Abstract
A whole-genome admixture scan in 1,597 African Americans identified a 3.8 Mb interval on chromosome 8q24 as significantly associated with susceptibility to prostate cancer [logarithm of odds (LOD) = 7.1]. The increased risk because of inheriting African ancestry is greater in men diagnosed before 72 years of age (P < 0.00032) and may contribute to the epidemiological observation that the higher risk for prostate cancer in African Americans is greatest in younger men (and attenuates with older age). The same region was recently identified through linkage analysis of prostate cancer, followed by fine-mapping. We strongly replicated this association (P < 4.2 × 10−9) but find that the previously described alleles do not explain more than a fraction of the admixture signal. Thus, admixture mapping indicates a major, still-unidentified risk gene for prostate cancer at 8q24, motivating intense work to find it.
Keywords: association, human genetics
Prostate cancer is the most common noncutaneous malignancy among U.S. men, with an estimated 234,460 new cases and 27,350 deaths in 2006 (1). African Americans have the highest incidence of prostate cancer in the United States, ≈1.6-fold higher than European Americans (http://jncicancerspectrum.oxfordjournals.org/cgi/statContent/cspectfstat;18). The higher risk (2–4) prompted the hypothesis that genetic factors in part account for this difference. If there are genetic risk variants that differ substantially in frequency across populations, admixture mapping should have power to detect them.
The idea of admixture mapping is to screen through the genome of populations of mixed ancestry such as African Americans (5), searching for regions where the proportion of DNA inherited from either the ancestral European or African population is unusual compared with the genome-wide average. Admixture mapping requires a relatively small number of markers for a whole-genome scan: a couple of thousand, rather than the hundreds of thousands estimated to be necessary in nonadmixed populations (5, 6). Because the mixture between European and West-African populations occurred within the past 15 generations (5), stretches of DNA with contiguous European and African ancestry have not had much time to break up because of recombination and typically extend millions of base pairs. Admixture mapping therefore studies highly selected SNPs every few million base pairs (Mb), rather than every few thousand as with linkage disequilibrium mapping.
Although admixture mapping was first proposed >50 years ago (7) and has good power to detect risk variants that are strikingly different in frequency across populations (6, 8), it has not been practical until recently. Appropriate panels of markers (5), combined with analytical methods (8–10), made possible the first admixture scans (11, 12) in 2005. Here, we describe a whole-genome admixture scan focusing on prostate cancer, a disease that has long been considered a test case for admixture mapping because of its marked difference in incidence rates across populations. We identify a highly significant association at 8q24. The same broad region has recently been implicated in prostate cancer by Amundadottir et al. (13). In addition to providing independent evidence of a locus at 8q24, the present study provides two pieces of information. First, we show an association with earlier age of diagnosis. Second, we show that the alleles identified in the previous study are insufficient to explain more than a small fraction of the admixture signal. Thus, the causative alleles remain to be identified.
Results
We studied 1,597 prostate cancer cases and 873 controls, the majority of which were participants in the Multiethnic Cohort study (14) (810 cases and 730 controls) (Table 5, which is published as supporting information on the PNAS web site). The other samples came from six studies, including studies that specifically ascertained cases with high-grade tumors, advanced- stage disease, diagnosis at a young age, or occurrence in a family with multiple affected individuals (15–17) (Table 1). The present study was designed to include more cases than controls, because admixture mapping works by comparing the proportion of ancestry in cases to the rest of their own genomes. In principle, controls are not needed (6, 8); however, we included controls because they are useful for follow-up analyses (8).
Table 1.
Characteristics of cases and controls from seven sources
| Source | Location | Cases | Controls | Cases, % Euro. ± 1 SE | Controls, % Euro. ± 1 SE | Mean age diagnosis (range) | % with Gleason score >7 | % with non-local tumors | % with prostate cancer in a first- degree relative | Decrease in peak LOD if these samples are removed* |
|---|---|---|---|---|---|---|---|---|---|---|
| Multiethnic Cohort | CA & HI | 810 | 730 | 23.57 ± 0.50 | 25.42 ± 0.57 | 68 (46–85) | 18 | 15 | 12 | 2.58 |
| L.A. County Men's Health Study | CA | 366 | 107 | 22.34 ± 0.83 | 26.37 ± 2.13 | 63 (42–88) | 28 | 39 | 21 | 1.37 |
| Study Early Onset Prostate Cancer | CA | 104 | — | 20.89 ± 1.37 | — | 60 (45–65) | 31 | 49 | 14 | 1.01 |
| PCGP | MI | 103 | — | 19.50 ± 1.01 | — | 55 (40–86) | 11 | 29 | 39 | 1.15 |
| Flint Men's Health Study | MI | 85 | — | 18.05 ± 1.21 | — | 65 (47–77) | 12 | 28 | 15 | 0.06 |
| Bay Area Men's Health Study | CA | 82 | 36 | 19.06 ± 1.52 | 20.13 ± 2.15 | 64 (44–78) | 25 | 94 | 28 | 1.16 |
| Genomics Collaborative | All U.S. | 47 | — | 16.16 ± 1.51 | — | 62 (39–81) | 14 | 38 | 28 | 0.57 |
| Combined samples | 1,597 | 873 | 22.11 ± 0.36 | 25.32 ± 0.55 | 65 (39–88) | 21 | 29 | 18 | 7.14 |
PCGP, Prostate Cancer Genetics Project; Euro., European.
*To assess how much each of the seven cohorts contributes to the signal of association, we removed each from the main admixture scan (run no. 9 in Table 2) and assessed how the peak LOD score at 8q24 changes. All seven cohorts contribute positively.
All 2,470 samples (1,597 cases and 873 controls) were genotyped by using one of two panels of markers chosen to be highly different in frequency between West Africans and European Americans (5). A total of 1,792 samples were genotyped in the “phase 1” panel [previously used in a scan for multiple sclerosis genes (12)] and 1,266 SNPs passed quality filters and were used in analysis (Table 6, which is published as supporting information on the PNAS web site). The remaining 678 samples were typed in a second-generation “phase 2” panel that extracts more information per SNP; 1,365 SNPs passed quality filters and were used in analysis. The analysis combines information from both panels into a single logarithm of odds (LOD) score statistic at each locus; observations >5 are considered strongly indicative of a disease locus (8). Formal significance is assessed by Bayesian methods. We take 10 to the power of the LOD score and average across points spaced every centimorgan across the genome. If the genome average is >100, then the Bayesian odds in favor of a disease locus is 100:1, and we interpret the data as showing significant evidence of a disease gene (8).
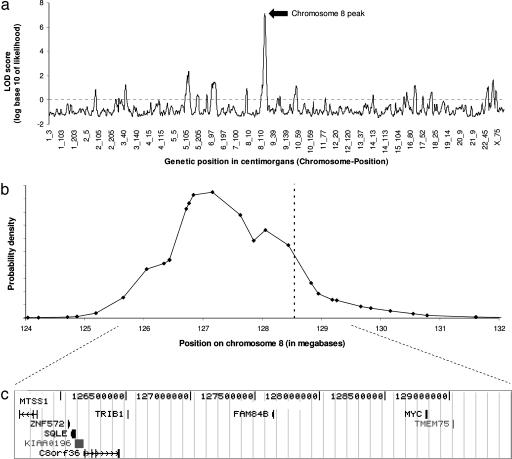
An initial admixture scan of 1,303 African-American prostate cancer cases produced a peak LOD score of 2.2 at 8q24. The signal was higher in a secondary analysis of individuals with a younger age at diagnosis, with the peak LOD score rising to 3.8 in the individuals who were <68 years of age (the threshold giving the strongest evidence of association). After genotyping 294 additional cases and 15 additional SNPs at 8q24 to obtain better local information about ancestry (see Materials and Methods), the peak LOD score increased to 4.1 in all cases and as high as 8.4 in the 1,176 who were diagnosed at <72 years of age. To correct for inflation of the score because of choosing the age threshold that gave the strongest significance, we integrated the evidence for association over an evenly spaced range of cutoffs (see Materials and Methods and Table 7, which is published as supporting information on the PNAS web site). This analysis yielded a peak LOD score of 7.1 (Fig. 1). Averaging 10 to the power of the LOD scores at equally spaced points genome-wide, we obtained a genome-wide average score of ≈19,000, exceeding the threshold of 100 for significance (8). After correcting for multiple hypothesis testing [by dividing by 4, because we tested four phenotypes (age, grade, stage, and familial disease) and focused on the one giving the strongest evidence], the odds in favor of a disease locus still greatly exceeded the threshold of 100 for significance.
Fig. 1.
Summary of results for the whole-genome admixture scan and characteristics of the 8q24 peak of association. (a) We present the LOD score at equally spaced points across the genome. The chromosome 8 peak is marked by a rise to 7.14. (b) We can use the data to calculate a probability distribution for the position of the peak. It aligns with the microsatellite and SNP recently associated with prostate cancer by Amundadottir et al. (13) (dashed line). (c) The 95% credible interval spans 3.8 Mb (125.68–129.48 Mb in build 35 of the human reference sequence) and contains nine known genes, including the c-MYC oncogene (diagram taken from http://genome.ucsc.edu) (data from the May 2004 genome assembly).
The analysis in the previous paragraph used age of diagnosis as a covariate but did not directly test whether men with younger age of diagnosis have higher risk at 8q24 than older men. To formally test this hypothesis, we exploited the fact that ANCESTRYMAP software (see ref. 8 and http://genepath.med.harvard.edu/∼reich) assigns scores for association to each individual separately (e.g., individual factors such as −0.02, 0.12) and then sums over all individuals to produce the total LOD score (Table 2). We rank-ordered the 1,588 cases in the scan for whom we had age information from youngest to oldest (Fig. 2). If the locus is not associated with age of diagnosis, the cumulative LOD should increase steadily to reach the total as additional samples are added. In fact, it rises to 5.4 LOD points above expectation at 71 years of age. To test whether this rise is significant, we permuted the data, reassigning ages of onset to different individuals (so that, in the randomized data, there could be no relationship between age of onset and allelic variation). In 1,000,000 permutations, only 318 showed a change in LOD score compared with the expectation exceeding the observed 5.4 (P < 0.00032). Repeating the analysis with a subset of samples obtained from a single prospective cohort [804 cases of African Americans with prostate cancer from the Multiethnic Cohort (MEC) Study], the association to age was also significant (P < 0.0011). These results indicate that there is a formally significant association of prostate cancer to ordering by age. We did not detect any associations when a similar analysis was applied to other subphenotypes: stage, grade, or family history (Supplemental Note 1 in Supporting Text, which is published as supporting information on the PNAS web site).
Table 2.
Admixture scan summaries
| No. of cases | No. of controls | Peak LOD score | |
|---|---|---|---|
| All prostate cancer cases | 1,597 | 873 | 4.07 |
| High grade (Gleason score >7) | 316 | 873 | 2.68 |
| Advanced stage (regional or metastatic cancer) | 414 | 873 | 2.81 |
| Family history (prostate cancer in a first-degree relative) | 281 | 873 | 1.86 |
| Age of diagnosis of <72 years and high-density genotyping at 8q24 | 1,176 | 873 | 8.39* |
| Drop out every even marker from run no. 5 to demonstrate independence of markers used | 1,176 | 873 | 8.95* |
| Drop out every odd marker from run no. 5 to demonstrate independence of markers used | 1,176 | 873 | 6.65* |
| Diagnosis at <72 years of age, high density and best model of 1.54-increased risk because of African ancestry | 1,176 | 873 | 9.39* |
| Integrating over age-of-diagnosis cutoffs as a formal test for statistical significance | 1,597 | 873 | 7.14* |
*Indicates a scan that meets formal criteria for genome-wide statistical significance.
Fig. 2.
To formally test for a relationship between age of onset and contribution to the chromosome 8 locus, we rank-ordered the individuals by age of onset and then calculated a score for increasing age cutoffs. The score rises to 5.40 above the expectation for 1,176 individuals diagnosed at <72 years of age. To evaluate whether this rise is unexpected, we permuted the data 1,000,000 times, randomizing scores with respect to individuals' ages of onset (guaranteeing that there is no relationship between age of diagnosis and contribution to the evidence of association). In only 318 of 1,000,000 permutations did we see a rise as high as in our data (P < 0.00032).
To explore how much of the increased incidence of prostate cancer in African-American men might be explained by African (as compared with European) ancestry at 8q24, we evaluated the risk for individuals carrying zero, one, and two chromosomes with African ancestry at the locus. Each African-derived chromosome is associated with ≈1.54-fold increased risk in younger individuals (90% credible interval 1.38–1.74) (Supplemental Note 2). We also estimated the proportion of control samples with zero, one, and two African-derived chromosomes, respectively (6.4%, 37.8%, and 55.8%, respectively). Extrapolating to the broader African-American population, the prostate cancer incidence in all African Americans IALL is higher than the incidence IEE in individuals who inherited two European-derived chromosomes at the locus by a factor of [(0.064) (1) + (0.378)(1.54) + 0.558(1.542)] = 1.969. Thus, the fraction of all prostate cancer incidence for African Americans <72 years of age that could be explained by ancestry at this locus is (IALL − IEE)/IALL = 1 − (1/1.969) = 49% (with a 90% credible interval of 39–59%). Thus, if it were possible to develop a treatment that reduced prostate cancer risk in the African-American population to the level that is seen in men who carry two copies of 8q24 inherited from recent European ancestors, the rate of prostate cancer would decrease by ≈49%. The total risk for prostate cancer that can be attributed to 8q24 in African-American men <72 years of age is still greater, because alleles at 8q24 increase prostate cancer risk even in chromosomes of entirely European origin (13). Thus, 8q24 has a major effect on population risk of prostate cancer, especially in younger African Americans.
Using the LOD scores at 8q24, we also calculated a posterior probability distribution to estimate the position of the disease-causing variants (see Fig. 1b and Materials and Methods). The 95% credible interval spans 3.80 Mb, from 125.68–129.48 Mb in build 35 of the human genome reference sequence (13.9 cM) and contains nine known genes (Fig. 1c). However, the admixture scan does not provide information about which gene or alleles within the locus confer risk.
Independently of this study, Amundadottir et al. (13) reported an SNP allele [A at rs1447295; odds ratio (OR) = 1.51; P < 1.0 × 10−11] and a microsatellite allele (−8 at DG8S737; OR = 1.62; P < 2.7 × 10−11) that map to the same region as the admixture peak (at 128.546 Mb and 128.554 Mb, respectively) and are highly associated with prostate cancer. The effect of the −8 allele was observed in European and African Americans, whereas the A allele effect was detected only in European-derived populations. The authors did not show, however, that either allele was causally involved in disease but instead suggested that they were both in linkage disequilibrium with an as-yet-unidentified causal variant. They also did not identify which gene in the region might be responsible for prostate cancer risk.
To directly compare the results of the two studies, we tested the previously associated alleles in the African-American cases and controls [excluding samples from Michigan, because they overlap those studied by Amundadottir et al. (13); see Materials and Methods]. The goal was to test whether the −8 allele at DG8S737 contributes to disease risk in African Americans beyond the risk that can be accounted for by the admixture signal. (Supplemental Note 3). We were concerned that the previously detected association in African Americans by Amundadottir et al. (13) (P < 0.0022, estimated OR of 1.60) might simply reflect an admixture signal across a large region (because of systematic differences in ancestry between cases and controls across several million base pairs of 8q24) and thus might not provide fine-mapping information in African Americans. Although Amundadottir et al. (13) tested for mismatching of cases and controls in overall proportion of ancestry, they did not control for a local rise in African ancestry throughout 8q24 in cases but not controls. Such a rise would be expected to cause thousands of alleles in the region that just happen to be more frequent in African Americans (including the microsatellite −8 allele) to show association with prostate cancer. When we correct for this effect in the African-American samples from the present study (Supplemental Note 3), we find that the contribution of the −8 allele to risk is nonsignificant (P = 0.22) (Table 3). The OR of 0.93–1.17 (95% credible interval) also rules out the OR = 1.60 reported in African-Americans (13).
Table 3.
Allelic association tests in African Americans adjusting for local rise in African ancestry
| Cases | Controls | P value | OR (95% CI) | |
|---|---|---|---|---|
| A allele at rs1447295 | 989 | 804 | 0.15 | 1.05 (0.95–1.16) |
| −8 allele at DG8S737 | 966 | 797 | 0.22 | 1.05 (0.93–1.17) |
| Haplotype of A and −8* | 902 | 776 | 0.31 | 1.03 (0.90–1.18) |
Cases diagnosed at <72 years of age and all controls. P values are one-tailed, testing for the previously associated allele (14) being more common (Supplemental Note 3). CI, confidence interval.
*For the haplotype test, we phased the cases and controls together, before carrying out the association analysis.
We next expanded the replication analysis to the four ethnicities in the MEC other than African Americans, by genotyping rs1447295 in 1,614 prostate cancer cases and 1,547 controls from these populations. The evidence for association is significant overall (P < 4.2 × 10−9), as well as separately in each group: Japanese Americans (P < 0.00034), Native Hawaiians (P < 0.00015), Latino Americans (P < 0.0014), and European Americans (P < 0.022) (Table 4). This analysis replicates the association identified by Amundadottir et al. (13), although we did not test for the possible confounding factor of population stratification. Interestingly, we do not replicate the association to tumor grade (Gleason ≥8 vs. Gleason <8; P = 0.47) reported by Amundadottir et al. (13).
Table 4.
rs1447295 association in the Multiethnic Cohort
| Group within the Multiethnic Cohort | No. of samples |
Frequency of A allele, % |
P value (one-tailed) | OR (95% confidence interval) | ||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||
| Native Hawaiians | 70 | 68 | 37.0 | 16.2 | 0.00015 | 3.02 (1.66–5.50)* |
| Japanese Americans | 449 | 465 | 23.8 | 17.2 | 0.00034 | 1.48 (1.18–1.86)* |
| Latino Americans | 640 | 567 | 13.5 | 9.5 | 0.0014 | 1.48 (1.14–1.91)* |
| European Americans | 455 | 447 | 13.1 | 10.0 | 0.022 | 1.35 (1.01–1.80)* |
| All samples together | 1,614 | 1,547 | 4.2 × 10−9 | 1.36 (1.22–1.51)† | ||
*OR estimated by using logistic regression adjusted for age.
†OR estimated by using logistic regression adjusted for age as well as ethnicity.
These results confirm the finding of Amundadottir et al. (13) that the 8q24 locus is important in prostate cancer. However, the alleles they reported do not explain the admixture signal (Supplemental Note 4 and Table 3). The specific variants causing increased risk for prostate cancer in African American because of 8q24 thus remain to be identified.
Discussion
We have used admixture mapping to identify a locus at 8q24 that substantially affects risk for prostate cancer. We highlight four findings.
First, this study shows that admixture mapping can be a powerful and practical way to map genetic variants for complex disease (5, 18). The results motivate the application of admixture mapping to other disorders, especially those like prostate cancer in which incidence varies across populations. These results also highlight the scientific value of studies to find disease genes in specific ethnic groups, such as African Americans.
Second, we show that the 8q24 locus contributes to a major increased risk for prostate cancer in African Americans with African ancestry at 8q24. The difference between these individuals and African Americans with European ancestry at 8q24 explains a large proportion of prostate cancer in younger African Americans. If one could intervene medically to reduce the risk for prostate cancer in African Americans <72 years of age to what would be expected if all African Americans had European ancestry at the locus, the incidence in men <72 years of age would decrease by approximately 49%. We also show that the admixture signal at 8q24 cannot be explained by the alleles identified by Amundadottir et al. (13); instead, there must be major, unmapped risk alleles at the locus.
Third, we detect a highly significant association of 8q24 with age. This finding is intriguing because it is known epidemiologically that the differential incidence of prostate cancer in African versus European Americans is greater at younger ages and is attenuated with older age (ref. 19; http://jncicancerspectrum.oxfordjournals.org/cgi/statContent/cspectfstat;18). Surveillance, Epidemiology, and End Results (SEER) Program registry data indicate that, for men diagnosed at <55 years of age, African Americans have a 2.27-fold higher rate than European Americans, but the ratio decreases to 1.48-fold for men diagnosed at ≥75 years of age (19). Genetic variation at 8q24 may be responsible for part of this effect.
Fourth, we identify a 3.8-Mb interval containing nine known genes that is likely to harbor variant(s) explaining the admixture peak. This is a tractable region for follow-up analysis. Somatic genetic data independently highlight the 8q24 region as one of the most frequently amplified regions in prostate cancer tumors (20, 21). The c-MYC oncogene, a key regulator in cellular proliferation, lies within the peak. Overexpression of c-MYC has been shown to induce tumors in mice and to create a cancer phenotype in benign prostatic epithelium (22, 23). It is possible that c-MYC could be the gene responsible for the prostate cancer risk, but no structural or regulatory variant has yet been identified.
Follow-up work will be necessary to identify the as-yet-undiscovered causal risk variant(s) at 8q24. Ultimately, discovering the causal gene(s) at 8q24 may translate into better understanding of prostate cancer and may play a role in strategies for screening of the population and identifying new targets for treatment and prevention.
Materials and Methods
Samples.
Samples were derived from seven sources (Table 1). The largest number came from the Multiethnic Cohort (MEC), a prospective cohort that began in 1993 and is still ongoing, which ascertains prostate cancer cases and controls by linking to databases from the California Cancer Registry, the Los Angeles County Cancer Surveillance Program, and the Hawaii Cancer Registry (14). The samples used in the admixture scan were all African-American cases and controls; however, for the validation genotyping of the rs1447295 SNP, we also genotyped prostate cancer cases and controls from four other ethnicities in the MEC: European Americans, Latino Americans, Japanese Americans, and Native Hawaiians. The second largest number of samples came from the Los Angeles County Men's Health Study (1999–2002), which was enriched for individuals with advanced-stage or high-grade prostate cancer, as identified through hospitals and private histopathology laboratories in Los Angeles County. The Bay Area Men's Health Study (15) (1997–2000) was enriched for individuals with regional- or distant-stage disease. The Study of Early Onset Prostate Cancer (1993–1995) was based in the San Francisco–Oakland Bay Area and included only individuals with histologically confirmed prostate cancer who were <66 years of age at diagnosis. The Genomics Collaborative, Ltd. samples were obtained from consenting individuals undergoing surgery for prostate cancer throughout the U.S. and were provided to this study at no cost by means of an academic collaboration. The Flint Men's Health Study samples (1996–2002) were obtained through a case-control study of prostate cancer in Genesee County, Michigan. The University of Michigan Prostate Cancer Genetics Project (PCGP) samples were obtained from an ongoing family-based study of prostate cancer susceptibility. PCGP cases have a family history of prostate cancer or early age at diagnosis defined as <55 years of age (we analyzed data only from the man with the youngest age of diagnosis in each family). We note that both the Flint Men's Health Study and PCGP samples (16, 17) overlap with those studied by Amundadottir et al. (13). The samples were provided by K.A.C. for replication purposes blinded to the locus under study. The results reported here, which also use a different type of information to localize disease genes (admixture linkage disequilibrium), are thus fully independent.
Genotyping.
The phase 1 and phase 2 panels of SNPs were both genotyped by using the Illumina BeadLab genotyping platform (24) [supplemented for phase 1 by Sequenom MassARRAY genotyping (25)]. At the 8q24 peak, we genotyped an additional 15 SNPs using Sequenom technology to extract maximal information about ancestry [these SNPs were chosen to have high frequency differentiation between the European and West-African populations (5) based on data from the Human Haplotype Map (26)]. We used previously described protocols to remove SNPs that did not perform well in genotyping, that were in linkage disequilibrium with each other in the ancestral European and West-African populations, or that did not seem to have appropriate intermediate frequencies in the African Americans compared with the ancestral populations (12). The rs1447295 genotyping was carried out by using the Applied Biosystems Inc. (ABI, Foster City, CA) Assay-on-Demand technology following the manufacturer's recommended protocol, and all of the African Americans were also genotyped at rs1447295 by using Sequenom technology. The DG8S737 genotyping was carried out by using ABI True Allele PCR Premix, with 5-pmol forward (5′-6FAM-TGATGCACCACAGAAACCTG-3′) and 5-pmol reverse (5′-GTTTCAAGGATGCAGCTCACAACA-3′) primers, and 60 ng of DNA per reaction. Reactions were analyzed on an ABI3730xl DNA Analyzer. Samples were scored by the ABI GeneMapper V3.7 software, with all genotypes confirmed by an experienced technician. To check the microsatellite genotyping results, we compared 168 samples that overlapped between this study and that of Amundadottir et al. (13) (data provided by K.A.C.); only five comparisons were inconsistent.
Admixture Analysis.
We used the ANCESTRYMAP software (8) to carry out the screens for association with prostate cancer. ANCESTRYMAP calculates a statistic for association at every position in the genome, under a prespecified family of risk models, calculating the likelihood of the data at the locus under an average of disease models versus the likelihood of the data if the locus has nothing to do with disease (the log base 10 of this is the LOD score). For most runs, we assume equally likely models of 0.3-, 0.4-, 0.6-, 0.7-, 0.8-, 1.2-, 1.5-, and 2-fold increased risk because of each copy of a European allele. This family of models reflects the hypothesis that African-derived alleles are more likely to confer risk but also tests for the alternative possibility. To obtain an overall assessment of the evidence for a disease locus anywhere in the genome, we average the factors for association at each point separately, providing a genome-wide assessment of whether there is a locus in the genome affecting risk.
Admixture Scan Accounting for Age of Diagnosis.
We carried out an admixture scan taking into account the possibility that individuals with a younger age of diagnosis contribute a more powerful admixture signal, while not inappropriately inflating the signal of association by picking the cutoff giving the strongest signal. We ran 22 independent scans for all individuals in the data set with diagnosis at <50, <53, <56, <57, <59, <60, <61, <62, <63, <64, <65, <66, <67, <69, <70, <71, <73, <74, <75, <76, and <78 years of age, as well as all cases (Table 7). Approximately 73 new samples were added in for each consecutive run. We then averaged the genome scores for association, which gives a statistically appropriate assessment of the evidence for association.
Permutation Analysis to Test Whether Some Phenotypes Contribute Unduly to the Signal of Association at 8q24.
To test whether the correlation of the 8q24 admixture association with a phenotype is significant, we carried out permutation analyses, considering separately the effect of stage of disease, grade of tumor, family history, and age of diagnosis (Fig. 2 and Supplemental Note 1). For each phenotype, we rank-ordered individuals by their values of the phenotype. We then calculated a cumulative LOD score at SNP rs780321 (used to mark the peak) for all individuals below each cutoff. We recorded the greatest excess or shortfall of the cumulative LOD score compared with the expectation if it increased linearly. We then wrote a PERL script to randomly permute the values of the phenotype over the samples, eliminating any relationship between the phenotype and score. A P value was calculated as the fraction of 1,000,000 permutations that produced a score for association as extreme as the data.
Inferring the Position of the Disease Locus.
To infer the position of the disease locus, we note that the LOD scores at each point of the genome can be taken to the power of 10 to give the relative probability of that locus containing the disease allele. After normalization, this calculation provides a probability distribution for the position of the locus. A 95% credible interval is obtained from the central area under the peak (Fig. 1c).
Supplementary Material
Acknowledgments
We thank the men with and without prostate cancer who participated in this study, Eric Lander and two reviewers for comments and criticism, Loreall Pooler and David Wong from the University of Southern California Genomics Laboratory for help with sample handling and genotyping, Courtney Montague at Harvard Medical School for assistance with genotyping, and the National Center for Research Resources Center for Genotyping and Analysis at the Broad Institute, without which this work would not have been possible. The genotyping for this work was supported by National Institutes of Health (NIH) Grant CA63464 (to B.E.H., C.A.H., D.A., and D.R.). M.L.F. was supported by a Department of Defense Health Disparity Training-Prostate Scholar Award (DAMD 17-02-1-0246), by a Howard Hughes Medical Institute physician postdoctoral fellowship, and by Dana–Farber/Harvard Partners Cancer Care Prostate Specialized Programs of Research Excellence (SPORE). N.P. was supported by NIH Career Transition Award HG02758. E.M.J. and S.A.I. were supported by California Cancer Research Program Grants 99-00527V-10182 and 99-00524V-10258, respectively. The Flint Men's Health Study was supported by the University of Michigan SPORE in Prostate Cancer (CA69568), the University of Michigan Department of Urology, and the University of Michigan Comprehensive Cancer Center. K.A.C. was supported by NIH Awards CA69568 and CA79596, and I.O.-G. and A.S.W. were supported by NIH Award CA67044. D.A. is a Charles E. Culpeper Scholar of the Rockefeller Brothers Fund and a Burroughs Wellcome Fund Clinical Scholar in Translational Research. D.R. is the recipient of a Burroughs Wellcome Career Development Award in the Biomedical Sciences.
Abbreviations
- LOD
logarithm of odds
- OR
odds ratio.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Bunker CH, Patrick AL, Konety BR, Dhir R, Brufsky AM, Vivas CA, Becich MJ, Trump DL, Kuller LH. Cancer Epidemiol Biomarkers Prev. 2002;11:726–729. [PubMed] [Google Scholar]
- 3.Freedland SJ, Isaacs WB. Prostate. 2005;62:243–252. doi: 10.1002/pros.20052. [DOI] [PubMed] [Google Scholar]
- 4.Simard J, Dumont M, Labuda D, Sinnett D, Meloche C, El-Alfy M, Berger L, Lees E, Labrie F, Tavtigian SV. Endocr Relat Cancer. 2003;10:225–259. doi: 10.1677/erc.0.0100225. [DOI] [PubMed] [Google Scholar]
- 5.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, De Jager PL, et al. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKeigue PM. Am J Hum Genet. 1997;60:188–196. [PMC free article] [PubMed] [Google Scholar]
- 7.Rife DC. Am J Hum Genet. 1954;6:26–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, et al. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montana G, Pritchard JK. Am J Hum Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, et al. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 12.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, et al. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 13.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, et al. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 14.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, Stram DO, Monroe KR, Earle ME, Nagamine FS. Am J Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John EM, Schwartz GG, Koo J, Van Den Berg D, Ingles S. Cancer Res. 2005;65:5470–5479. doi: 10.1158/0008-5472.CAN-04-3134. [DOI] [PubMed] [Google Scholar]
- 16.Cooney KA, Strawderman MS, Wojno KJ, Doerr KM, Taylor A, Alcser KH, Heeringa SG, Taylor JM, Wei JT, Montie JE, et al. Urology. 2001;57:91–96. doi: 10.1016/s0090-4295(00)00873-6. [DOI] [PubMed] [Google Scholar]
- 17.Cooney KA, McCarthy JD, Lange E, Huang L, Miesfeldt S, Montie JE, Oesterling JE, Sandler HM, Lange K. J Natl Cancer Inst. 1997;89:955–959. doi: 10.1093/jnci/89.13.955. [DOI] [PubMed] [Google Scholar]
- 18.Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L, et al. Hum Genet. 2002;110:553–560. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- 19.Whittemore AS, Keller JB, Betensky R. J Natl Cancer Inst. 1991;83:1231–1235. doi: 10.1093/jnci/83.17.1231. [DOI] [PubMed] [Google Scholar]
- 20.van Duin M, van Marion R, Vissers K, Watson JE, van Weerden WM, Schroder FH, Hop WC, van der Kwast TH, Collins C, van Dekken H. Genes Chromosomes Cancer. 2005;44:438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
- 21.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- 22.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 23.Williams K, Fernandez S, Stien X, Ishii K, Love HD, Lau YF, Roberts RL, Hayward SW. Prostate. 2005;63:369–384. doi: 10.1002/pros.20200. [DOI] [PubMed] [Google Scholar]
- 24.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al. Cold Spring Harbor Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 25.Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Koster H. Proc Natl Acad Sci USA. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.