Department of Energy Genomes-to-Life award, and A.M.D. also is supported by a Genome Scholar/Faculty Transition award (National Institutes of Health/National Human Genome Research Institute).
Abbreviations
- DEG
differentially expressed gene
- eQTL
expression quantitative trait loci
- PGV
proportion of the genetic variance.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. Nature. 2005;437:1299–1320. [Google Scholar]
- 2.Hirschhorn JN, Daly MJ. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 3.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 4.Brem RB, Storey JD, Whittle J, Kruglyak L. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, Castellani LW, Suh M, Armour C, Edwards S, Lamb J, et al. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 6.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doss S, Schadt EE, Drake TA, Lusis AJ. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 9.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 10.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, et al. Nat Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 12.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 14.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Proc Natl Acad Sci USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Brem RB, Kruglyak L. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 18.Utz HF, Melchinger AE, Schon CC. Genetics. 2000;154:1839–1849. [PMC free article] [PubMed] [Google Scholar]
- 19.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 20.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein BE, Tong JK, Schreiber SL. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 23.Sudarsanam P, Iyer VR, Brown PO, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieb JD, Liu X, Botstein D, Brown PO. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 25.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wotton D, Shore D. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 27.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, et al. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.