Abstract
A wide range of biomarkers, reflecting activity in a number of biological systems (e.g., neuroendocrine, immune, cardiovascular, and metabolic), have been found to prospectively predict disability, morbidity, and mortality outcomes in older adult populations. Levels of these biomarkers, singly or in combination, may serve as an early warning system of risk for future adverse health outcomes. In the current investigation, 13 biomarkers were examined as predictors of mortality occurrence over a 12-year period in a sample of men and women (n = 1,189) 70–79 years of age at enrollment into the study. Biomarkers examined in analyses included markers of neuroendocrine functioning (epinephrine, norepinephrine, cortisol, and dehydroepiandrosterone), immune activity (C-reactive protein, fibrinogen, IL-6, and albumin), cardiovascular functioning (systolic and diastolic blood pressure), and metabolic activity [high-density lipoprotein (HDL) cholesterol, total to HDL cholesterol ratio, and glycosylated hemoglobin]. Recursive partitioning techniques were used to identify a set of pathways, composed of combinations of different biomarkers, that were associated with a high-risk of mortality over the 12-year period. Of the 13 biomarkers examined, almost all entered into one or more high-risk pathways although combinations of neuroendocrine and immune markers appeared frequently in high-risk male pathways, and systolic blood pressure was present in combination with other biomarkers in all high-risk female pathways. These findings illustrate the utility of recursive partitioning techniques in identifying biomarker combinations predictive of mortal outcomes in older adults, as well as the multiplicity of biological pathways to mortality in elderly populations.
Keywords: older adults, catecholamines, inflammation, recursive partitioning
A substantial diversity of biomarkers, reflecting possible dysregulation in multiple biological systems (e.g., cardiovascular, neuroendocrine, immune, and sympathetic nervous systems) have been used to predict downstream morbidity and mortality in elderly populations (1–7). The prediction rules are usually based on indicators of health risks that are combined in additive scoring algorithms, some of which have been interpreted as operationalizations of the concept of allostatic load (8–10). Such additive formulations are responsive to the reality that many older people have multiple co-occurring biological risk factors (4, 11–14). However, they may also obscure understanding of multisystem dysregulation by creating a kind of “black box” summary score, the contents of which may vary considerably from person to person. Indeed, examination of what is in the box often reveals no single combination of risk factors that occurs with high frequency in large population samples (15). Instead, there seem to be many different combinations of biomarkers associated with high risk of adverse health outcomes in older adults, a reflection of the multiple biological pathways to disease, disability, and mortality in elderly populations.
The primary objective of this article is to identify potentially diverse biological pathways to mortality in a high-functioning cohort of older (70–79 years of age) adults from the MacArthur Study of Successful Aging, a prospective epidemiological investigation of factors associated with healthy aging. Specifically, our aims are to (i) identify combinations of biomarkers and their zones of values associated with high levels of mortality risk in older men and women; (ii) examine whether biomarkers most predictive of mortality differ between men and women; and (iii) introduce prediction rules with high levels of sensitivity and specificity that are based on conjunctions of biomarker conditions. A secondary aim is to present recursive partitioning (16, 17) as an analytical methodology that allows for the identification of such heterogeneous combinations of biomarkers and their value zones. Throughout, the focus is on identifying subclinical levels of biomarkers that characterize high-risk (HR) conditions, because such knowledge has the potential to contribute to preventive interventions that might prolong life beyond what is expected on the basis of current clinical risk criteria.
We selected for examination thirteen biomarkers that represent various regulatory systems in the body, including the cardiovascular [systolic blood pressure (SBP) and diastolic blood pressure (DBP)], neuroendocrine [epinephrine (EPI), norepinephrine (NE), cortisol, and dehydroepiandrosterone (DHEA)], metabolic [high-density lipoprotein (HDL) cholesterol, total/HDL cholesterol, glycosylated hemoglobin (HbA1c)], and immune [IL-6, fibrinogen, C-reactive protein (CRP), and albumin] systems. Biomarkers were selected for use in analyses if the biomarker was a primary mediator of a biological regulatory system responsive to internal or external challenges (e.g., sympathetic nervous system hormones and inflammatory cytokines, such as IL-6), or if the biomarker was known to exhibit change in response to interaction with a primary mediator (e.g., CRP production in response to IL-6). The remaining measures were selected to represent secondary outcomes of these mediating processes. Many of the biomarkers have been examined individually in previous research as predictors of disability, morbidity, and mortality in older adults, but they are less often examined in combination as predictors of health outcomes. This practice has led to a limited understanding of the potential utility of information from multiple biological systems in the prediction of health outcomes in older adults.
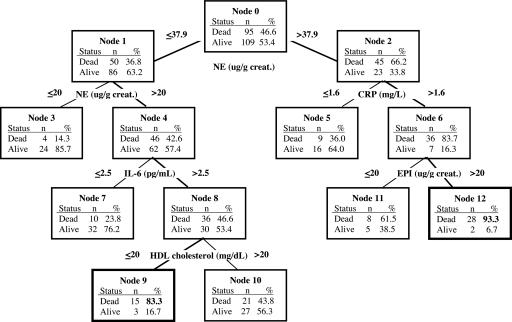
Our general analytic strategy was to use repeated subsamples of male and female participants from a larger sample of older adults to develop recursive partitioning (RP) trees (16, 17) for each male and female subsample. Each produced tree identified multiple combinations of biomarkers and their value ranges, what we refer to as “pathways,” which led to a subgroup of participants within the tree with high or low rates of mortality. The tree depicted in Fig. 1 provides an example of the various HR pathways (i.e., biomarker combinations) that can be identified with such a technique. For example, a combination of high levels of NE, CRP, and EPI led to a subgroup of 30 male participants (terminal node 12 in Fig. 1) with a mortality rate of 93.3% within the group. A second group of male participants (terminal node 9) with a high mortality rate (83.3%) is characterized by a combination of biomarkers that includes NE levels in a moderate range, high levels of IL-6, and low levels of HDL cholesterol.
Fig. 1.
Example of a single tree produced from RP analyses of male subsample. Thirteen biomarkers were entered as candidate predictors of the occurrence of mortality over a 12-year period. The biomarker predictor selected as the splitting variable at a particular node in the tree is depicted at the top of two branched lines beneath it. The numerical value in each branch signifies the zone of biomarker values associated with a particular mortality rate depicted in the box below the branch. Boxes at the end of a branch chain are terminal nodes, and the combination of biomarkers and cut point values within a branch chain leading to a terminal node forms a biomarker pathway. Those pathways with terminal nodes that have a ≥70% rate of mortality in men and a ≥60% rate of mortality in women were defined as HR pathways.
It is important to emphasize that our use of the word “pathway” in describing routes to terminal (end) nodes in trees does not imply a time ordering of events (or a process) leading to a particular state (dead or alive). All pathways simply represent logical AND statements describing combinations of biomarker conditions that are associated with degrees of mortality risk (as indicated by the mortality rate in a terminal node). To ensure stable trees, we restricted the number of levels at which the tree could be split to five (restricting the potential number of biomarkers in a pathway to a maximum of five), and we required that each pathway represent the biomarker combinations predicting a given mortality rate for a minimum of 10–15 individuals (to limit the possibility that we were identifying idiosyncratic pathways). Our tree-growing strategy diverged slightly from common RP practices in three respects. First, in contrast to the typical use of RP to identify a single tree produced from the selection of the optimal splitting predictor at each node in a tree, we also explored the production of additional trees when using second, third, fourth, or even fifth best splits at the top three nodes within a tree. Our justification for the use of these seemingly suboptimal splits is that we frequently find that two or more biomarkers have nearly equivalent goodness-of-split scores at a given node. Thus, many additional HR pathways are identified than would be the case if we insisted on optimal splits at every point in the tree generation process [see Zhang et al. (18) for a discussion of the benefits of exploring suboptimal splits and multiple trees in RP analyses]. Second, we used our multitree generation method on repeated subsamples of a larger data set. This subsampling technique allowed us to identify HR biomarker combinations that may predominate in specific subsets of the larger analytic sample, while also allowing for examination of the predictive performance of observed biomarker pathways in predicting mortality in the larger sample. Third, from the multiple trees grown in multiple subsamples of the larger data set, we selected a smaller collection of trees, a forest, that was used to characterize the multiple HR pathways across the selected trees. Thus, we obtained a set of varying biomarker combinations (pathways) predictive of a high rate of mortality in this sample of older adults. We then examined the consequences, in terms of mortality, of individuals' representation in varying numbers of these HR pathways across the forest of trees.
Results
Descriptive statistics for each biomarker for male and female participants are presented in Table 1. In terms of mean levels, men and women are, overall, fairly similar. However, women have higher levels of HDL cholesterol, cortisol, NE, and EPI, whereas males have higher DHEA and IL-6 and a higher total to HDL cholesterol level (all P ≤ .01 for mean comparisons of log-transformed normalized variables).
Table 1.
Descriptive statistics and high-risk cut point values derived from RP analyses of biomarker predictors in male and female samples
| Descriptive statistics |
HR cut points |
|||||||
|---|---|---|---|---|---|---|---|---|
| Males (N = 328) |
Females (N = 339) |
Males | Females | Conv.* | ||||
| Range | Mean (SD) | Range | Mean (SD) | |||||
| SBP, mmHg | 86.7–192.7 | 138.2 (18.6) | 98.0–204.7 | 137.6 (18.9) | >158.7 | >141 | >140 | |
| DBP, mmHg | 43.3–108.7 | 77.3 (10.5) | 50.0–109.3 | 76.4 (10.1) | <69, 73.3 | >91 | <75, >90 | |
| HDL chol., mg/dl | 10.0–118.0 | 42.3 (13.1) | 22.0–111.0 | 51.7 (15.6) | <27.5, 32.5 | <40 | ||
| Total/HDL chol. | 1.7–22.6 | 5.3 (2.0) | 2.1–11.9 | 4.9 (1.7) | >5 | |||
| CRP, mg/liter | 0.2–45.4 | 3.2 (4.8) | 0.2–65.6 | 3.3 (6.1) | <1.3, 1.6 | >3.6 | >3 | |
| IL-6, pg/ml | 0.4–40.4 | 4.9 (6.0) | 0.6–36.5 | 4.1 (5.1) | >1.9, 2.5, 2.6, 2.7 | >4.5 | ||
| Fibrinogen, mg/dl | 98.0–893.0 | 289.8 (86.5) | 130.0–694.0 | 288.5 (82.6) | >228.5, 235, 235–289 | |||
| DHEA, mg/dl | 5.0–324.0 | 83.1 (55.9) | 5.0–222.0 | 55.9 (36.1) | <43.5, 55 | <17.5 | ||
| Cortisol, ug/g creat. | 0.7–139.3 | 20.2 (16.8) | 6.1–126.5 | 23.4 (16.1) | >9.1, 14.6, 29, 37.3 | |||
| NE, ug/g creat. | 1.7–220.0 | 37.8 (23.8) | 4.0–145.7 | 42.8 (19.3) | >20, 20–37.9, 27.2, 35.9, 37.9, 64.2 | >26.5 | ||
| EPI, ug/g creat. | 0.8–21.8 | 3.5 (2.2) | 1.1–15.6 | 4.5 (2.2) | >1.7, 1.7–3.6, 2.8, 3.4, 4.3 | |||
| HbA1c, % | 4.1–20.2 | 6.9 (2.0) | 3.6–15.8 | 6.8 (1.8) | >7 | >7.5 | >7 | |
| Albumin, mg/dl | 2.6–6.1 | 4.1 (0.3) | 3.3–4.9 | 4.1 (0.3) | <3.5, >5 | |||
Recursive Partitioning Forests and Mortality Prediction.
Men.
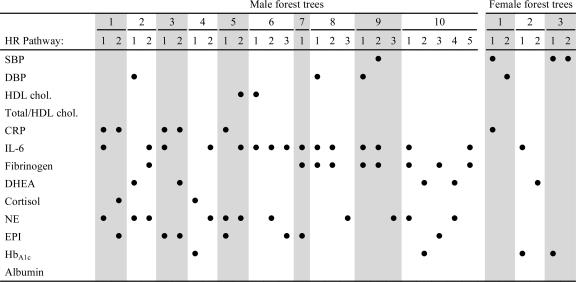
As detailed in Table 2, 11 of the 13 biomarkers enter into HR pathways (≥ 70% dead) for the ten trees selected for the male forest. These biomarkers are cortisol, CRP, IL-6, fibrinogen, NE, EPI, HbA1c, HDL cholesterol, DHEA, and SBP and DBP. Each pathway contained one to three biomarker values, with most pathways composed of a combination of three biomarkers. Inflammatory and hormone biomarkers occurred most frequently in high-risk pathways, with blood pressure and HbA1c appearing less frequently. The cut points defining the boundaries of high-risk zones are given in Table 1. In some pathways, biomarker value zones associated with high risk for mortality are comparable with conventional clinical values associated with increased risk for disease and death; in others, risk zone values fell notably above or below conventional cutoffs (e.g., SBP and CRP values). Conventional risk values are not available for all examined biomarkers.
Table 2.
Biomarkers present in HR mortality pathways in the male and female forests
A ● indicates that the biomarker in the row of the dot is present in the HR pathway represented by the column of the dot. Shading is provided to assist readability.
Sensitivity and specificity of mortality prediction was calculated for each of the trees in the male forest based on a training sample and then upon their use in a test sample. The results for each individual tree are shown in Table 3. To exploit the diversity of HR pathways across the forest, we introduced a hierarchy of prediction rules: predict dead within 12 years of baseline if the individual is on an HR pathway in at least k trees, where 1 ≤ k ≤ 10. The sensitivity and specificity of these rules, as a function of k, are displayed in Table 3. The best choices, which maintain both high sensitivity and specificity, are for k = 2 or 3.
Table 3.
Sensitivity and specificity of mortality prediction for HR tree pathways
| Tree | Sensitivity/specificity of pathways from individual trees |
Sensitivity/specificity of representation in multiple HR pathways |
|||||
|---|---|---|---|---|---|---|---|
| Training sample |
Testing sample |
k+ (no. of HR pathways) | Sensitivity, % | Specificity, % | |||
| Sensitivity, % | Specificity, % | Sensitivity, % | Specificity, % | ||||
| Males | |||||||
| 1 | 52.4 | 58.1 | 37.5 | 38.2 | 1 | 92.8 | 42.9 |
| 2 | 57.3 | 40.9 | 46.7 | 20.6 | 2 | 85.0 | 56.5 |
| 3 | 66.0 | 59.1 | 46.9 | 39.7 | 3 | 77.2 | 70.2 |
| 4 | 43.2 | 56.9 | 29.2 | 46.2 | 4 | 62.3 | 81.4 |
| 5 | 45.3 | 51.4 | 15.3 | 36.5 | 5 | 51.5 | 87.6 |
| 6 | 51.6 | 57.8 | 33.3 | 48.1 | 6 | 36.5 | 92.5 |
| 7 | 54.9 | 54.5 | 41.5 | 43.1 | 7 | 27.5 | 97.5 |
| 8 | 45.1 | 45.5 | 33.8 | 33.3 | 8 | 21.0 | 98.8 |
| 9 | 45.1 | 52.7 | 33.8 | 41.2 | 9 | 13.2 | 100 |
| 10 | 81.7 | 71.9 | 59.5 | 54.2 | 10 | 7.8 | 100 |
| Females | |||||||
| 1 | 33.3 | 95.5 | 17.2 | 84.7 | 1 | 53.7 | 79.1 |
| 2 | 33.9 | 95.4 | 17.9 | 85.7 | 2 | 22.1 | 96.7 |
| 3 | 27.9 | 95.4 | 25.9 | 89.0 | 3 | 7.4 | 99.6 |
k+, indicates membership in at least the designated number of pathways (e.g., k+ = 3 indicates representation in at least 3 HR pathways). An individual can be in only up to one HR pathway within any single tree, but can be in multiple HR pathways across the 10 trees.
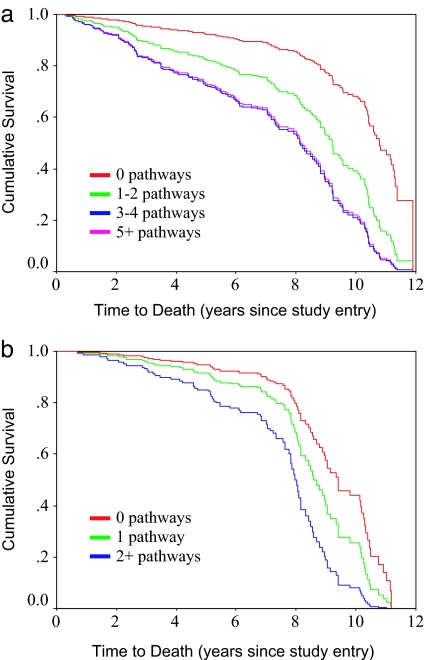
One would anticipate that the probability of death within 12 years should be a monotone increasing function of the number of HR pathways on which a given individual is located. This behavior is shown in the nested family of survivor curves in Fig. 2a.
Fig. 2.
Survival over the 12-year follow-up period as a function of representation in varying numbers of HR pathways in the male (a) and female (b) forests.
Women.
Only six of the 13 biomarkers enter into HR pathways (≥60% dead) in the three trees selected for the female forest (see Table 2). These biomarkers are SBP, DBP, HbA1c, CRP, IL-6, and DHEA. HR female pathways contained one or two biomarkers, with blood pressure biomarkers represented in the majority of trees, and inflammatory, hormone, and HbA1c biomarkers also represented in one or more trees. As documented in Table 1, HR biomarker zones derived from analyses were similar to established conventional risk zones, although the cut-points for CRP and HbA1c were slightly higher than conventional values.
Sensitivity and specificity of each of the individual trees and of representation in HR paths in multiple trees, k, across the forest, are shown in Table 3. As was the case with men, the probability of death within 12 years was an increasing function of membership in an increasing number of HR pathways in the forest (see Fig. 2b).
Combinations of HR Conditions in Male and Female Forests.
Men.
Closer examination of those men (n = 106) who were in HR pathways in at least half of the trees in the forest (i.e., 5+), indicated that there is a cluster of five biomarkers that occur together at elevated levels: CRP, IL-6, fibrinogen, NE, and EPI. In this subgroup of men, 71.7% have all five of these biomarkers at elevated-risk levels, 97.2% have four or more of the five biomarkers, and 100% have three or more of the five biomarkers at elevated-risk levels.
Women.
For women in HR pathways in two or more trees out of the forest (n = 29), a cluster of four biomarkers occurred frequently: SBP, CRP, IL-6, and HbA1c. All four of these biomarkers were present for 17.2% of the women, and two smaller biomarker clusters of SBP, HbA1c, and CRP or IL-6 occurred in 34.5% or 37.9% of the women, respectively. SBP was present at elevated risk levels in all (100%) of the women with two or more HR pathways.
Gender Comparisons.
Elevated SBP occurs in 100% of the HR female pathways and in only 17% of the HR male pathways. Fibrinogen, NE, and EPI, individually and in combination, dominate male pathways but do not even occur in female pathways. CRP and IL-6 occurred frequently in both male and female HR pathways.
Discussion
The present analyses demonstrated that there are multiple routes, characterized by combinations of biological variables, that lead to mortality in older adults. In most instances, HR mortality pathways are characterized by the interacting presence of biomarkers from multiple regulatory systems. In men, markers of the endocrine and immune systems were commonly represented in HR mortality pathways, with a lesser role for indicators of the cardiovascular and metabolic systems. Fewer HR pathways were identified in women, but a range of biomarkers was present, including blood pressure, inflammatory markers, DHEA, and HbA1c.
The predominance of inflammatory biomarkers in HR pathways in both men and women points to a central role of these primary mediators in mortality outcomes in older adults. The concomitant presence of a number of neuroendocrine variables, especially NE in male pathways, points to the potential interacting influence of neuroendocrine and inflammatory biomarkers in affecting health in elder adults. Biomarkers of these systems regulate the functions of a wide range of biological systems, including metabolic, reproductive, and cardiovascular systems, as well as other neuroendocrine and immune processes (5, 27, 28). As these findings illustrate, the combined presence of a number of these biomarkers at HR levels may serve as an early warning sign of subsequent mortality.
The cut points for specific biomarkers in observed risk pathways also serve to identify the zones of biomarker values that are associated with high mortality risk. For a number of biomarkers, the range of values in HR pathways is comparable with values identified in the biomedical literature as associated with increased risk of disease and death (e.g., see values for HbA1c, SBP (females), and DBP in Table 1). However, in a number of HR pathways, biomarker cut points varied substantially from conventional clinical criteria, with threshold values above or below established clinical criteria (e.g., see values for CRP in Table 1). These findings highlight the utility of RP techniques in identifying not only clusters of biomarkers that predict health outcomes in older adults, but also specific and interacting zones of biomarker values associated with negative outcomes, including those that may vary in men and women.
An important feature of our analysis was the utilization of population subsampling and RP tree construction methodology to develop a forest and, thereby, identify multiple combinations of biomarkers and their ranges defining elevated-risk conditions for mortality. The variation in risk zone boundaries (see Table 1) is a further reflection of the heterogeneity in subpopulations regarding preclinical levels of biomarkers that define elevated risk conditions. The construction of distinct trees on different subsamples of a larger data set was the principal analytical step that exposed this variability. This process is consistent with the philosophy of bagging classification trees (29, 30). However, our formation of a prediction rule diverges from bagging algorithms. The bagging method would require an individual to be in an HR pathway in a majority of trees across a forest (e.g., ≥5 in the 10-tree male forest) to qualify for mortality prediction. In contrast, we used a less-stringent requirement: membership in an HR pathway in 2+ or 3+ trees. This alternative ensured higher sensitivity and specificity of the prediction rule than would have been obtained with the standard bagging method.
Our findings may have implications for clinical practice and future clinical research. With a focus on prevention, it may be useful to include assays on biomarkers such as CRP, IL-6, fibrinogen, EPI, and NE as part of a standard physical examination. It is important to note that a growing literature documents linkages between these biomarkers and more macro upstream factors, such as environmental, behavioral, psychosocial, and sociodemographic factors (10, 31). Thus, values on these markers may represent the biological signature of individuals' life experiences and may represent the key pathways through which such experiences are transduced into positive or negative states of health.
One limitation of this study is that these pathways were explored in a sample of older adults who were recruited for participation on the basis of high levels of cognitive and physical functioning. Thus, our findings may not generalize to older adults with lower levels of baseline functioning. Another limitation is that biomarker information was limited to a single measurement point at the beginning of the study. Greater sensitivity in the prediction of mortality might be achieved by using information on biomarker values from multiple time points or change in biomarker levels over time. Our analyses also did not incorporate other risk (e.g., psychosocial or environmental stressor experience) or protective (e.g., health promoting behaviors) factors that might interact with biological variables to affect health. The inclusion of these other variables in analyses is an important aim of future work in this area.
Finally, although the focus here has been on the prediction of high risk for mortality, it is also worth investigating what combinations of biomarkers confer low risk for mortality. Such efforts may point to important protective processes (e.g., the role of DHEA-S in down-regulating cortisol), and as such advance knowledge of interacting regulatory systems that promote healthy aging.
Materials and Methods
Participants were from the MacArthur Study of Successful Aging, a longitudinal investigation of high functioning older adults. Participants were sampled on the basis of age (70–79 years of age) and cognitive and physical functioning levels (those in the top third of their age group on two measures of cognitive and four measures of physical functioning) from three community-based cohorts (Durham, NC; East Boston, MA; and New Haven, CT) that were a part of the Established Populations for Epidemiological Studies of the Elderly (32, 33).
Of the 4,030 age-eligible adults, a cohort of 1,313 met screening criteria and were invited to participate; 1,189 (530 men, 659 women) agreed to participate and provided informed consent. As part of baseline data collection, participants completed face-to-face and phone interviews and provided blood (80.3%) and overnight urine (85.8%) samples. Baseline data collection occurred in 1988 and 1989, with follow-up interviews in 1991 and 1995.
Measures.
Biological measures.
SBP and DBP were measured as the average of the second and third of three seated blood pressure readings. Blood and urine samples were collected on the morning following participants' baseline interview. Blood samples were used to assay HDL and total cholesterol, CRP, IL-6, fibrinogen, DHEA, albumin, and HbA1c. HDL cholesterol level was assessed by the direct homogeneous method (Genzyme Diagnostics, Cambridge, MA), and total cholesterol was measured by using colorimetric, enzymatic methods (34). CRP and IL-6 were measured by high-sensitivity ELISA (High Sensitivity Quantikine Kit, R & D Systems, Minneapolic, MN). Fibrinogen was assessed by an automated clotrate assay based on the original method of Clauss (35), with the ST4 instrument (Diagnostica Stago, Parsippany, NJ). DHEA was measured by a one-site chemiluminescence immunometric assay on the Nichols Advantage. Levels of albumin were assessed by automated Sequential Multiple Analyzer. HbA1c levels were assayed by affinity chromatography methods (36) and are expressed as percentage (%) of blood plasma. Cortisol, NE, and EPI were assayed from overnight (12-hour) urine samples with HPLC (37, 38) by Nichols Laboratories (San Juan Capistrano, CA). Values are reported in micrograms per gram (μg/g) creatinine to adjust for body size.
Mortality.
Deaths were identified through contact with next of kin at the 1991 and 1995 interviews and through the National Death Index. As of 2000, approximately half (49.8%; n = 492, 284 males, 208 females) of the participants were identified as deceased. Date of death information was available for all but three of the deceased participants whose death was confirmed by next of kin contact at the 1995 interview. Because the actual date of death was unknown, these participants were assigned a date of death that occurred approximately halfway in between their previous (1991) examination date and the follow-up (1995) examination date.
Missing data.
A significant number of participants were missing information on one or more biomarkers. Complete data for all 13 biomarkers were available for 339 females (51.4%) and 328 males (61.9%). Four participants were missing data on all biomarkers and were therefore excluded from analyses; the remaining participants were missing data on 1 to 11 biomarkers. Primary analyses were conducted by using subsamples of male and female participants with complete data. However, because tree-growing algorithms can use data with incomplete information, analyses were repeated with data sets that included participants with missing data on some biomarker predictors to compare the content and predictive performance of trees obtained from such analyses with those obtained from analyses with complete data. Results are detailed in supporting information, which is published on the PNAS web site.
Participants with complete data did not differ from those with incomplete data in terms of educational attainment, marital status, ethnicity, number of chronic health conditions, self-rated health, or smoking behavior. However, those with complete data did report greater alcohol use in the previous month and more physical activity and had higher cognitive and physical functioning scores than those with incomplete data.
Recursive Partitioning Forests and Mortality Prediction.
Generating trees for mortality prediction.
Gender-stratified analyses were conducted to identify combinations of biomarker predictors that may vary between men and women and because gender itself is a primary determinant of mortality rate. In preliminary RP analyses, which included gender as a predictor, in all cases gender was the primary splitting predictor variable at the root node in each tree. This finding led to subsequent biomarker combination pathways that were specific for men and women; thus, gender-stratified analyses were conducted for ease of analysis and interpretation.
We drew four training samples with replacement from complete male (n = 328) and female (n = 339) data sets; each contained ≈60% of the original sample (males: n1 = 196, n2 = 204, n3 = 212, n4 = 182; females: n1 = 199, n2 = 209, n3 = 221, n4 = 226). On each training sample, we fit up to 15 trees using the commercially available software program AnswerTree v3.1 (SPSS, Chicago, IL), according to the following strategy. First, splits were specified, at each level of the tree, by using the variable and cut point that resulted in the best Gini goodness-of-split value among all candidate variables. Then, trees with second, third, fourth, and fifth best splits at the root node were grown, followed by trees in which the second best split at one child node in the second level was considered for each of the five trees generated in the previous step from allowing the first to the fifth best split at the root node. The rationale for this multiplicity of trees was as follows: (i) there was frequently a small numerical difference in the goodness of split statistic between two variables, whereas there could possibly be a considerable difference in ultimate predictive performance of a tree using a slightly less than optimal split at a given point in it; and (ii) there was a priori evidence in the literature that elevated levels of particular biomarkers, either alone or in combination, were predictive of later life mortality and, thus, should be allowed as a candidate predictor in a tree. Additional tree growing parameters included the following: (i) a node could be split only if it contained 20 or more participants; (ii) a terminal node had to include 10 or more participants; and (iii) the maximum number of levels to which a tree could be grown was five. These parameters ensured that a given risk pathway described combinations of biomarkers that indicated high mortality risk for a significant number of participants (for at least ≈5% of the subsample) and that produced tree pathways were not too complex (i.e., did not have more than five biomarkers in a risk pathway).
In a given tree, a pathway into a terminal node (i.e., a specific combination of biomarker cut points) with a mortality rate ≥70% in males and >60% in females was defined as a set of HR conditions. A cutoff of 70% for males was selected because it was substantially above the overall mortality rate for men at 12 years beyond baseline (50.9%); a lower cutoff of 60% for females was selected because of the lower mortality rate (28%) in women. A prediction rule for mortality, using a single tree, was specified as follows: predict dead within 12 years of baseline if the individual has biomarker conditions as specified by a pathway into a terminal node with mortality rate ≥70% (males) or ≥60% (females).
Selection of trees to build a forest.
Trees were selected from each subsample and retained for possible inclusion in a final prediction instrument if (i) a substantial proportion of participants (at least 20% of males and 10% of females) were represented in HR terminal nodes (actual range of proportion of males = 23.5–51.1%; actual range of proportion of females = 11.8–14.1%); (ii) a diversity of biomarkers entered at least once in the pathways to HR nodes; (iii) a chosen tree was not a mere duplicate of another tree chosen to be included in the forest; and (iv) there were very few, if any, biomarker conditions implying low-risk according to conventional biomedical criteria in a HR pathway. Although our tree-growing strategy allowed for up to 60 trees to be produced across the four training samples for males and females, a smaller number of trees that met tree-growing parameters were produced in the male and female samples. A total of 52 trees were grown in the four training samples for males. A set of 10 trees, with at least one coming from each of the four training sample sets of trees, was selected to represent the male forest. A smaller set of 20 trees was grown from the four training samples for females. Only 3 trees were selected to represent the final female forest. The small size of the female forest is due to the low number of trees whose HR pathways were interpretable as such, and for which there were substantial numbers of women in the HR terminal nodes. The difficulty in identifying many acceptable trees for the women's forest is a consequence of the fact that the 12-year mortality rate is low (28%), whereas we are simultaneously trying to identify women with multiple HR conditions (a relatively rare phenomenon in this population).
Supplementary Material
Acknowledgments
Work on this article was supported by National Institute on Aging Grants P01-AG020166, AG010415, AG-17056, and AG-17265 and by the MacArthur Research Networks on Successful Aging and Socioeconomic Status and Health through grants from the John D. and Catherine T. MacArthur Foundation.
Abbreviations
- CRP
C-reactive protein
- DHEA
dehydroepiandrosterone
- DBP
diastolic blood pressure
- EPI
epinephrine
- HbA1c
glycosylated hemoglobin
- HDL
high-density lipoprotein
- HR
high-risk
- NE
norepinephrine
- RP
recursive partitioning
- SBP
systolic blood pressure
Footnotes
The authors declare no conflict of interest.
References
- 1.Danesh J, Collins R, Appleby P, Peto R. J Am Med Assoc. 1998;279:1477–1482. doi: 10.1001/jama.279.18.1477. [DOI] [PubMed] [Google Scholar]
- 2.Harris TB, Ferrucci L, Tracy RP, Corti MC, Wacholder S, Ettinger WH, Jr, Heimovitz H, Cohen HJ, Wallace R. Am J Med. 1999;106:506–512. doi: 10.1016/s0002-9343(99)00066-2. [DOI] [PubMed] [Google Scholar]
- 3.Seeman TE, Gruenewald TL. In: Medical and Psychiatric Comborbidity over the Course of Life, Eaton WW, editor. Washington, DC: American Psychiatric Publishing; 2006. pp. 179–196. [Google Scholar]
- 4.Seeman TE, Crimmins E, Singer B, Bucur A, Huang M, Gruenewald TL, Berkman LF, Reuben DB. Soc Sci Med. 2004;58:1985–1997. doi: 10.1016/S0277-9536(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 5.McEwen BS. Metabolism. 2003;52:10–16. doi: 10.1016/s0026-0495(03)00295-6. [DOI] [PubMed] [Google Scholar]
- 6.de Bruin VM, Vieira MC, Rocha MN, Viana GS. Brain Cognit. 2002;50:316–323. doi: 10.1016/s0278-2626(02)00519-5. [DOI] [PubMed] [Google Scholar]
- 7.Ershler WB, Keller ET. Annu Rev Med. 2000;51:245–270. doi: 10.1146/annurev.med.51.1.245. [DOI] [PubMed] [Google Scholar]
- 8.Seeman TE, Singer BH, Rowe JW, Horwitz RI, McEwen BS. Arch Intern Med. 1997;157:2259–2268. [PubMed] [Google Scholar]
- 9.Seeman TE, McEwen BS, Rowe JW, Singer BH. Proc Natl Acad Sci USA. 2001;98:4770–4775. doi: 10.1073/pnas.081072698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwen BS, Seeman T. Ann NY Acad Sci. 1999;896:30–47. doi: 10.1111/j.1749-6632.1999.tb08103.x. [DOI] [PubMed] [Google Scholar]
- 11.Straub RH, Miller LE, Scholmerich J, Zietz B. J Neuroimmunol. 2000;109:10–15. doi: 10.1016/s0165-5728(00)00296-4. [DOI] [PubMed] [Google Scholar]
- 12.Crimmins EM, Johnston M, Hayward M, Seeman T. Exp Gerontol. 2003;38:731–734. doi: 10.1016/s0531-5565(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 13.Kubzansky LD, Berkman LF, Glass TA, Seeman TE. Psychosom Med. 1998;60:578–585. doi: 10.1097/00006842-199809000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Reed DM, Foley DJ, White LR, Heimovitz H, Burchfiel CM, Masaki K. Am J Public Health. 1998;88(10):1463–8. doi: 10.2105/ajph.88.10.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer B, Ryff CD, Seeman TE. In: Allostasis, Homeostasis, and the Costs of Physiological Adaptation, Schulkin J, editor. Cambridge, UK: Cambridge Univ Press; 2004. pp. 113–149. [Google Scholar]
- 16.Zhang H, Singer B. Recursive Partitioning in the Health Sciences. New York: Springer; 1999. [Google Scholar]
- 17.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. 2nd Ed. Belmont, CA: Wadsworth; 1984. [Google Scholar]
- 18.Zhang HP, Yu CY, Singer B. Proc Natl Acad Sci USA. 2003;100:4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones W, Materson BJ, Oparil S, Wright JT, et al. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 20.Taylor JO, Cornoni-Huntlery J, Curb JD, Manton KG, Ostfeld AM, Scherr P, Wallace RB. Am J Epidemiol. 1991;134:489–501. doi: 10.1093/oxfordjournals.aje.a116121. [DOI] [PubMed] [Google Scholar]
- 21.Langer RD, Ganiats TG, Barrett-Connor E. Am J Epidemiol. 1991;134:29–38. doi: 10.1093/oxfordjournals.aje.a115990. [DOI] [PubMed] [Google Scholar]
- 22.Toth PP. Am J Cardiol. 2005;96:50K–58K. doi: 10.1016/j.amjcard.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Sempos CT, Cleeman JI, Carroll MD, Johnson CL, Bachorik PS, Gordon DJ, Burt VL, Briefel RR, Brown CD, Lippel K, et al. J Am Med Assoc. 1993;269:3015–3023. doi: 10.1001/jama.269.23.3009. [DOI] [PubMed] [Google Scholar]
- 24.Pearson TA, Bazzarre TL, Daniels SR, Fair JM, Fortmann SP, Franklin BA, Goldstein LB, Hong Y, Mensah GA, Sallis JF, et al. Circulation. 2003;107:645–651. doi: 10.1161/01.cir.0000054482.38437.13. [DOI] [PubMed] [Google Scholar]
- 25.American Diabetes Association Diabetes Care. 2005;28(Suppl 1):S4–S36. [PubMed] [Google Scholar]
- 26.Vincent JL, Dubois MJ, Navickis RJ, Wilkes MM. Ann Surg. 2003;237:319–334. doi: 10.1097/01.SLA.0000055547.93484.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black PH. Brain Behav Immun. 2003;17:350–364. doi: 10.1016/s0889-1591(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 28.Reichlin S. N Engl J Med. 1993;329:1246–1253. doi: 10.1056/NEJM199310213291708. [DOI] [PubMed] [Google Scholar]
- 29.Breiman L. Machine Learning. 1996;24:123–140. [Google Scholar]
- 30.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference and Prediction. New York: Springer; 2001. [Google Scholar]
- 31.Seeman TE, Crimmins E. Ann NY Acad Sci. 2001;954:88–117. doi: 10.1111/j.1749-6632.2001.tb02749.x. [DOI] [PubMed] [Google Scholar]
- 32.Cornoni-Huntley J, Ostfeld AM, Taylor JO, Wallace RB, Blazer D, Berkman LF, Evans DA, Kohout FJ, Lemke JH, Scherr PA, et al. Aging (Milan) 1993;5:27–37. doi: 10.1007/BF03324123. [DOI] [PubMed] [Google Scholar]
- 33.Berkman LF, Seeman TE, Albert M, Blazer D, Kahn R, Mohs R, Finch C, Schneider E, Cotman C, McClearn G, et al. J Clin Epidemiol. 1993;46:1129–1140. doi: 10.1016/0895-4356(93)90112-e. [DOI] [PubMed] [Google Scholar]
- 34.Siedel J, Hagele EO, Ziegenhorn J, Wahlefeld AW. Clin Chem. 1983;29:1075–1080. [PubMed] [Google Scholar]
- 35.Clauss A. Acta Haematol. 1957;17:237. doi: 10.1159/000205234. [DOI] [PubMed] [Google Scholar]
- 36.Little RR, England JD, Wiedmeyer HM, Goldstein DE. Clin Chem. 1983;29:1113–1115. [PubMed] [Google Scholar]
- 37.Canalis E, Reardon GE, Caldarella AM. Clin Chem. 1982;28:2418–2420. [PubMed] [Google Scholar]
- 38.Krstulovic AM. J Chromatogr. 1982;229:1–34. doi: 10.1016/s0378-4347(00)86033-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.