Abstract
Head and neck squamous cell carcinoma (HNSCC) is a leading cause of cancer mortality worldwide. Recent reports have associated a subset of HNSCC with high-risk human papillomaviruses (HPVs), particularly HPV16, the same subset of HPVs responsible for the majority of cervical and anogenital cancers. In this study we describe a mouse model for HPV-associated HNSCC that employs mice transgenic for the HPV16 oncogenes E6 and E7. In these mice, E6 and E7 induce aberrant epithelial proliferation and, in the presence of a chemical carcinogen, they increase dramatically the animal's susceptibility to HNSCC. The cancers arising in the HPV16-transgenic mice mirror the molecular and histopathological characteristics of human HPV-positive HNSCC that distinguish the latter from human HPV-negative HNSCC, including overexpression of p16 protein and formation of more basaloid cancers. This validated model of HPV-associated HNSCC provides the means to define the contributions of individual HPV oncogenes to HNSCC and to understand the molecular basis for the differing clinical properties of HPV-positive and HPV-negative human HNSCC. From this study, we identify minichromosome maintenance protein 7 (MCM7) and p16 as potentially useful biomarkers for HPV-positive head and neck cancer.
Keywords: head and neck squamous cell carcinoma (HNSCC), human papillomavirus 16 (HPV16), minichromosome maintenance protein 7 (MCM7), p16, transgenic
Head and neck cancers are the sixth most common malignancy in the world (1), and recent advances in treating them are not reflected in improved survival. Head and neck squamous cell carcinoma (HNSCC) is etiologically associated with tobacco use; alcohol consumption; and, in Southeast Asia, chewing of betel nut (2–6). However, more recent studies have implicated high-risk human papillomaviruses (HR-HPVs), the same viruses recognized as the causative agents of cervical and most other anogenital malignancies, in the etiology of a subset of HNSCC (7).
HR-HPV DNA has been detected in ≈20% of HNSCC, particularly in malignancies of the oropharynx, where ≈50% of the cancers have been found to harbor viral DNA. HPV16, the type associated with the majority of cervical carcinomas, is the HR-HPV linked with the overwhelming majority of HPV-positive HNSCC (95%) (8–10). Other HR-HPV genotypes have been detected, including HPV18, HPV31, and HPV33. Viral oncogenes E6 and E7, the papillomaviral genes considered largely responsible for the onset as well as persistence of cervical cancer, have been detected in both integrated and extrachromosomal HPV genomes in HNSCC (11). E6 and E7 are best known for their ability to bind and inactivate the tumor suppressors p53 and pRb (12–17), and these respective properties have been associated with their oncogenic properties (18, 19). Consistent with E6 and E7 contributing to HPV-positive HNSCC is the absence of genetic or epigenetic alterations in the p53 and pRb pathways in HPV-positive HNSCC, in stark contrast to what is observed in HPV-negative HNSCC (11). Furthermore, different sets of chromosomal aberrations have been shown to be associated with HPV-positive and -negative disease (20, 21), implying that HPV-positive and HPV-negative HNSCC represent distinct diseases.
In the studies described below we have developed an animal model for HPV-associated HNSCC. This model is based on the use of keratin 14 (K14) E6- and K14E7-transgenic mice, in which the expression of the HPV16 oncogenes E6 and E7, respectively, is targeted to stratified epithelia. Both E6 and E7 can induce a variety of acute phenotypes studied previously in the epidermis of these mice (22, 23). Furthermore, both K14E6 and K14E7 mice can develop spontaneous skin tumors in their adult life, and in these mice E6 and E7, respectively, can contribute to distinct stages of skin carcinogenesis, as investigated in a model for chemically induced, multistage carcinogenesis (24). Doubly transgenic mice (harboring both the K14E6 and K14E7 transgenes) show a greater propensity for tumors than the singly transgenic mice, and these tumors arise earlier in life (24). These HPV-transgenic mice have also been used to develop a model in which both E6 and E7 have been shown to contribute to cervical carcinogenesis (25).
We have now successfully generated a mouse model for HPV-associated HNSCC by using these K14E6- and K14E7-transgenic mice. The properties of the tumors and underlying progressive disease arising in this animal model are remarkably similar to those seen in human patients who develop HNSCC. Because tobacco use is implicated in HNSCC, we monitored cancers in K14E6/K14E7 doubly transgenic mice treated with the chemical carcinogen, 4-nitroquinoline 1-oxide (4NQO). 4NQO causes a spectrum of DNA damage similar to that caused by tobacco-associated carcinogens (26–30), and it induces cancers of the oral cavity in rodents when it is supplied in their drinking water (31). 4NQO-treated, HPV16-transgenic mice developed HNSCC at a much higher frequency than nontransgenic mice. Histopathological analyses of these mice demonstrated the presence of a progressive neoplastic disease in the oral cavity and esophagus with remarkable similarities to the progressive disease associated with human HNSCC. The cancers arising in the HPV-transgenic mice display properties distinct from the cancers arising in nontransgenic mice. Importantly, these differences are the same as those seen between HPV-positive and HPV-negative human HNSCC, providing further validation to this mouse model for HPV-associated HNSCC. Using this mouse model, we identify minichromosome maintenance protein 7 (MCM7) to be a biomarker that distinguishes between HPV-positive and HPV-negative HNSCC.
Results and Discussion
HPV Oncogenes E6 and E7 Induce Aberrant Proliferation in Oral and Esophageal Epithelia.
K14E6 and K14E7 mice, transgenic for the HPV16 oncogenes, were previously characterized for the tumorigenic phenotypes of E6 and E7 in the skin (22–24) and the cervix (25). The transgenes in these mice use the human K14 transcriptional promoter to drive expression of the HPV16 E6 or E7 gene in the basal layer of the stratified epithelium, the natural host tissue of papillomavirus infection in humans. In this study we characterized the tumorigenic properties of HPV16 E6 and E7 in the oral mucosa, another natural site of infection by HPVs in humans.
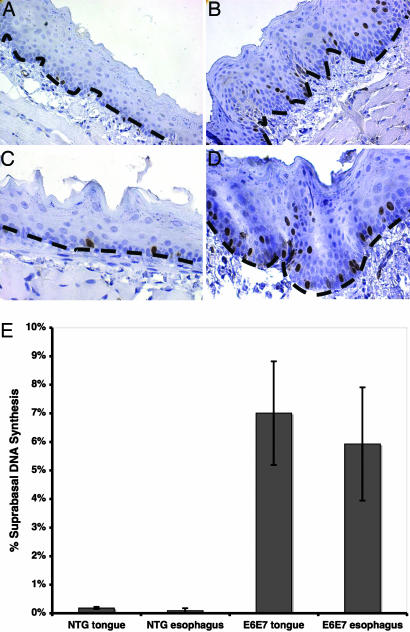
In other stratified epithelia evaluated in HPV-transgenic mice, i.e., the epidermis of the skin and stratified epithelia of the cervix, HPV16 E6 and/or E7 induces epithelial hyperplasia, which is characterized by an induction of DNA synthesis in the suprabasal compartment of the stratified epithelia (23, 32, 33). We therefore assessed whether E6 and E7 together induced a similar phenotype in the context of the oral and esophageal epithelia. Young adult K14E6/K14E7 mice transgenic for both E6 and E7, obtained from a cross between K14E6 and K14E7 mice, were injected i.p. with the nucleotide analog BrdU 1 h before being killed, and BrdU-specific antibodies were used to detect its incorporation into newly synthesized DNA on histological sections. As expected, in nontransgenic control mice, nuclei undergoing DNA synthesis were only observed in the basal layer of the epithelium. The percentage of suprabasal cells actively synthesizing DNA was <1% in both the tongue and the esophagus of these mice. In mice expressing E6 and E7, however, nuclei undergoing DNA synthesis were observed not only in the basal but also in the suprabasal compartment. The percentage of suprabasal cells supporting DNA synthesis in the tongue and esophagus of the K14E6/K14E7 mice was quantified as 7% and 6%, respectively. The difference in the frequency of suprabasal cells supporting DNA synthesis between the transgenic and control groups was statistically significant in both the tongue and esophagus (P = 0.02) (Fig. 1). This result confirms that, as seen in other tissues, E6 and E7 can reprogram cells in a normally quiescent compartment of the epithelium to reenter the cell cycle. Importantly, this short-term effect of E6 and E7 correlates with their oncogenic potential in other tissues, leading us to predict that HPV16 E6 and E7 would be tumorigenic in these oral sites as well.
Fig. 1.
HPV16 oncogenes can induce DNA synthesis in the normally quiescent compartment of oral and esophageal epithelia. Adult mice (7 weeks old) were injected with the nucleotide analog BrdU 1 h before euthanasia. The tongue and esophagus were harvested from each mouse, fixed, and embedded in paraffin. BrdU-specific immunohistochemistry was performed on slides containing sections of tongue (A and B) and esophagus (C and D) from nontransgenic (A and C) and K14E6/K14E7-transgenic (B and D) mice. The results from three animals of each genotype were quantified (suprabasal positive nuclei/total nuclei) (E). In both the tongue and esophagus of HPV16-transgenic mice (E6E7), the increase in suprabasal DNA synthesis compared with that seen in nontransgenic (NTG) counterparts was statistically significant (P = 0.02; Wilcoxon rank-sum test).
K14E6/K14E7-Transgenic Mice Are More Susceptible to 4NQO-Induced Carcinogenesis.
To assess the contributions of HPV16 E6 and E7 in HNSCC, we employed a strategy previously used to induce oral and esophageal cancers in nontransgenic mice (31). This strategy makes use of 4NQO, a carcinogen that, although not a natural tobacco derivative, causes a spectrum of DNA damage similar to that caused by tobacco-derived carcinogens (26–30). Groups of HPV16-transgenic (K14E6/K14E7) mice and nontransgenic mice were treated for 16 weeks with 4NQO dissolved in their drinking water at a dose 1/10th of that used in the previous study (31). As in the prior study, mice were taken off 4NQO treatment for a period of 8 weeks and then killed. Overt tumors in the tongue and esophagus were counted, and the tissues were collected for further histopathological analyses. The results of our study clearly indicated that E6 and E7 synergize with 4NQO to induce tumors in the oral cavity and esophagus. Specifically, the K14E6/K14E7 doubly transgenic mice were significantly (P = 0.0001) more susceptible to tumors than their nontransgenic counterparts (Table 1). Although nearly all K14E6/K14E7 mice (20 of 21) had macroscopic tumors, only a few of their nontransgenic counterparts (3 of 19) had macroscopically identifiable tumors at the same end point. Furthermore, a number of the 4NQO-treated K14E6/K14E7 mice (≈40%) became runted and had to be killed ahead of the predetermined end point because of tumors obstructing the oropharynx or the esophagus. In contrast, all of the 4NQO-treated nontransgenic mice, including the few with tumors observed at necropsy, appeared healthy until the scheduled end point. Untreated age-matched mice of both genotypes did not develop tumors.
Table 1.
Frequency and pathology of tumors arising in the tongue and esophagus of 4-NQO-treated mice
| Genotype | Frequency of overt tumors | Histopathological classification of disease |
|||||
|---|---|---|---|---|---|---|---|
| Normal | Hyper/dysplasia | Papilloma | Carcinoma grade |
||||
| I | II | III | |||||
| Nontransgenic | 3/19 | 0 | 4 | 2 | 0 | 2 | 0 |
| K14E6/K14E7 | 20/21 | 0 | 0 | 1 | 0 | 1 | 6 |
At the time of euthanasia, animals treated with 4-NQO were subjected to necropsy. Macroscopic tumors (invasive and noninvasive) in the tongue and esophagus of mice were scored (eight specimens per cohort), and the results were tabulated. HPV16-transgenic (K14E6/K14E7) mice were significantly more susceptible to the development of macroscopic tumors (two-sided Fischer's exact test; P < 0.0001). A subset of the mice scored for overt pathology (including, in the treated nontransgenic group, all three mice that had overt tumors, and, in the transgenic group, the one mouse with no overt tumors) were embedded, sectioned, and stained with H&E. Every 20th section was evaluated histopathologically for disease. Mice were scored for the most severe phenotype found in any of the sections evaluated. The analysis was also done with a set of age-matched mice not treated with carcinogen (untreated), none of which showed any signs of neoplastic disease (not shown). Statistical analysis confirmed that HPV-transgenic (K14E6/K14E7) mice are more likely to develop invasive cancers than nontransgenic mice (two-sided Fisher's exact test; P = 0.04). Furthermore, the HPV16-transgenic mice developed cancers of a higher grade, a result that is statistically significant (two-sided Wilcoxon rank-sum test; P = 0.002).
Studies have reported that smoking has a stronger association with HPV-negative than HPV-positive HNSCC patients (34). This aspect of the human disease is not necessarily reflected in our model, which could be partly accounted for by the low dose of carcinogen used in this work. One could imagine that human HPV-negative patients with a low tobacco exposure might have a lower incidence of HNSCC than HPV-positive patients with a comparable exposure; however, those cases would be harder to report in epidemiological studies because the effect would be masked by the cases with the highest exposure.
Individuals who are HR-HPV-seropositive and who smoke tobacco have been found to be at greater risk for HNSCC than individuals who are positive for either one of these factors alone (35). The nature of any cooperation between these two carcinogenic agents is a source of debate, with clinical studies that support either a synergistic or an additive relationship between them (35, 36). Transformation studies using oral keratinocytes argue for a synergy between the viral oncogenes and tobacco carcinogens. Even though HPV16 E6 and E7 are sufficient to immortalize human oral keratinocytes, exposure to tobacco carcinogens is required for these cells to become tumorigenic in nude mice (37, 38). Synergy between a carcinogen and a papillomavirus to induce tumors in the esophagus of animals has also been shown with bovine papillomavirus 4-infected cattle that were fed a diet of bracken fern, which contains the flavonoid carcinogen quercetin (39). In our animal model, the severity of the tumorigenic phenotypes observed in the 4NQO-treated HPV16-transgenic mice compared with that observed in the 4NQO-treated nontransgenic mice and the untreated mice supports there being a synergy between the HPV oncogenes and this chemical carcinogen. Thus, our study, although using a synthetic carcinogen instead of tobacco smoke, strengthens the argument for a synergistic carcinogenic effect in patients who are exposed to both etiological factors, HR-HPVs and tobacco.
Neoplastic Disease in K14E6/K14E7 Mice Is More Aggressive than in Nontransgenic Mice.
To examine the pathology of the overt tumors as well as the disease state of the overall tissue, a complete histopathological examination of a subset of the mice was performed. After scoring the overt tumor phenotypes, the entire esophagus and tongue of eight 4NQO-treated and three untreated K14E6/K14E7 and nontransgenic mice were embedded in paraffin and sectioned, and histopathological analysis was performed on every 20th section of each tissue. The entire epithelium on each section was scored for the presence of disease, from simple hyperplasia (epithelial thickening) to dysplasia (hyperplasia accompanied by nuclear atypia and abnormalities of the basement membrane), and for the presence of papillomas (noninvasive tumors) and carcinomas (invasive tumors). Carcinomas were further subclassified from grades I to IV, based on the degree of keratinization within the cancer (Fig. 2). Each mouse was scored based on the most severe degree of disease found in any of the sections from its tissues, and the results were tabulated (Table 1).
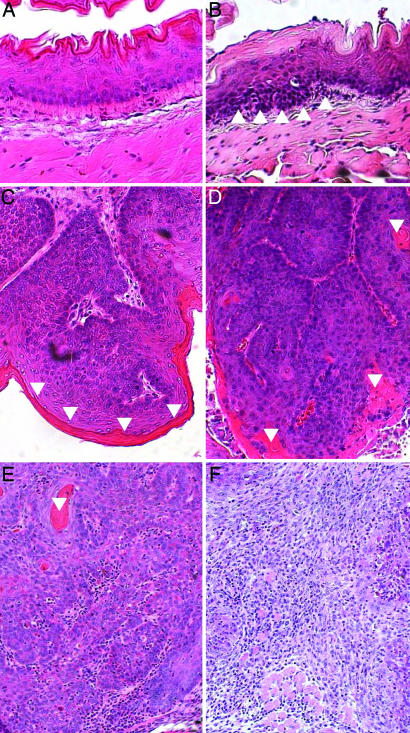
Fig. 2.
Carcinogen-treated mice develop a progressive neoplastic disease. Shown are sections of tissues harvested from 4NQO-treated mice. The tissues were fixed, embedded in paraffin, sectioned, and stained with H&E. (A–C) Nontransgenic animals. (D–F) Bitransgenic animals. (A) The epithelium in tissues from untreated animals remained normal. (B) Histopathologic analysis of the treated mice indicated that they developed areas of hyperplasia with embedded dysplasia (an example of dysplastic epithelium is demarcated by arrowheads). Hyperplasia was not observed in the absence of dysplasia. This phenotype was focal in nontransgenic mice but profuse in HPV-transgenic mice. More severe neoplastic disease was also noted, ranging from benign papillomas or polyps (C) that had not invaded the underlying stroma to grade I (D), grade II (E), and grade III (F) invasive squamous-cell carcinomas. Note that the grade I cancer is well differentiated, showing areas of keratinization (keratin in C–E is demarcated by inverted arrowheads). In contrast, most of the cells within the grade III cancer have a basaloid morphology, with few differentiated cells evident and virtually no keratin observed.
Whereas only a subset of the 4NQO-treated nontransgenic mice developed papillomas or low-grade carcinomas, there was a complete penetrance of tumors in 4NQO-treated K14E6/K14E7 mice, including the one 4NQO-treated K14E6/K14E7 mouse that had no overt macroscopic tumors at the time of necropsy (during detailed histopathological scoring, a microscopic papilloma was found in the tissues from this mouse) (Table 1). This statistically significant (P = 0.04; Fisher's exact test) difference confirmed the conclusion drawn from the overt assessment of disease, i.e., that E6 and E7 cooperate with the carcinogen 4NQO to induce squamous-cell carcinoma of the oral cavity and upper digestive tract.
Histological assessment revealed two other striking differences between the disease arising in the nontransgenic versus HPV-transgenic mice. First, the grade of carcinoma seen in 4NQO-treated K14E6/K14E7 mice was significantly more severe (Table 1) (P = 0.002; Wilcoxon rank-sum test) than that observed in the 4NQO-treated nontransgenic mice. Second, very few, if any, areas of the epithelium lining the esophagus or tongue remained normal in the 4NQO-treated K14E6/K14E7 mice. The vast majority of the tissue was highly dysplastic, with tumors arising in the context of this near-total disease state. In contrast, the 4NQO-treated nontransgenic mice possessed normal epithelium throughout their esophagus and tongue with the exception of focal areas of disease.
The more basaloid morphology and decreased keratinization seen in cancers from HPV-transgenic mice correlate with human HNSCC, in which the HPV-positive cancers are reported to have a more basaloid pathology (8, 40, 41). In humans this histopathological difference as well as positivity for p16, discussed later in the text (42), correlate with a greater responsiveness of HPV-positive cancers to radiation treatment. The underlying reason for this therapeutic response is unknown. A possible explanation may be that the more proliferative nature associated with tumors of this specific histopathology makes them more responsive to radiation treatment. The availability of a relevant animal model now provides the basis for further addressing the differences in response to therapy seen in humans and in developing more effective therapeutic interventions.
Tumors in tissues other than those of interest were not detected by visual inspection at the time of necropsy, with the exception of one 4NQO-treated K14E6/K14E7 mouse, which, in addition to an esophageal tumor, also had a carcinoma in the forestomach (an organ normally lined by stratified epithelium) and a tumor on the surface of the liver. Histopathological and immunohistochemical analyses indicated that the tumor on the liver was a squamous-cell carcinoma (Fig. 3). This tumor mass on the liver stained positive for K14, a keratin not normally expressed in hepatocytes. We infer that this mouse likely suffered from metastatic disease, presumably originating from the cancer found in either the esophagus or forestomach.
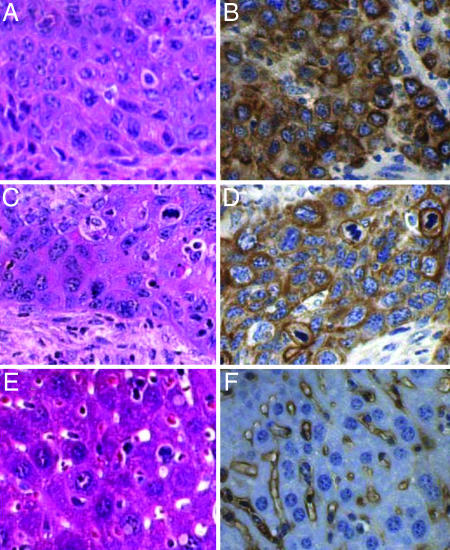
Fig. 3.
Evidence for metastatic disease. Shown are cross-sections of tumors collected from a 4NQO-treated K14E6/K14E7-transgenic mouse that had tumors in the esophagus (A and B), forestomach (not shown), and liver (C and D). Normal liver is shown in E and F. (A, C, and E) Sections were stained with H&E. (B, D, and F) Sections were stained with antibodies specific to K14. Note the similarity in cellular morphology of the tumor from the esophagus (A) and liver (C), which is distinct from the morphology of the liver hepatocytes (E). Note the abundant cytoplasmic staining for K14 in the tumor from the esophagus (B) and liver (D) and the absence of cytoplasmic staining for K14 in the surrounding hepatocytes in the liver (F).
In humans, HPV-associated HNSCC is strongly associated with the oropharyngeal sites, particularly the base of the tongue and the tonsil (11, 41, 43) in addition to other sites in the head and neck (44–46), including the esophagus (47). In the case of esophageal carcinoma, increasing reports of detection of HPV DNA and serological evidence of virus infection in these cancers are putting forth the notion that HPVs are a risk factor for this cancer, at least in those geographical regions in which there is a high incidence of this malignancy. In our mice, cancers arose throughout the oral cavity and the esophagus. Because the incidence of HPV-positive cancers is highest in the human oropharynx, we took entire heads from a subset of the 4NQO-treated HPV-transgenic mice, and we sectioned them sagittally to allow us to examine the oropharyngeal area (not shown). Tumors were present in this anatomical location. The high penetrance of dysplasia and tumors throughout the mucosal epithelium in this area obscured our ability to define clearly the origin of the tumors within this portion of the oral cavity. In addition, the mouse equivalent to the human tonsil is anatomically less well defined, making it more difficult to identify the specific substructures from which the tumors originate. Regardless, the range of sites within the head and neck region in which we observe tumors arising in our HPV-transgenic mice is similar to that seen in humans, and therefore these mice are a useful experimental model in which to study the role of HPV in malignancies throughout the head and neck region.
Tumors in Transgenic and Nontransgenic Mice Have Molecular Differences Analogous to Those of Human HPV-Positive and HPV-Negative HNSCC.
Molecular differences between human HPV-positive and HPV-negative head and neck cancers have been identified that are indicative of these cancers evolving by different mechanisms, even though they arise at the same anatomical sites. We therefore investigated whether in our mouse model for head and neck cancers we observed similar molecular differences between tumors arising in the nontransgenic versus HPV-transgenic mice. First, we assessed whether there were any gross differences in the proliferation indices of the tumors arising in the 4NQO-treated nontransgenic versus the HPV-transgenic mice because such differences could confound interpretation of differential staining patterns that might be observed with other biomarkers. We found that the proliferation indices in tumors from nontransgenic mice were similar to those from HPV16-transgenic mice as assessed by immunohistochemistry using an antibody specific to the proliferation antigen Ki67 (3 tumors from nontransgenic and 12 tumors from transgenic mice tested). Cells with positive nuclear staining for Ki67 were found throughout the thickness of both tumors that arose in the 4NQO-treated nontransgenic and K14E6/K14E7-transgenic mice at comparable levels (Fig. 4 A and B).
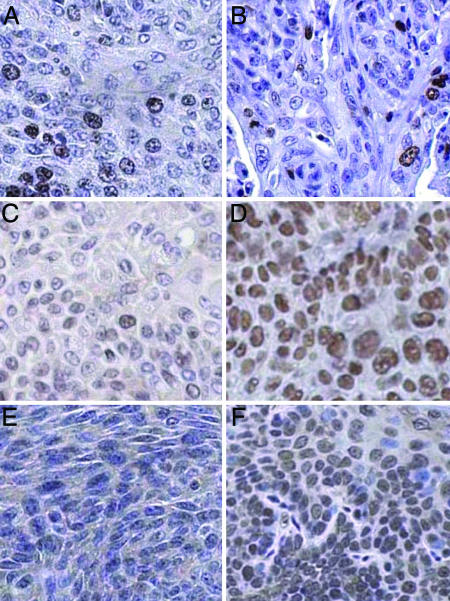
Fig. 4.
Cancers from nontransgenic and K14E6/K14E7-transgenic mice express different markers, results consistent with those seen in human HPV-negative versus HPV-positive HNSCC. Shown are histological sections of tumors from 4NQO-treated nontransgenic (A, C, and E) and K14E6/K14E7-transgenic (B, D, and F) mice that were immunohistochemically stained for Ki67 (A and B), p16 (E and F), or MCM7 (C and D). Note the similar frequency of Ki67-positive cells in the tumors from nontransgenic and HPV-transgenic mice. In contrast, note the increased frequency and intensity of staining for p16 and MCM7 in the tumor from the HPV-transgenic mice compared with the tumor from the nontransgenic mouse.
A robust biomarker for use in distinguishing between HPV-positive and HPV-negative head and neck carcinomas in humans is the cyclin-dependent kinase inhibitor p16. This gene is frequently inactivated epigenetically in HPV-negative head and neck cancers by means of hypermethylation, an event that results in minimal, if any, p16 protein being detectable by immunohistochemistry (48, 49). In contrast, levels of the p16 protein are usually elevated in HPV-positive tumors of the head and neck and anogenital tract. We performed p16-specific immunohistochemistry on head and neck tumors from nontransgenic (n = 3) and HPV-transgenic mice (n = 12). As seen in human tumors, there was almost no nuclear staining for p16 in any of the three tumors tested from nontransgenic mice, whereas p16-nuclear staining was robust in 11 of 12 tumors arising in the HPV-transgenic mice (Fig. 4 E and F).
The cumulative parallels that distinguish HPV-positive from HPV-negative head and neck cancers in humans and our mouse model validate the latter as a useful experimental system in which to gain insights into the biology of HPV-positive HNSCC. One such insight has been gained through our identification of MCM7 as an informative biomarker preferentially overexpressed in the tumors arising in the HPV-transgenic mice (Fig. 4). We previously demonstrated that MCM7 is highly induced in human cervical cancers and in cervical cancers arising in our HPV-transgenic mice (32). To address whether MCM7 is a marker for HPV-positive HNSCC, tumors from 4NQO-treated mice were stained with antibodies specific to MCM7. MCM7 was more abundant in tumors from the HPV-transgenic mice than in tumors from the nontransgenic mice (Fig. 4 C and D). All three nontransgenic tumors showed modest levels of staining, comparable with the amount of staining seen with Ki67 in the same tumors. Of the 15 stained tumors from transgenic mice, all had more nuclei with positive staining than any of the nontransgenic tumors. In 10 of these 15 tumors, positive staining was observed in nearly all nuclei. These findings indicate that MCM7 is a robust marker that distinguishes between HPV-positive and HPV-negative head and neck cancers.
MCM7 overexpression likely reflects the specific ability of HPV16 E7 to inactivate pRb and other pocket proteins, which directly leads to increased E2F transcriptional activity. The Rb–E2F pathway is likewise thought to be deregulated in HPV-negative HNSCC by virtue of the decreased expression of the cyclin-dependent kinase inhibitor p16. However, as demonstrated by the difference in expression of the E2F-responsive gene MCM7, even though the same pathway is deregulated by alternative means the consequences to the cell are quantitatively different. We conclude that the up-regulation of MCM7 in HPV-positive lesions is a robust indicator of E7 activity, and it may find use in the future as a biomarker that helps in distinguishing HPV-positive from HPV-negative disease in human HNSCC and, perhaps, in understanding underlying differences in the pathogenesis of the HPV-positive versus HPV-negative HNSCC. Consistent with this possibility, MCM7 was recently identified to be specifically up-regulated in HPV-positive HNSCC (D. Pyeon, E. M. Smith, L. P. Turek, T. Haugen, and P. Ahlquist, personal communication). This finding further substantiates the validity of our mouse model and the conclusions we have drawn from its analysis.
Materials and Methods
Mice.
The generation of K14E6 and K14E7 has been previously described (22, 23). To obtain bitransgenic K14E6/K14E7 mice, female K14E6 mice were crossed with male K14E7 mice. Tail clippings were collected from all of the offspring at the time of weaning, and DNA was isolated for genotyping by PCR. Mice were then genotyped for the presence of both transgenes. All mice were maintained on the FVB/N inbred genetic background. Mice were housed in the Association for Assessment of Laboratory Animal Care-approved McArdle Laboratory Animal Care Unit at the University of Wisconsin Medical School. All protocols for animal work were approved by the University of Wisconsin Medical School Institutional Animal Care and Use Committee.
4NQO-Induced Oral Carcinogenesis Study.
Adult K14E6/K14E7-transgenic and nontransgenic control mice (between 6 and 7 weeks of age) were treated with the carcinogen 4NQO (Sigma, St. Louis, MO) in their drinking water at a concentration of 10 μg/ml for a period of 16 weeks. The mice were then returned to a normal water supply for 8 weeks. At the end of this period, mice were euthanized, overt tumors on the tongue and esophagus were recorded, and the tissues were collected for histological analysis.
Histological Analysis.
The tongue and esophagus were fixed in 10% (vol/vol) buffered formalin, embedded in paraffin, and thin-sectioned (5 μm). Every 20th section of the tissues was stained with H&E and examined for the presence of hyperplasia, dysplasia, papillomas, and carcinomas.
Immunohistochemistry.
Procedures for immunohistochemical staining of histological sections were performed as described in ref. 50. Incubations with primary antibodies were performed as follows: BrdU (1:50 dilution; Calbiochem, La Jolla, CA) in blocking solution at 4°C; Ki67 (1:25 dilution; Dako, Carpinteria, CA); MCM7 (1:200 dilution; Neomarkers, Fremont, CA); pRb, (G3-245, 1:50 dilution; Pharmingen, San Diego, CA); p16 (M156, 1:100 dilution; Santa Cruz Biotechnology, Santa Cruz, CA). After overnight incubation with primary antibody at 4°C, sections were washed in PBS (Invitrogen, Carlsbad, CA) and incubated in a 1:100 dilution in PBS of biotinylated anti-rat Ig (Pharmingen) for Ki67 or 1:100 Vectastain universal elite secondary antibody for all other antigens (Vector Laboratories, Burlingame, CA) for 30 min at room temperature. The signal was amplified and developed as previously described.
Acknowledgments
We thank members of the Lambert laboratory for useful discussions, Amy Liem and the University of Wisconsin Comprehensive Cancer Center Histology facility for technical assistance, Dr. Ruth Sullivan for pathology consultations, and Drs. Bill Sugden and Paul Ahlquist for critical reading of this article. This work was supported by National Institutes of Health Grants CA022443, CA098428, CA113297, and DE017315 (to P.F.L.).
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- HR-HPV
high-risk human papillomavirus
- K14
keratin 14
- MCM7
minichromosome maintenance protein 7
- 4NQO
4-nitroquinoline 1-oxide.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hunter KD, Parkinson EK, Harrison PR. Nat Rev Cancer. 2005;5:127–135. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 2.Chen JK, Katz RV, Krutchkoff DJ. Cancer. 1990;66:1288–1296. doi: 10.1002/1097-0142(19900915)66:6<1288::aid-cncr2820660632>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Cullen JW, Blot W, Henningfield J, Boyd G, Mecklenburg R, Massey MM. Public Health Rep. 1986;101:355–373. [PMC free article] [PubMed] [Google Scholar]
- 4.Poschl G, Seitz HK. Alcohol Alcohol. 2004;39:155–165. doi: 10.1093/alcalc/agh057. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez T, Altieri A, Chatenoud L, Gallus S, Bosetti C, Negri E, Franceschi S, Levi F, Talamini R, La Vecchia C. Oral Oncol. 2004;40:207–213. doi: 10.1016/j.oraloncology.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Seitz HK, Matsuzaki S, Yokoyama A, Homann N, Vakevainen S, Wang XD. Alcohol Clin Exp Res. 2001;25:137S–143S. doi: 10.1097/00000374-200105051-00024. [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 8.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 9.Niedobitek G, Pitteroff S, Herbst H, Shepherd P, Finn T, Anagnostopoulos I, Stein H. J Clin Pathol. 1990;43:918–921. doi: 10.1136/jcp.43.11.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snijders PJ, Cromme FV, van den Brule AJ, Schrijnemakers HF, Snow GB, Meijer CJ, Walboomers JM. Int J Cancer. 1992;51:845–850. doi: 10.1002/ijc.2910510602. [DOI] [PubMed] [Google Scholar]
- 11.Gillison ML. Semin Oncol. 2004;31:744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 13.Werness BA, Levine AJ, Howley PM. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 14.Huibregtse JM, Scheffner M, Howley PM. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer SN, Wazer DE, Band V. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 16.Dyson N, Howley PM, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 17.Jones DL, Thompson DA, Munger K. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen M, Song S, Liem A, Androphy E, Liu Y, Lambert PF. J Virol. 2002;76:13039–13048. doi: 10.1128/JVI.76.24.13039-13048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heck DV, Yee CL, Howley PM, Munger K. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, Brakenhoff RH. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 21.Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR, Brakenhoff RH. Oncogene. 2006;25:2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 22.Song S, Pitot HC, Lambert PF. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herber R, Liem A, Pitot H, Lambert PF. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song S, Liem A, Miller JA, Lambert PF. Virology. 2000;267:141–150. doi: 10.1006/viro.1999.0106. [DOI] [PubMed] [Google Scholar]
- 25.Riley RR, Duensing S, Brake T, Munger K, Lambert PF, Arbeit JM. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 26.Venkat JA, Shami S, Davis K, Nayak M, Plimmer JR, Pfeil R, Nair PP. Environ Mol Mutagen. 1995;25:67–76. doi: 10.1002/em.2850250110. [DOI] [PubMed] [Google Scholar]
- 27.Ramotar D, Belanger E, Brodeur I, Masson JY, Drobetsky EA. J Biol Chem. 1998;273:21:489–496. doi: 10.1074/jbc.273.34.21489. [DOI] [PubMed] [Google Scholar]
- 28.Kranendonk M, Pintado F, Mesquita P, Laires A, Vermeulen NP, Rueff J. Mutagenesis. 1996;11:327–333. doi: 10.1093/mutage/11.4.327. [DOI] [PubMed] [Google Scholar]
- 29.Panigrahi GB, Walker IG. Biochemistry. 1990;29:2122–2126. doi: 10.1021/bi00460a023. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DC, Husain I, Chaney SG, Panigrahi GB, Walker IG. Nucleic Acids Res. 1991;19:365–370. doi: 10.1093/nar/19.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Clin Cancer Res. 2004;10:301–313. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 32.Brake T, Connor JP, Petereit DG, Lambert PF. Cancer Res. 2003;63:8173–8180. [PubMed] [Google Scholar]
- 33.Song S, Gulliver GA, Lambert PF. Proc Natl Acad Sci USA. 1998;95:2290–2295. doi: 10.1073/pnas.95.5.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindel K, Beer KT, Laissue J, Greiner RH, Aebersold DM. Cancer. 2001;92:805–813. doi: 10.1002/1097-0142(20010815)92:4<805::aid-cncr1386>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz SM, Daling JR, Doody DR, Wipf GC, Carter JJ, Madeleine MM, Mao EJ, Fitzgibbons ED, Huang S, Beckmann AM, et al. J Natl Cancer Inst. 1998;90:1626–1636. doi: 10.1093/jnci/90.21.1626. [DOI] [PubMed] [Google Scholar]
- 36.Herrero R, Castellsague X, Pawlita M, Lissowska J, Kee F, Balaram P, Rajkumar T, Sridhar H, Rose B, Pintos J. J Natl Cancer Inst. 2003;95:1772–1783. doi: 10.1093/jnci/djg107. [DOI] [PubMed] [Google Scholar]
- 37.Li SL, Kim MS, Cherrick HM, Doniger J, Park NH. Carcinogenesis. 1992;13:1981–1987. doi: 10.1093/carcin/13.11.1981. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Han S, Baluda MA, Park NH. Oncogene. 1997;14:2347–2353. doi: 10.1038/sj.onc.1201078. [DOI] [PubMed] [Google Scholar]
- 39.Campo MS, O’Neil BW, Barron RJ, Jarrett WF. Carcinogenesis. 1994;15:1597–1601. doi: 10.1093/carcin/15.8.1597. [DOI] [PubMed] [Google Scholar]
- 40.Poetsch M, Lorenz G, Bankau A, Kleist B. Head Neck. 2003;25:904–910. doi: 10.1002/hed.10301. [DOI] [PubMed] [Google Scholar]
- 41.Wilczynski SP, Lin BT, Xie Y, Paz IB. Am J Pathol. 1998;152:145–156. [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J, Sasaki C, Joe J, Camp RL, Rimm DL, Psyrri A. J Clin Oncol. 2006;24:736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 43.Dahlgren L, Dahlstrand HM, Lindquist D, Hogmo A, Bjornestal L, Lindholm J, Lundberg B, Dalianis T, Munck-Wikland E. Int J Cancer. 2004;112:1015–1019. doi: 10.1002/ijc.20490. [DOI] [PubMed] [Google Scholar]
- 44.Loning T, Ikenberg H, Becker J, Gissmann L, Hoepfer I, zur Hausen H. J Invest Dermatol. 1985;84:417–420. doi: 10.1111/1523-1747.ep12265517. [DOI] [PubMed] [Google Scholar]
- 45.Syrjanen K, Syrjanen S, Lamberg M, Pyrhonen S, Nuutinen J. Int J Oral Surg. 1983;12:418–424. doi: 10.1016/s0300-9785(83)80033-7. [DOI] [PubMed] [Google Scholar]
- 46.Syrjanen KJ, Syrjanen SM, Lamberg MA, Happonen RP. Arch Geschwulstforsch. 1983;53:537–546. [PubMed] [Google Scholar]
- 47.Syrjanen K. In: Papillomavirus Research: From Natural History to Vaccines and Beyond, Saveria Campo M, editor. Norfolk, UK: Caister Academic; 2006. pp. 229–254. [Google Scholar]
- 48.Begum S, Cao D, Gillison M, Zahurak M, Westra WH. Clin Cancer Res. 2005;11:5694–5699. doi: 10.1158/1078-0432.CCR-05-0587. [DOI] [PubMed] [Google Scholar]
- 49.Gillison ML, Lowy DR. Lancet. 2004;363:1488–1489. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 50.Balsitis SJ, Sage J, Duensing S, Munger K, Jacks T, Lambert PF. Mol Cell Biol. 2003;23:9094–9103. doi: 10.1128/MCB.23.24.9094-9103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]