Abstract
EBV is a paradigm for human tumor viruses because, although it infects most people benignly, it also can cause a variety of cancers. Both in vivo and in vitro, EBV infects B lymphocytes in G0, induces them to become blasts, and can maintain their proliferation in cell culture or in vivo as tumors. How EBV succeeds in these contrasting cellular environments in expressing its genes that control the host has not been explained. We have genetically dissected the EBV nuclear antigen 1 (EBNA1) gene that is required for replication of the viral genome, to elucidate its possible role in the transcription of viral genes. Strikingly, EBNA1 is essential to drive transcription of EBV's transforming genes after infection of primary B lymphocytes.
Keywords: transcriptional regulation, high-mobility group A protein, HMGA1a
Epstein-Barr virus (EBV) is a human pathogen causally associated with multiple lymphomas and carcinomas. EBV differs from other human tumor viruses, such as the papillomaviruses, in that it initially infects nonproliferating B lymphocytes and induces and maintains their proliferation. EBV, therefore, needs to express its transforming genes in markedly different cellular environments. We sought to determine how EBV accomplishes these early phases of its life cycle and found that one viral protein, EBV nuclear antigen 1 (EBNA1), is essential for regulating the transcription of EBV's transforming genes.
In general, neither cells infected with EBV in vitro, nor its associated tumor cells, support the productive cycle of the virus. Rather, the viral genome is maintained in these cells stably as a plasmid and expresses few of its genes. In all infected cells that proliferate in vitro or in vivo, EBNA1 is expressed and maintains replication of the viral plasmid by (i) fostering origin recognition complexes that bind specifically to plasmid origins of DNA synthesis and (ii) retaining replicated plasmids in the proliferating host cell (1). EBNA1 also can affect transcription. Some of the analyses that have identified EBNA1 as a regulator of transcription may reflect EBNA1's ability to bind introduced DNA templates and conduct them into the nucleus (2); other analyses do measure either EBNA1's increase in transcription of integrated templates (3) or an inhibition of a transfected template (4). What is unknown is whether EBNA1's transcriptional activities contribute to the EBV life cycle. We have addressed this uncertainty by introducing defined mutations into EBNA1 within EBV's 165-kbp genome and analyzing B cells infected with these recombinant viruses. One such mutation, which supports plasmid replication as does wild-type EBNA1, fails to support the use of a promoter required to express EBV's transforming genes. Our analyses identify EBNA1 as a regulator of transcription of EBV's viral genes and, thus, the transformed phenotypes they induce.
Results
A Deletion of UR1 in EBNA1 Is Defective for Maintaining Proliferation of Infected B Cells.
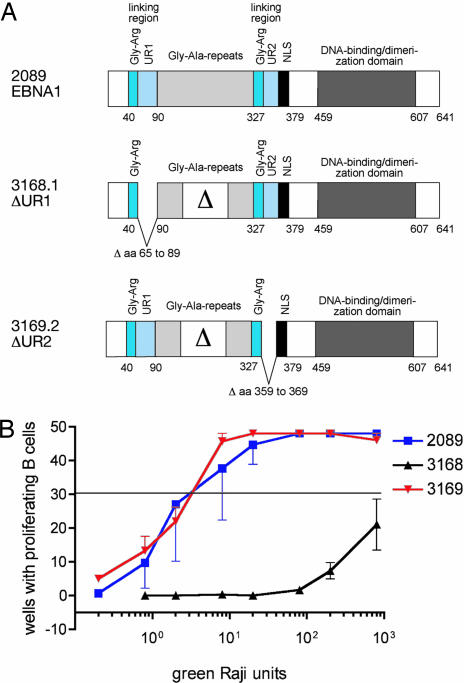
Two deletions were introduced into EBNA1 in maxi-EBVs, which are F-plasmids containing the intact viral genome in Escherichia coli. These deletions separately removed the unique sequences within the two linking regions of EBNA1. Previous analyses have implicated these linking regions as contributors to both the transcriptional and replicational activities of EBNA1 (5). These linking regions both contain runs of glycine and arginine residues and two unique stretches, UR1 (residues 65–89) and UR2 (residues 359–369). We deleted the unique sequences within EBNA1's linking regions to ascertain their possible contribution to EBNA1's functions (Fig. 1A). The mutated maxi-EBVs, as F-plasmids, along with one encoding wild-type EBV, were introduced into 293 D cells to generate viral stocks (6). All three plasmids were maintained in these human cells in culture, indicating that the mutant derivatives of EBNA1 were similarly competent in their support of EBV's plasmid replication (data not shown). Stocks of the three viral strains, 3168 (ΔUR1-EBNA1), 3169 (ΔUR2-EBNA1), and 2089 (wild-type EBV), were generated, titered on Raji cells because each strain was capable of encoding GFP, and used to infect primary B cells. Strain 3169 (ΔUR2-EBNA1) was identical to 2089 (wild-type) in its efficient induction and maintenance of proliferation of B cells, indicating that neither a deletion of UR2 nor the shortened glycine/alanine repeats (shared by both the mutant viruses) affect transformation. Strain 3168 (ΔUR1-EBNA1), however, had 0.1% of wild-type EBV's transforming ability (Fig. 1B).
Fig. 1.
Deletion of UR1 from EBNA1 inhibits transformation of B cells. (A) Shown are the alleles of EBNA1 engineered into strains of EBV: 2089, wild-type EBV (22); or mutant viruses 3168 ΔUR1 and 3169 ΔUR2. The EBNA1 allele present in the two mutant EBVs lacks about half of the internal glycine/alanine repeats as indicated (Δ), with no functional consequences (28). (B) Efficiency of transformation of B cells. In limiting dilution assays, the numbers of green Raji units (GRUs) of 2089 wild-type EBV and the two EBNA1 mutant EBVs (3168 ΔUR1-EBNA1 and 3169 ΔUR2-EBNA1) necessary to yield proliferating clones were measured. The horizontal line at ≈30 wells positive (of 48 wells plated) indicates the zero term of the Poisson equation and identifies the average number of GRUs of the different virus stocks required to yield one proliferating clone. Approximately three GRUs were sufficient with both 2089 wild-type and 3169 ΔUR2-EBNA1 EBVs to establish one growing clone. One thousand or more GRUs were required by the 3168 ΔUR1-EBNA1 mutant.
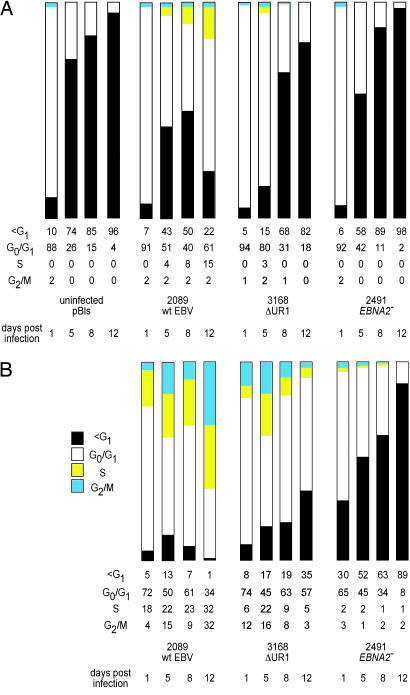
To characterize the defect in 3168 (ΔUR1-EBNA1), we analyzed newly infected B cells for their ability to undergo DNA synthesis, as measured by incorporation of BrdU, and for their viability, as measured by their forward and side light-scattering characteristics. Three viral strains were used in these analyses at a multiplicity of infection of 0.1: 3168 (ΔUR1-EBNA1), 2089 (wild-type EBV), and 2491 (EBNA2−). The 2491 (EBNA2−) strain serves as a negative control because B cells infected with this strain, as well as uninfected B cells, fail to enter the cell cycle and die by apoptosis as early as 5 days post-infection (p.i.) (7). The analyses measuring incorporation of BrdU followed both uninfected and infected cells in each population and demonstrated that the populations infected with 3168 (ΔUR1-EBNA1) and 2089 (wild-type EBV) had cells in S phase as early as 5 days p.i., whereas the uninfected cells or cells infected with 2491 (EBNA2−) never had cells in S phase (Fig. 2A). The analyses measuring forward and side light-scattering focused on the GFP+ infected cells and revealed that cells infected with 2089 (wild-type EBV) began and remained in a proliferating state, those infected with 2491 (EBNA2−) failed to proliferate, and those infected with 3168 (ΔUR1-EBNA1) paralleled those infected with wild-type EBV through day 5 p.i. but gradually withdrew from the proliferative cycle thereafter (Fig. 2B and Fig. 6A, which is published as supporting information on the PNAS web site). The fraction of cells infected with 3168 (ΔUR1-EBNA1) and 2491 (EBNA2−) strains having a subG1 content of DNA increased to 35% and 89%, respectively, by day 12 p.i., indicating that these infected cells were likely dying apoptotically (Fig. 2B). Thus, B cells infected with EBV in which EBNA1 lacks UR1 do enter the cell cycle but soon withdraw from it and die.
Fig. 2.
Primary B cells infected with the ΔUR1-EBNA1 mutant EBV cease to proliferate 1 week p.i. (A) The cell cycle status of the entire population of infected and uninfected primary B cells was determined with the thymidine analogue BrdU, which was added for 2 h before FACS analysis. The cells were stained with an APC-coupled BrdU-specific antibody, and the cellular DNA was counterstained with the DNA intercalating dye 7-AAD. Cells were analyzed for G1, S, and G2/M phases of the cell cycle and for subG1 DNA content. FACS analyses were performed until a total of 3 × 104 cells were collected and set to 100%. Uninfected cells and cells infected with the EBNA2− mutant 2491 EBV did not enter the cell cycle and became apoptotic by day 5 p.i., as indicated by their subG1 DNA content. Cells infected with the ΔUR1-EBNA1 mutant EBV initially entered the cell cycle, as indicated by a small fraction of cells in S phase, but later ceased to proliferate. (B) To exclude cellular debris, the EBV-infected cells were gated for forward and sideward scatter, and only GFP+ cells within the lymphocyte gate were considered for analysis. Wild-type 2089 EBV induced progression in the cell cycle as early as 24 h p.i., and the fraction of cells in S or G2/M phase increased from that time point onward. Primary B cells infected with 3168 (ΔUR1-EBNA1) were indistinguishable from wild-type EBV-infected cells until day 5 p.i. At days 8 and 12 p.i., the majority of ΔUR1-EBNA1 EBV-infected cells ceased to proliferate, and the fraction of cells with a subG1 DNA content increased considerably. Cells infected with the EBNA2− mutant 2491 EBV did not enter the cell cycle and became increasingly apoptotic, as indicated by their subG1 DNA content. One representative experiment of three is shown.
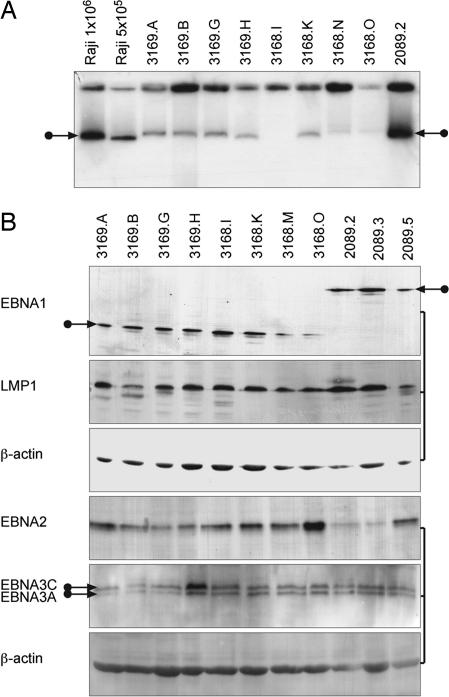
To reveal mechanistically the defect in cells infected with 3168 (ΔUR1-EBNA1), we tested the ability of the ΔUR1-EBNA1 derivative to support plasmid replication and the ability of the rare (0.1% of wild-type levels) cells that do proliferate after infection with 3168 (ΔUR1-EBNA1) to express viral proteins. ΔUR1-EBNA1 supports the stable replication of plasmids with EBV-derived plasmid origins oriP and 8× Rep* as efficiently as does ΔUR2-EBNA1 (Table 1). Eight clones of B cells infected with 3168 (ΔUR1-EBNA1) or 3169 (ΔUR2-EBNA1), after proliferating for more than 25 generations, maintained EBV DNA extrachromosomally (Fig. 3A) and expressed those viral proteins characteristic of EBV's latent phase of infection, as did clones infected with wild-type EBV (Fig. 3B). Although clones of cells infected with 3168 (ΔUR1-EBNA1) had fewer viral plasmids per cell, all did maintain viral DNAs extrachromosomally. Clearly, the ΔUR1 derivative can support plasmid replication, and cells infected with 3168 (ΔUR1-EBNA1) can, under some circumstances, express all of the EBV proteins required for the initiation and maintenance of proliferation of infected B cells.
Table 1.
ΔUR1-EBNA1 and ΔUR2-EBNA1 support stable replication of OriP plasmids
| EBNA1 allele | Test plasmid | Fraction of transfected cells supporting long-term plasmid replication |
|---|---|---|
| ΔUR1 | OriP | 0.11 ± 0.7 |
| FR + 8 × Rep* | 0.05 ± 0.01 | |
| FR | 0.002 ± 0.0002 | |
| ΔUR2 | OriP | 0.12 ± 0.06 |
| FR + 8 × Rep* | 0.06 ± 0.03 | |
| FR | 0.001 ± 0.0002 |
BJAB cells stably expressing different alleles of EBNA1 were electroporated with the different test vectors, each expressing resistance to puromycin and one expressing GFP. After 48 h, different numbers of GFP-positive cells were plated per well, along with 1 μg/ml puromycin, in 96-well plates. The number of wells negative for proliferating cells after 3 weeks was counted and used to calculate the fraction of transfected cells supporting long-term plasmid replication, based on the Poisson distribution. The fractions determined for the ΔUR1 and ΔUR2 alleles are within 2-fold of those measured in parallel for wild-type EBNA1 (31).
Fig. 3.
EBNA1 lacking UR1 can support extrachromosomal DNA replication and latent gene expression. B cell clones obtained with 2089 wild-type EBV, 3168 (ΔUR1-EBNA1), or 3169 (ΔUR2-EBNA1) from limiting dilution assays were expanded and analyzed for genomic extrachromosomal DNA molecules and latent gene expression. (A) Extrachromosomal viral DNA was detected by Gardella gel hybridization. The autoradiogram of the blotted Gardella gel hybridized with a specific, 32P-labeled plasmid probe revealed signals typical of extrachromosomal copies of EBV, as indicated by arrows (29, 30). Signals from 1 × 106 cells per lane with clones infected with 2089 wild-type or 3169 (ΔUR2-EBNA1) gave signals similar to Raji cells, which contain ≈50 copies of EBV per cell as a reference. Clones obtained with 3168 (ΔUR1-EBNA1) had a lower copy number, as indicated by weaker signals, with one case (3168.I) not visible in this exposure of the autoradiogram. (B) The expression of latent EBV proteins such as EBNA1, EBNA2, EBNA3A and -C, and LMP1 is similar among clones of B cells established with wild-type EBV and EBNA1 mutant viruses, as detected by Western blot analysis.
ΔUR1-EBNA1 Is Defective for Supporting Transcription from the Bam C Promoter (Cp).
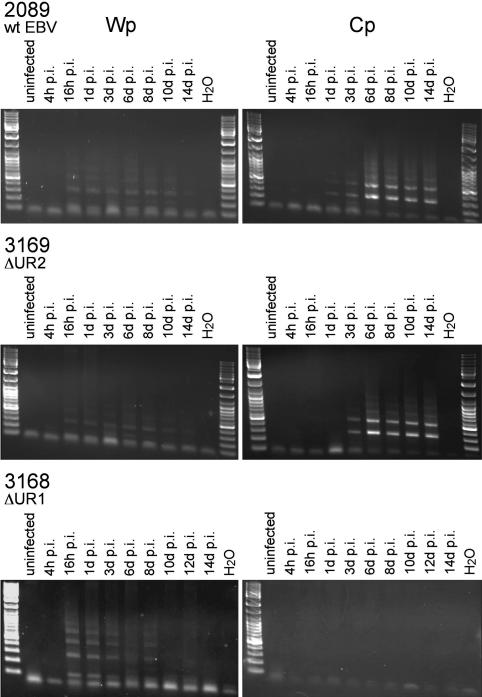
RNAs encoding EBV's six nuclear antigens are known to be expressed immediately after infection from the Bam W promoter (Wp). This promoter usually ceases to function within days as the cells differentiate into blasts and divide, and a second promoter, the Bam Cp, begins to support expression of these genes (8–10). It is not known how Wp is regulated in infected primary B cells, but transfection experiments in EBV-negative B cell lines indicate that CREB/ATF factors regulate Wp positively (11). Similarly, it is not known how Cp is regulated in infected primary B cells, but transfection experiments in both EBV-negative and -positive B cell lines indicate that multiple cellular factors affect Cp and that EBNA1, cellular Sp1, and NFY may positively regulate Cp together (12–15). We assayed whether this change in promoter function occurs in cells infected with 3168 (ΔUR1-EBNA1), 3169 (ΔUR2-EBNA1), and 2089 (wild-type EBNA1). Primary B cells were infected with the different viruses at a multiplicity of infection of 0.025. RNAs were isolated from these cells at different times, reverse-transcribed, normalized to the levels of GFP RNAs expressed from the human cytomegalovirus immediate early promoter present in each of the recombinant EBVs, and assayed for the promoters from which they were expressed. B cells infected with all three viruses expressed viral RNAs originating from Wp within 16 h p.i.; RNA expression diminished to undetectable levels by 8–10 days p.i. (Fig. 4). B cells infected with 3169 (ΔUR2-EBNA1) and 2089 (wild-type EBV) began to express detectable levels of RNAs from Cp 3 days p.i. and continued to do so throughout the course of the experiment (Fig. 4). Cells infected with 3168 (ΔUR1-EBNA1) failed to express RNAs from Cp. The UR1 domain of EBNA1 appears critical for expression of viral transcripts from Cp in most newly infected, proliferating B cells. In those rare cells infected with 3168 (ΔUR1-EBNA1) that survived to proliferate, both Wp and Cp were found to support transcription of EBV's transforming genes (data not shown), indicating that these promoters were competent to function given the appropriate cellular environment.
Fig. 4.
Primary B cells infected with 3168 (ΔUR1-EBNA1) fail to activate the Cp. Primary B cells were infected with wild-type 2089 EBV, 3168 (ΔUR1-EBNA1), or 3169 (ΔUR2-EBNA1), and RNA was harvested at the indicated time points. cDNA was synthesized and analyzed by real-time PCR for the level of GFP-specific transcripts that served as an internal control for normalization of cDNAs specifically derived from the virally infected B cells. cDNA preparations that contained the same number of GFP cDNA molecules were amplified by conventional PCR with primer pairs specific for transcripts originating from Cp or Wp. Wp-specific transcripts could be detected as early as 16 h p.i. and could no longer be detected by 10–12 days p.i. Transcripts originating from Cp became apparent at ≈3 days p.i. in primary B cells infected with 2089 wild-type or 3169 (ΔUR2-EBNA1) mutant EBV. Cells infected with 3168 (ΔUR1-EBNA1) showed comparable levels of Wp transcription but failed to activate the Cp.
UR1 Restores Transformation to EBV Carrying a Defective Derivative of EBNA1.
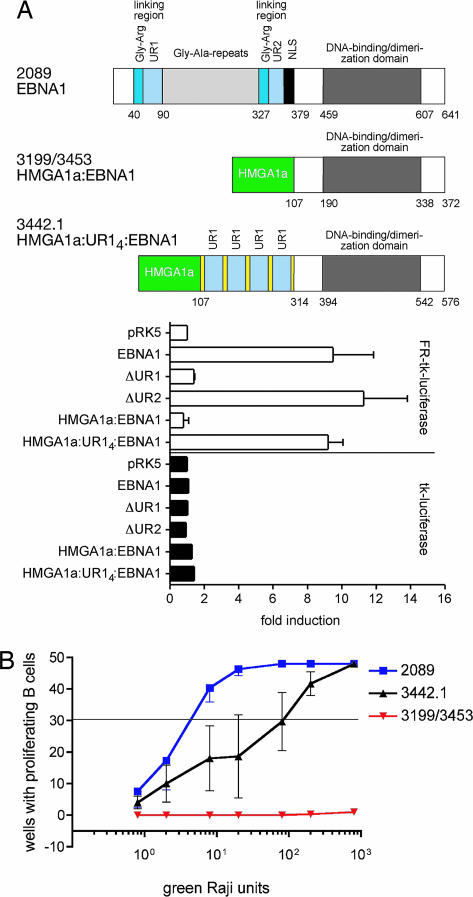
A derivative of EBNA1 in which the first 378 of its residues are substituted with HMGA1a has been found to support the replication of plasmids carrying EBV's origin of plasmid replication, oriP (16), but not the transcription of reporter plasmids regulated by wild-type EBNA1 (Fig. 5A). We introduced this derivative of EBNA1 into two independent recombinant maxi-EBVs, 3199 (HMGA1a:EBNA1) and 3453 (HMGA1a:EBNA1), and found that they replicated as plasmids in 293 cells (data not shown). Virus stocks were generated from these 293 clones and used to infect primary B cells. Neither virus stock supported the initiation and maintenance of proliferation of the infected cells (Fig. 5B). Four copies of UR1 were inserted into the HMGA1a:EBNA1 derivative. This fusion, which supports the transcription of reporter plasmids, as does wild-type EBNA1 (Fig. 5A), was introduced into a maxi-EBV, 3442.1 (HMGA1a:UR14:EBNA1) (Fig. 5A). This recombinant was introduced into 293 cells, and virus stocks were generated and used to infected primary B cells. The introduction of four copies of UR1 into the otherwise transcriptionally defective derivative of EBNA1 restored the ability of the recombinant EBV to transform primary B cells (Fig. 5B). Thus we conclude that the UR1 domain of EBNA1 is essential for expressing EBV's transforming genes that allow infected B cells to maintain their proliferation (Fig. 6B).
Fig. 5.
UR1 rescues the null phenotype of an HMGA1a:EBNA1 mutant EBV. (A Upper) Wild-type EBNA1 and the fusion derivatives HMGA1a:EBNA1 and HMGA1a:UR14:EBNA1 are shown schematically. The numbers to the left identify the different EBV viruses: 2089 wild-type EBV or mutant viruses 3199/3453 (HMGA1a:EBNA1) and 3442 (HMGA1a:UR14:EBNA1). (A Lower) Also shown are measurements of luciferase activity induced by transfecting DG75 cells with vectors expressing the different alleles tested for EBNA1 that support the transcription of a reporter having EBNA1-binding sites (FR) in cis or lacking them. The introduction of four copies of UR1 into HMGA1a:EBNA1 restores transcriptional activity to HMGA1a:EBNA1. (B) Limiting dilution assays were used to measure the numbers of green Raji units (GRUs) of 2089 wild-type EBV and the two mutant EBVs 3199/3453 (HMGA1a:EBNA1) and 3442 (HMGA1a:UR14:EBNA1) required to induce clonal outgrowth of B cells. Approximately three GRUs were sufficient with 2089 wild-type EBV; no clonal outgrowth could be observed when the two isogenic 3199 or 3453 virus stocks were analyzed. The 3442 (HMGA1a:UR14:EBNA1) mutant virus was slightly impaired, as indicated by ≈100 GRUs being required per proliferating B cell.
Discussion
EBV is recognized for the efficiency with which it infects resting human B cells in vitro and induces and maintains their proliferation. These in vitro activities likely contribute to the malignant B cell proliferative diseases with which EBV is associated in vivo (17). How EBV can express its functions necessary to subvert the growth control of a resting B cell host and continue to express those functions required to maintain proliferation in a differentiated, proliferating blast has remained enigmatic. Our directed mutational study of EBNA1 has now identified it as being essential for ensuring the transcription of EBV's transforming genes in proliferating B cells.
EBNA1 binds to two clusters of sites in the EBV genome, which together constitute an origin of viral plasmid replication oriP. EBNA1's binding of this origin is needed to recruit the origin recognition complex, in order to initiate licensed DNA synthesis and maintain the viral plasmid in proliferating cells (1). Our experiments demonstrate that EBNA1 has an additional, transcriptional role in the EBV life cycle. Deletion of the 25 residues that make up a unique sequence (UR1) within linking region 1 of EBNA1 (Fig. 1A) compromises neither EBNA1's support of replication of small plasmids (Table 1) nor of intact EBV (Fig. 3A), but abrogates EBNA1's ability to support transcription from EBV's BamHI Cp (Fig. 4) and thus the ability of EBV to transform resting B cells into long-term proliferating blasts (Figs. 1B and 6). UR1 is necessary for this transcriptional function of EBNA1 (Fig. 5A). Studies with chimeric derivatives of EBNA1's DNA-binding domain and the cellular protein HMGA1a, plus or minus UR1, indicate that UR1 also is sufficient for this function when directed to sites in DNA bound by EBNA1 (Fig. 5B).
These findings provide a framework for describing the regulation of EBV's transforming genes from the initial infection through the establishment of long-term proliferation of its host B cell. EBV, upon infecting a resting primary B lymphocyte, encounters a transcriptional environment in which its BamHI Wp supports transcription. It is possible that the virus particle brings with it a protein that contributes to this environment, EBNA1 for example. The viral DNA template is regulated by this environment as well and must be bound by histones not present in the particle to establish the appropriate chromatin. Wp may function before formation of the chromatin that mediates transcription from Cp to yield the transcripts expressed initially (Fig. 4). These viral transcripts encode EBV's transforming proteins, including EBNA1 and EBNA2, or encode viral proteins (e.g., EBNA2) that drive expression of other viral transforming genes such as LMP1 and LMP2 (reviewed in ref. 18). EBNA2 can also promote expression from Cp (19). LMP1 and other viral transforming genes foster the transcription of a bevy of cellular genes, including c-myc, and additional transcriptional regulators that define the phenotypes of a proliferating B cell (20). Either EBNA1 when first synthesized binds FR to activate the BamH1 Cp, or other newly expressed cellular proteins bind UR1 of EBNA1 and support transcription at Cp either directly or indirectly by allowing expression of other factors that act at Cp. This change in usage of promoters is required because Wp becomes inactive shortly after infection, presumably because one or more factors expressed in undifferentiated, primary B cells needed for Wp to function are no longer expressed in the differentiated proliferating blast or because alterations in the chromatin state of the viral genome abrogate Wp activity.
The recognition that EBNA1 is required both for replication of the EBV genome and for expression of its transforming genes links EBV's DNA synthesis and transcription mechanistically and positionally. This one viral protein, whose cis-acting elements are clustered in 3% of the viral genome, contributes to these activities. EBV shares this mechanistic and positional linkage with human tumorigenic papillomaviruses, which also contain a cluster of cis-acting sites required for both viral DNA replication and transcriptional control (reviewed in ref. 21). Human tumorigenic papillomaviruses encode two proteins, E1 and E2, that mediate these activities, whereas EBNA1 combines these activities within itself. This combination makes EBNA1 a particularly desirable target for the development of antiviral, antitumor drugs.
Materials and Methods
Plasmids.
Recombinant plasmids, which carry the different EBNA1 alleles, were cloned by conventional techniques. The coding sequence of HMGA1a was amplified from a cDNA library and fused in phase to amino acid coordinate 379 of EBNA1 to construct the HGMA1a:EBNA1 chimera in the plasmid p2679.3. To design the HMGA1a:UR14:EBNA1 triple fusion, a 159-bp oligonucleotide fragment containing the UR1 domain of EBNA1, flanked by artificial linking domains [(SG4)2], was cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to yield the plasmid p3329.2. A tetramer of this UR1 fragment was cloned between the HMGA1a domain and the DNA-binding domain of EBNA1 in the plasmid p3437.14. All critical parts of the coding regions of the EBNA1 alleles were confirmed by sequencing, and DNA sequences of all plasmids are available upon request.
Construction of Viral Mutants.
The EBNA1 mutants were constructed based on the wild-type B95.8 EBV strain cloned into the F-factor plasmid p2089 (22). The different EBNA1 alleles were introduced with the chromosomal building strategy (23), as described in a recent review (24). The 2491 EBNA2− and the 2828 EBNA1− mutant EBVs are published in refs. 7 and 25. The genetic compositions of the modified EBV genomes were verified by restriction enzyme analysis, Southern blot hybridization, and partial DNA sequencing. Details for the generation of all EBV mutants are available upon request.
Preparation and Quantification of Infectious Viral Vector Stocks.
Cell lines were established after individual transfection of the maxi-EBV DNAs into 293 cells and subsequent selection with hygromycin, as described in ref. 26. To obtain virus stocks, the cell lines were transiently transfected with expression plasmids encoding BZLF1 (27) and BALF4 (6) to induce EBV's lytic cycle. Four days p.i., supernatants were harvested and filtered through 1.2-μm pore filters. The different EBV vector stocks were quantified by infection of Raji cells, as described in ref. 7. Briefly, 3 × 105 Raji cells were incubated at 37°C in 24-well cluster plates with different dilutions of the virus stocks to be analyzed. The absolute number of GFP+ cells was determined by UV microscopy in a defined fraction of the total Raji cell population 4 days p.i., to yield green Raji cell units (GRUs).
Isolation, Separation, and Infection of Primary B Lymphocytes.
Human primary mononuclear cells were isolated from adenoids, depleted of T cells by rosetting with sheep erythrocytes, and purified by Ficoll–Hypaque density-gradient centrifugation. The cell population was further depleted of monocytes by plastic adherence for 1 h. To quantify the efficiency of growth transformation with the different virus stocks, 3 × 105 human primary B lymphocytes per well were seeded in 96-well cluster plates on lethally irradiated WI38 human fibroblast feeder cells and infected, using serially diluted virus stocks, with 2089 wild-type EBV and the different viral mutants in a total volume of 100 μl, as described in ref. 7. Forty-eight wells were infected with each single virus dilution. A 50-μl portion of medium was exchanged every week for fresh medium, and the number of wells with proliferating cells was determined 6 weeks p.i. The infection experiments with different viral mutants were performed independently at least in triplicate.
Gardella Gel and Immunostaining.
Gardella gel hybridizations were done as described in ref. 25. Antibodies directed against LMP1 (CS1–4; DAKO, Carpenteria, CA); EBNA3A, C (a kind gift of Martin Allday, Imperial College London, London, U.K.); EBNA1, LMP2A, and EBNA2 (1H4, 14B6, and R3 + 1E6; a kind gift of Elisabeth Kremmer, GSF-Institute of Molecular Immunology, Munich, Germany); and β-actin (I-19, sc-1616; Santa Cruz Biotechnology, Santa Cruz, CA) were used for immunostaining of Western blot membranes, which were developed by using peroxidase-coupled secondary antibodies (Promega, Madison, WI) and ECL reagent (Amersham Pharmacia, Piscataway, NJ).
RT-PCR Analysis.
RNA was isolated and cDNA prepared as described in ref. 7. To monitor cellular DNA contamination of the RNA preparation, PCRs were performed with primers for the abundant cellular β-actin transcripts. PCRs for β-actin were as follows: initial template denaturation 4 min at 94°C, amplification for 25 cycles (1 min at 94°C, 1 min at 61°C, 1 min at 72°C), final elongation for 10 min at 72°C. PCRs for cDNA detection of transcripts originating from Cp and Wp were as follows: Cp, 5 min at 94°C, 30 cycles (1 min at 94°C, 1 min at 63°C, 1 min 72°C), 10 min at 72°C; Wp, 5 min at 94°C, 30 cycles (1 min at 94°C, 1 min at 65°C, 1 min at 72°C), 10 min at 72°C. The primer sequences for Wp-specific cDNA detection were 5′-GGAGTCCACACAAATGGG-3′ and −5′-ACCGGTGCCTTCTTAGGA-3′. Primer sequences for Cp-specific cDNA detection were 5′-TGTAGATCTGATGGCATAGAGAC-3′ and −5′-ACTGAAGCTTGACCGGTGCCTTCTTAGGAG-3′. β-Actin cDNA was amplified with the primers 5′-CACCCTGTGCTGCTCACCGAGGCC-3′ and 5′-ACCGCTCGTTGCCAATAGTGATGA-3′. GFP cDNA was amplified with the primers 5′-GCAGTGCTTCAGCCGCTACC-3′ and 5′-GCTTGTGCCCCAGGATGTTG-3′.
Proliferation Kinetics and Cell Cycle Analysis.
Primary B cells (4 × 105) were infected with 4 × 104 GRUs of the different virus stocks in 20 ml of culture medium to obtain a multiplicity of infection of 0.1. At the given time points, a defined fraction of the culture was harvested and further analyzed for the number of living GFP+ cells and their cell cycle status. Infection experiments were carried out with 2089 EBV, the EBNA2− deletion mutant 2491, and the different mutant virus stocks (see Figs. 2 and 6); for a negative control, the cells were left uninfected. To determine the fraction of apoptotic cells, Annexin-V staining was performed with the Annexin-V APC kit (BioVision, Mountain View, CA). Cells were harvested, spun, and stained with 5 μl of APC-coupled Annexin-V and 5 μl of propidium iodide (PI) at a final concentration of 10 μg/ml, and the GFP+ cells were characterized by three-parameter FACS analysis. As an internal volume standard for FACS analysis, calibration beads (BD CaliBRITE beads; Becton Dickinson, Palo Alto, CA) were added at a final concentration of 2 × 104 beads/ml to permit measurement of the absolute numbers of GFP+, AnnexinV−/PI− cells at a given time point p.i. To analyze the cell cycle status of the infected primary B cells vs. the uninfected controls, the samples were incubated with the thymidine analog BrdU for 2 h before FACS analysis at each time point, and the incorporation of BrdU was analyzed (BrdU flow kit; BD Biosciences Pharmingen, San Diego, CA). The cells were stained with an APC-coupled BrdU-specific antibody after fixation and permeabilization, and the cellular DNA was counterstained with the DNA-intercalating dye 7-AAD (Becton Dickinson), in accordance with the manufacturer's protocol. FACS analysis was performed until 3 × 104 cells were analyzed. The recorded data were gated for cells in the G1, S, and G2/M phases of the cell cycle and for cells with a subG1 DNA content. The total of all events was set to 100%.
Supplementary Material
Acknowledgments
We thank Sibille Humme and Aloys Schepers for their contributions. This work was supported by Deutsche Forschungsgemeinschaft Grants SFB455, Ha1354/3, and Ha1354/4 (to W.H.) and Public Health Service Grants CA70723 and CA22443 (to B.S.). B.S. is an American Cancer Society Research Professor.
Abbreviations
- EBNA
EBV nuclear antigen
- p.i.
postinfection
- Wp
W promoter
- Cp
C promoter
- GRU
green Raji unit.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hammerschmidt W, Sugden B. In: DNA Replication and Human Disease. DePamphilis ML, editor. Woodbury, New York: Cold Spring Harbor Lab Press; 2006. Chap 34. [Google Scholar]
- 2.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. J Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy G, Sugden B. Mol Cell Biol. 2003;23:6901–6908. doi: 10.1128/MCB.23.19.6901-6908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sample J, Henson EB, Sample C. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackey D, Sugden B. Mol Cell Biol. 1999;19:3349–3359. doi: 10.1128/mcb.19.5.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuhierl B, Feederle R, Hammerschmidt W, Delecluse HJ. Proc Natl Acad Sci USA. 2002;99:15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altmann M, Hammerschmidt W. PLoS Biology. 2005;3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woisetschlaeger M, Jin XW, Yandava CN, Furmanski LA, Strominger JL, Speck SH. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woisetschlaeger M, Strominger JL, Speck SH. Proc Natl Acad Sci USA. 1989;86:6498–6502. doi: 10.1073/pnas.86.17.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woisetschlaeger M, Yandava CN, Furmanski LA, Strominger JL, Speck SH. Proc Natl Acad Sci USA. 1990;87:1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby H, Rickinson A, Bell A. J Gen Virol. 2000;81:1057–1066. doi: 10.1099/0022-1317-81-4-1057. [DOI] [PubMed] [Google Scholar]
- 12.Sugden B, Warren N. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puglielli MT, Woisetschlaeger M, Speck SH. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson T, Zetterberg H, Wang YC, Rymo L. J Virol. 2001;75:5796–5811. doi: 10.1128/JVI.75.13.5796-5811.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borestrom C, Zetterberg H, Liff K, Rymo L. J Virol. 2003;77:821–829. doi: 10.1128/JVI.77.2.821-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung SC, Kang MS, Kieff E. Proc Natl Acad Sci USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammerschmidt W, Sugden B. Trends Mol Med. 2004;10:331–336. doi: 10.1016/j.molmed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Kieff E, Rickinson AB. In: Fields Virology. 4th Ed. Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2511–2573. [Google Scholar]
- 19.Yoo L, Speck SH. J Virol. 2000;74:11115–11120. doi: 10.1128/jvi.74.23.11115-11120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dirmeier U, Hoffmann R, Kilger E, Schultheiss U, Briseno C, Gires O, Kieser A, Eick D, Sugden B, Hammerschmidt W. Oncogene. 2005;24:1711–1717. doi: 10.1038/sj.onc.1208367. [DOI] [PubMed] [Google Scholar]
- 21.Howley PM, Lowy DR. In: Fields Virology. 4th ed. Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2197–2230. [Google Scholar]
- 22.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 24.Neuhierl B, Delecluse HJ. Methods Mol Biol. 2005;292:353–370. doi: 10.1385/1-59259-848-x:353. [DOI] [PubMed] [Google Scholar]
- 25.Humme S, Reisbach G, Feederle R, Delecluse HJ, Bousset K, Hammerschmidt W, Schepers A. Proc Natl Acad Sci USA. 2003;100:10989–10994. doi: 10.1073/pnas.1832776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dirmeier U, Neuhierl B, Kilger E, Reisbach G, Sandberg ML, Hammerschmidt W. Cancer Res. 2003;63:2982–2989. [PubMed] [Google Scholar]
- 27.Hammerschmidt W, Sugden B. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 28.Leight ER, Sugden B. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 29.Delecluse HJ, Schüller S, Hammerschmidt W. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardella T, Medveczky P, Sairenji T, Mulder C. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Lindner SE, Leight ER, Sugden B. Mol Cell Biol. 2006;26:1124–1134. doi: 10.1128/MCB.26.3.1124-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.