Abstract
Dengue virus, the causative agent of dengue fever and its more serious manifestation dengue hemorrhagic fever, is widespread throughout tropical and subtropical regions. The virus exists as four distinct serotypes, all of which have cocirculated in Bangkok for several decades with epidemic outbreaks occurring every 8–10 years. We analyze time-series data of monthly infection incidence, revealing a distinctive pattern with epidemics of serotypes 1, 2, and 3 occurring at approximately the same time and an isolated epidemic of serotype 4 occurring in the intervening years. Phylogenetic analysis of virus samples collected over the same period shows that clade replacement events are linked to the epidemic cycle and indicates that there is an interserotypic immune reaction. Using an epidemic model with stochastic seasonal forcing showing 8- to 10-year epidemic oscillations, we demonstrate that moderate cross-protective immunity gives rise to persistent out-of-phase oscillations similar to those observed in the data, but that strong or weak cross-protection or cross-enhancement only produces in-phase patterns. This behavior suggests that the epidemic pattern observed in Bangkok is the result of cross-protective immunity and may be significantly altered by changes in the interserotypic immune reaction.
Keywords: cross-immunity, mathematical model, phylogenetics, rsv, phase pattern
Understanding the factors that drive epidemiological patterns is central to the effective management of infectious disease. The recent explosion in pathogen gene sequence data provides a unique opportunity to link mechanisms at immunological and genetic levels with large-scale epidemiological patterns. In particular, the marriage of epidemiological theory with the analysis of phylogenetic data from pathogen populations has the potential to greatly advance our understanding of infectious disease dynamics at both the intra- and interhost levels (1). In this study, we employ such an integrated approach to study the epidemiology of dengue virus (DENV), one of the most important emerging pathogens of humans, and show that epidemic patterns in Bangkok may be the result of cross-protective immunity between serotypes.
DENV is a positive-sense RNA virus (genus Flavivirus) with a characteristic phylogenetic structure comprising four serotypes, DENV-1 to DENV-4. All four serotypes occur throughout tropical and subtropical regions, and an estimated 2.5 billion people are at risk from infection (2). The majority of infections lead to dengue fever, a mild febrile disease. However, the last 50 years has seen a growing number of cases of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), more severe and potentially fatal forms of the disease. A popular hypothesis is that DHF/DSS results from antibody-dependent enhancement, whereby antibodies from a previous infection facilitate subsequent infection with a heterogeneous serotype (3). However, the exact mechanisms of pathogenesis remain contentious, and the potential for enhancement also may depend on the particular strain type (genotype) within the serotype and immunological differences within the host (4–7). Theoretical studies of the potential impact of antibody-dependent enhancement on the epidemiology of dengue have shown that enhancement of transmission may generate cycles or chaotic oscillations in infection numbers (8), leading to an optimum degree of enhancement to prevent extinction (9), whereas enhancement of mortality may prevent immunologically similar serotypes from coexisting (10).
Our study focuses on Bangkok, where all four DENV serotypes have cocirculated since at least the 1950s (3), creating a reservoir from which dengue epidemics spread through the rest of Thailand (11). First, we reanalyze a 20-year time series of serotypically differentiated DENV incidence in Bangkok, revealing a distinctive epidemic pattern between the serotypes. We then present a phylogenetic analysis of samples collected over the same period. This analysis suggests that the extinction and replacement of whole clades within a serotype is coupled to the epidemic pattern and that there is an immunological reaction between serotypes. To study the implications of such an immune reaction further, we employ a mathematical model. By varying the interserotypic immune reaction, we demonstrate that moderate cross-protective immunity can account for the most striking pattern observed the data, but strong or weak cross-protective immunity and cross-enhancement do not. Together, the time series data, phylogenetic analysis, and mathematical modeling offer fresh insight into factors potentially governing the epidemiology and evolution of DENV and, more broadly, the role of immune cross-reaction in parasite strain dynamics.
Results
Incidence of Dengue in Bangkok.
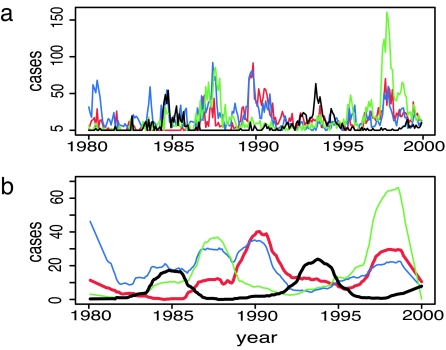
Transmission of DENV in Bangkok occurs throughout the year, although there are strong seasonal fluctuations in incidence related to vector population density (12). Records of the annual number of confirmed cases of dengue at the Queen Sirikit National Institute of Child Health in Bangkok between 1977 and 2000 show that large-scale epidemics have occurred every 3–5 years, with an epidemic of each serotype approximately every 8–10 years (13). Because severe disease is sometimes, but not always, associated with secondary infections, the number of hospitalized cases may depend in a complex way on the exact strains of each serotype in circulation and the sequence of multiple infections (14, 15). Therefore, infection numbers in the data may not be directly proportional to the actual number of infections with each serotype in the wider population but should nevertheless reflect the general pattern of incidence. To obtain a more detailed description of this pattern, we reanalyzed the finer-scale monthly records of these data. Removing the seasonal component and smoothing the time series (see Methods) reveals that the four DENV serotypes have an interesting pattern of changing prevalence, as shown in Fig. 1. Large outbreaks of serotypes 1, 2, and 3 are approximately coincident (in-phase), whereas outbreaks of serotype 4 tend to occur between the major epidemics of the other serotypes (out-of-phase). Cross-correlation analysis confirms this pattern. At lag 0, there is significant positive correlation between DENV-1, -2, and -3 and significant negative correlation between all of these serotypes and DENV-4. This out-of-phase pattern is particularly striking for DENV-1 and DENV-4, and we will focus on these two serotypes.
Fig. 1.
Monthly confirmed cases of each dengue serotype at Queen Sirikit National Institute of Child Health, Bangkok 1980 to 2000. (a) Incidence of each serotype after adjusting as if the serotype was isolated in 100% of cases every month. (b) Adjusted data after removing the seasonal effect and smoothing. Red, DENV-1; blue, DENV-2; green, DENV-3; black, DENV-4.
It seems unlikely that this highly distinctive epidemiological pattern is the result of chance or a transitory phenomenon. We therefore need to understand what processes lead to the phase shift in DENV-4 or, alternatively, the absence of a phase shift in the other serotypes. One possibility is interserotypic immune reaction. The actual degree of cross-reaction associated with a primary dengue infection is uncertain. There is clearly not complete protection against other serotypes as sequential (secondary) infections with different serotypes are observed. Antibody-dependent enhancement also may occur in some cases (3, 16, 17), although no consistent pattern of cause and effect has yet been established. Antibody titer concentration may determine whether an immune reaction is protective or enhancing (5). Experiments using human cell lines have found that, although cross-reactivity in T cells is common, cross-neutralization is more specific (4, 18). It is not clear that similar reactions occur in vivo, and it is difficult to quantify the actual degree of cross-reaction in the population at large from such experiments. However, in the following sections, we will consider phylogenetic evidence, suggesting that there is indeed some form of interserotypic immune reaction. We will then explore how this interaction may affect the epidemiology of DENV by including cross-reaction as a free parameter in a mathematical model.
Phylodynamics of Dengue in Bangkok.
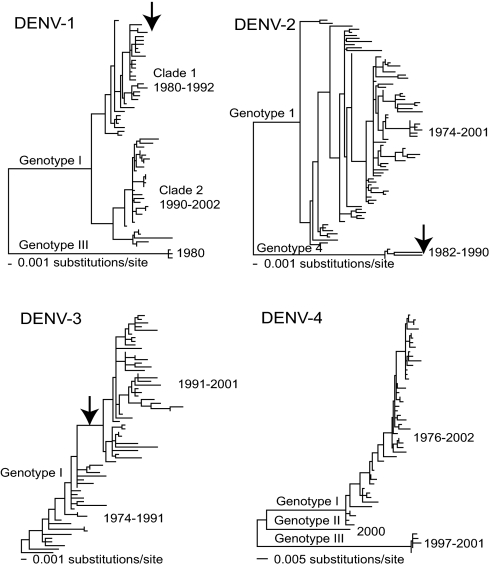
At the phylogenetic level, each viral serotype is subdivided into a number of distinct clades (such as genotypes) that have differing frequencies within populations and experience episodes of extinction and replacement (19–23). What causes these disjunct phylogenetic patterns is unclear, although the lack of definitive evidence for fitness differences among strains has led most workers to conclude that clade extinction results from random population bottlenecks and stochastic fadeout (20, 22, 24–26). We analyzed gene sequence data for all DENV serotypes circulating in Bangkok during the period 1973–2002 (see Methods) and found a concordance between episodes of clade extinction and changing levels of serotype abundance, with this process occurring on an ≈10-year time scale (Fig. 2).
Fig. 2.
Phylogenetic relationships of dengue virus in Bangkok, Thailand, sampled from 1973–2001. All four DENV serotypes are shown. Major genotypes and clades within genotypes are indicated, as are the range of sampling times within in each group. The phylogenetic events in DENV-1, DENV-2, and DENV-3 that coincide with the changing prevalence of DENV-4, particularly clade turnover, are indicated by arrows. All horizontal branch lengths are drawn to scale.
In DENV-1, genotype III viruses are sampled only in 1980 and never subsequently, with the majority of viruses assigned to genotype I. Within genotype I, a clade sampled from 1990–2002 coexists with, and eventually replaces, a clade sampled from 1980–1992. Strikingly, the extinction of the 1980–1992 clade and the rise of the 1990–2002 clade exactly coincides with a decline in prevalence of DENV-1 and an increase in prevalence of DENV-4 (27). All of these data come from the same population. If extinction simply correlated with the end of the epidemic cycle, then we would expect a loss of genetic diversity across the DENV-1 phylogeny as a whole. However, we observe that only specific clades suffer extinction, with those surviving experiencing no loss of diversity. This pattern strongly suggests that a selective process, such as immunological cross-reaction, rather than a general reduction in serotype prevalence is responsible for the clade extinction events. Further, the surviving 1990–2002 clade is more genetically distinct from DENV-4 than the 1980–1992 clade, which suffered extinction (27). Similarly, in DENV-2, viruses assigned to genotype IV are sampled only in Bangkok from 1982–1990, with all later viruses assigned to genotype I (28). Again, the extinction of genotype IV of DENV-2 coincides with the beginning of a decline in the frequency of this serotype and an increase in the prevalence of DENV-4. Although there are no such dramatic events in DENV-3, a phylogenetic shift occurred in 1991, corresponding to a long branch at the center of the tree, and again corresponding to a trough in the prevalence of DENV-3. Finally, three genotypes of DENV-4 were observed in the Bangkok data, all of which were present during 2000, with one first sampled in 1997 and another represented by only a single isolate. Although it is not yet possible to determine the precise mechanistic basis of these episodes of clade extinction and replacement, all involve amino acid changes in the E (envelope) gene, the major viral antigen, again supporting the view that they involve immunological cross-reaction between DENV-4 and the other serotypes.
Epidemic Model.
The epidemiological data with its distinctive phase pattern indicates that some kind of interaction is causing DENV-4 epidemics to be out of phase with those of the other serotypes, particularly DENV-1. The phylogenetic data further suggest that there is a clade-specific immunological reaction between DENV-4 and DENV-1. To ascertain whether, and how, these observations are connected, it is necessary to determine how interactions among viral serotypes cocirculating within the same population may be mediated by complex patterns of cross-reaction. In particular, we need to understand how different degrees of immune reaction between serotypes affect the phase pattern of epidemics. To study this effect, we used a mathematical model. This model is based on a standard susceptible–infected–recovered formulation (29) forced with an annually periodic transmission rate β′ = β(1 + δcos(2πt)), representing seasonal fluctuations in the vector population and extended to represent two pathogen serotypes interacting by immune cross-reaction (30, 31). This cross-reaction is encapsulated by a single parameter σ, which modifies the probability of secondary infection. Values of σ between 0 and 1 represent cross-protection with σ = 0 giving complete protection against secondary infection and σ = 1 no protection. Values of σ > 1 represent increasingly strong cross-enhancement. The model is parsimonious, containing just five parameters (see Methods for full details). Of these parameters, host life expectancy and the duration of infection are well established for dengue. The base transmission rate can be determined only indirectly from estimates of the basic reproductive number R0, whereas the degree of seasonal fluctuation is difficult to measure empirically. However, the main results that we will present hold when these parameters take a range of values. Thus, cross-reaction is the only parameter considered as free, and examining the behavior of the model as this is changed is our key objective.
Deterministic Model Behavior.
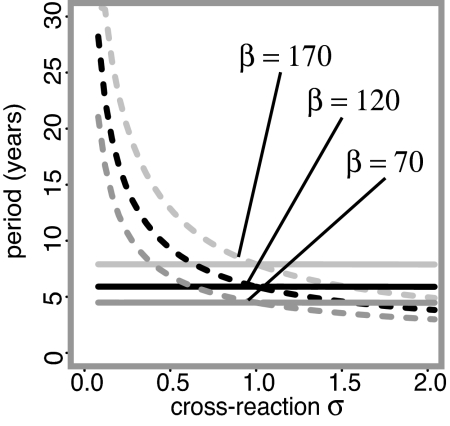
It has been shown that the seasonally forced susceptible–infected–recovered model may display a range of behavior depending on both the strength of the seasonality (δ in our model) and, in the two serotype model, the degree of cross-reaction between the serotypes (31, 32). In addition to solutions oscillating with the same frequency as the forcing (in our case annually), regular lower frequency solutions may occur as the result of subharmonic resonance (33–35). An idea of the resonance periods likely to be observed can be constructed by calculating the “natural” or “intrinsic” period (36, 37) from the eigenvalues of the linearized system (31). For the two-serotype system, there are two natural periods. One is independent of the degree of cross-reaction σ and corresponds to the in-phase mode. The other is strongly dependent on the degree of cross-reaction and corresponds to the out-of-phase mode. Fig. 3 shows how the two natural periods for our system vary with the base transmission rate β. This guide is not definitive to the resonance states we might expect because the actual dynamics result from a complex interaction of the two natural periods. Nevertheless, it shows that as β increases, the period of in-phase oscillations increases and the degree of cross-immunity at which out-of-phase period 10 oscillations occur becomes weaker and weaker, eventually switching to cross-enhancement. Based on the data, we are interested in solutions with a relatively long epidemic period, and to excite resonance solutions with a period many times higher than the forcing period, the amplitude of the seasonal oscillation δ must be large. However, such large fluctuations also tend to have a destabilizing effect on the system, leading to irregular solutions wildly oscillating with no discernible pattern. As a result of this instability, solving the system for a large set of randomly generated initial conditions failed to reveal any resonance solutions with periods comparable with those observed in the data. A deterministic seasonally forced resonating susceptible–infected–recovered model cannot therefore explain the distinctive pattern of dengue epidemics.
Fig. 3.
Natural periods, depending on the degree of cross-reaction σ, calculated from eigenvalues of the unforced, linearized deterministic system. Solid lines correspond to solutions with an in-phase pattern; dashed lines to solutions with an out-of-phase pattern. Three values of β are shown: β = 170 (pale gray), β = 120 (black), and β = 70 (dark gray). Other parameter values are γ = 52 and μ = 1/60.
Stochastic Model.
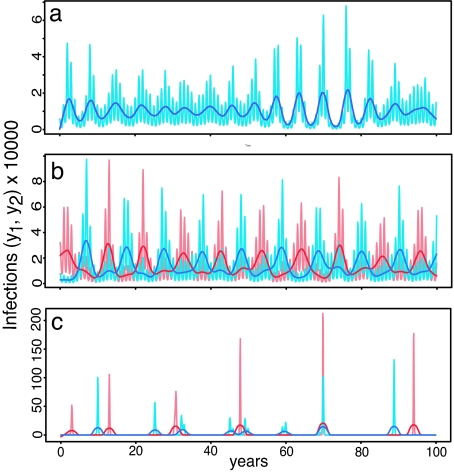
Oscillations at the natural period of the system, however, may persist if random perturbations repeatedly disrupt convergence to a lower period equilibrium state. Because the vector population, among other epidemiological parameters, is likely to be sensitive to variations in environmental conditions, stochasticity was introduced to the model by random variation in the magnitude of the transmission rate fluctuations (δ). Numerical solutions of this stochastic version of the model for different degrees of cross-reaction produce three distinct patterns. In general, for strong or weak cross-immunity, the two serotypes are always perfectly in-phase as shown in Fig. 4a. The period is approximately as predicted by the in-phase mode eigenvalue described above. For intermediate cross-immunity, there is generally an out-of-phase pattern as shown in Fig. 4b, with the period of each serotype approximately as predicted by the out-of-phase mode eigenvalue for σ ≈ 0.5. Because of the stochasticity in the system, some switching may occur between this state and the in-phase state, particularly at the edges of the parameter region for which it occurs. For cross-enhancement, some solutions show the two serotypes perfectly in-phase, but most solutions have a quite different structure as shown in Fig. 4c. This solution is characterized by very sharp epidemic spikes interspersed by long intervals of extremely low incidence and no discernable epidemic period or phase structure. This behavior is likely to be related to the complex periodicity and chaotic oscillations that enhancement has been shown to generate in the unforced deterministic system (8, 9) but exacerbated by the inherent oscillations introduced by the sinusoidal transmission rate. Repeated switching between this state and the in-phase state was not observed. Of these three solutions states, only the smooth, relatively regular out-of-phase state shown in Fig. 4b resembles the pattern observed in the data. This result indicates that cross-immunity can account for the phase structure observed between DENV-1 and DENV-4, but only if it is of moderate intensity.
Fig. 4.
Sample solutions for the infected proportion of the population (y1, y2) generated from the stochastic model for different degrees of cross-reaction. Pale lines are model output; darker lines are the same data after removing seasonal components and smoothing. Red, serotype 1; blue, serotype 2. (a) σ = 0.1, strong cross-immunity, in-phase with approximate period 6. (b) σ = 0.4, intermediate cross-immunity, out-of-phase with approximate period 10. (c) σ = 1.6, cross-enhancement, very large, irregular epidemics with no clear pattern. Numerical solution was by fourth-order Runge–Kutta method. Initially, one serotype was introduced to a naïve population. After 500 years, the second serotype was introduced at a random time in a 50-year interval. The system then was solved for an additional 4,500 years to ensure a quasi-equilibrium state. Parameter values were δ = 0.1, β = 120, γ = 52, and μ = 1/60.
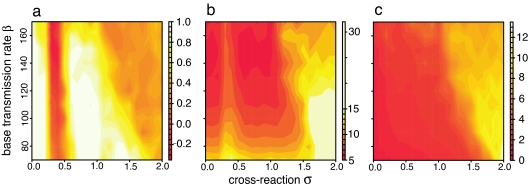
The occurrence of these three different patterns is robust to the value of the base transmission rate β. Twenty separate runs of the model were made at each point of a parameter space with σ ranging from 0 to 2 and β from 70 to 170. These β values correspond to basic reproductive numbers R0 of 1.3 to 3.3, which are at the lower end of empirical estimates for dengue (from 1.3 to 7.7; refs. 38 and 39), but higher values lead to rather short natural periods. Fig. 5a shows the mean value of the cross-correlation function at lag 0 for all of the runs. At σ ≈ 0.5, there is a well defined band of negative cross-correlation indicative of the out-of-phase pattern. For other values of σ < 1, cross-correlation is high, indicating that most solutions are in-phase. When σ > 1, intermediate cross-correlation suggests either a mixture of patterns or no pattern at all. Fig. 5b shows the mean period of the solutions. For the in-phase region, the period is approximately as predicted by the in-phase mode eigenvalue. For the out-of-phase region, the period is always longer and approximately corresponds to the value predicted by the out-of-phase mode eigenvalue for σ = 0.5. In the mixed region, the period is very high, suggesting that the majority of solutions are irregular. Fig. 5c shows the mean epidemic size. This component shows little variation except in the region of mixed patterns where it is very large, again indicating that the majority of these solutions are irregular.
Fig. 5.
Indicators of the distribution of solution patterns as σ and β vary. (a) Average cross-correlation at lag 0 between time series for the number of infections with each serotype (y1, y2). (b) Average epidemic period for serotype 1. Calculated by dividing the length of the series in years by the total number of epidemic peaks in that time. (c) Average epidemic size for serotype 1, calculated by dividing the sum of the epidemic peaks by the total number of peaks. All diagrams are based on 100 years of model output, after removing seasonal effects and smoothing so that epidemics are clearly distinguishable, with 20 separate model runs for each value of β and σ. Other numerical details as in Fig. 4 legend. Parameter values were δ = 0.1, γ = 52, and μ = 1/60.
Values for the magnitude of seasonal variation in transmission between 0.4 and 1.2 also produce similar results to the base value of 0.1 used throughout this paper. As δ becomes smaller, the no-pattern state begins at higher values of σ, and the out-of-phase state spans a narrower range of σ values. For δ < 0.4, only the in-phase solution occurs. As δ becomes larger, the no-pattern state begins at lower values of σ and, for δ > 1.2, the large oscillations are so destabilizing that only the no-pattern state occurs. In general, the type of solutions that occur depend in a complex way on β and δ. However, the relationship between the solution pattern and cross-immunity described here is relatively robust and occurs in a similar way across a range of values for the transmission parameters.
Discussion
We have presented a phylogenetic analysis revealing that DENV-4 has an immunological cross-reaction with DENV-1 and, possibly, other serotypes, with the resultant selection pressure leading to clade extinctions and replacements. We also have examined the possible influence of such an immune interaction by using a parsimonious mathematical model with seasonal forcing and environmental stochasticity. This model demonstrated that the immune interaction must be moderately cross-protective to produce the distinctive out-of-phase pattern observed in the epidemiological data. Although it has been suggested that there is either weak cross-protection or cross-enhancement in dengue, in our model, neither of these lead to the characteristic out-of-phase pattern that we observe between DENV-1 and -4. That DENV-1, -2, and -3 are approximately in-phase suggests that the immune reaction between them may be weakly cross-protective or enhancing. In theory, strong cross-protection also would cause these three serotypes to be in-phase, although significant secondary infection rates indicate that it is unlikely. However, the time series is relatively short, and only the out-of-phase pattern between DENV-1 and DENV-4 is clear. As more data become available, this situation may change, and it will be possible to examine the epidemic patterns of the other serotypes in more detail. A further implication of the model is that cross-reaction influences the epidemic period by adjusting the phase structure. For instance, when the out-of-phase solution generates epidemics of each serotype approximately every 10 years, an epidemic of one strain or the other occurs every 5 years. However, the associated in-phase solution would generate coincident epidemics of both strains every 6 years. At present, more than three-quarters of infections in Bangkok are caused by serotypes 1, 2, and 3, with epidemics every 10 years. If DENV-4 is indeed responsible for this epidemic pattern, then changes in its immune interaction with the other serotypes could result in reversion to an in-phase pattern with epidemics of all four serotypes occurring more frequently. Such changes could arise, for instance, if medical intervention modifies the immune structure of the host population or the selection pressure noted from the phylogenetic analysis leads to sufficient accumulation of antigenic differences.
Throughout the modeling work, we have assumed that the serotypes are identical because there is no consistent data on epidemiological differences between DENV serotypes. We also have modified our original model to include such differences but found no evidence that they could account for the observed patterns in the absence of immune cross-reactions. There are many other possible extensions to be considered in future work, particularly if new data become available to inform the development and parameterization of a more detailed model. Based on the observation that antibody-dependent enhancement may depend on antibody titer, it would be interesting to investigate a model in which the cross-reaction changes with time from enhancement to weakening protection. A model with more than two interacting serotypes also is likely to be interesting, although the difficulty of interpretation and the absence of a clear method for determining the immune response to a tertiary challenge may be significant problems.
The phylogenetic analysis presented here suggests that there is an interserotypic immune reaction and immune selection for strains or clades within each serotype. Our model suggests that this immune reaction could be responsible for the epidemic pattern, although we cannot determine whether the epidemic pattern also is responsible for immune selection. The basic reproductive number of a serotype is increased by weaker cross-immunity, or stronger cross-enhancement, with a different serotype. Simple numerical experiments with our model confirmed that introducing a mutant strain of one of the serotypes with a preferential immune response always leads to the extinction of the other strain. Because changes in the immune composition of the host population also are governed by the epidemic cycle, it seems likely that mutant strains arising at certain points of the cycle are more likely to be subject to immune selection than those arising at other points. Experiments with our model did not reveal any correlation between the time to extinction and the phase structure of the epidemic cycle or the point of the cycle at which the mutant strain was introduced. However, to test the link between the epidemiological dynamics and the phylogeny in detail will require a large-scale individual-based model such as that previously used to construct model phylogenies for influenza (40).
Even if the epidemic cycle does not directly influence immune selection, our analysis reveals that the interserotypic immune reaction and epidemic cycle are closely interwoven. If the interserotypic cross-immunity we propose can be confirmed, both in Bangkok and more generally, it would have important implications for management of dengue. Furthermore, cross-immunity also may be influencing the epidemic phase structure of other diseases. Respiratory syncitial virus occurs as two main subtypes, A and B, and has seasonal oscillations in infection incidence. Data from Finland show a two-year epidemic cycle composed of out-of-phase four-year epidemic cycles of each subtype (41). The theory presented here suggests that these two subtypes are interacting via moderate strength cross-immunity. As more data become available, similar patterns may be revealed for other pathogens, and it may be possible to connect radical changes in their epidemiology to changes in the intensity of immune reactions or the composition of serotypes.
Methods
Time-Series Analysis.
The data were monthly records of patients diagnosed with dengue in the Queen Sirikit National Institute of Child Health, Bangkok, previously published in annual form (13). As given, these data are biased because each month, the serotype was determined for between 0% and 90% of cases and, because of improved technology, there has been a steady increase in the number of isolations since 1990. To account for this bias, the data were scaled to represent 100% sampling each month. The years 1977–1979 were omitted because there were too many months with incomplete records. Values for the few other months in which the serotype was identified for zero cases were assigned by linear interpolation. A LOESS smoother was then applied to remove the seasonality and smooth the data. The cross-correlation functions for each serotype pair of the smoothed data were then calculated by using R.
Evolutionary Analysis.
We collected E (envelope) gene sequence data for all virus serotypes circulating in Bangkok, Thailand, during the period 1973–2002 (27, 28, 42). Each sequence was taken from a patient admitted to Queen Sirikit National Institute of Child Health. This resulted in data sets of the following size: DENV-1 (61 sequences, 1,485 bp), DENV-2 (79 sequences, 1,485 bp), DENV-3 (66 sequences, 1,479 bp), DENV-4 (53 sequences, 1,485 bp). Sequence alignment was undertaken manually. Maximum likelihood phylogenetic trees were then estimated for all three data sets. In all cases, we used the most general GTR+I+Γ4 model of nucleotide substitution available in the PAUP* package (43) and undertook successive rounds of branch swapping.
Mathematical Model.
Fundamentally, the two serotype susceptible–infected–recovered model is composed of nine compartments covering all possible infection states of an individual in the host population. However, some compartments can be combined to reduce the system to five compartments dividing the population into those susceptible to primary infection by either serotype (x), susceptible to secondary infection by each serotype (z1, z2) and infected by each serotype (y1, y2) (31). This simplification results in some of the compartments overlapping (see supporting information, which is published on the PNAS web site, for more details). The two serotypes interact through a cross-reaction parameter (0 ≤ σ ≤ 2) acting on susceptibility. After being infected by one serotype, the probability of being infected by the other serotype is decreased by cross-immunity (0 < σ < 1 with σ = 0 being complete protection and σ = 1 being no protection) or increased by cross-enhancement (σ > 1 with higher values of σ causing greater enhancement). The transmission rate β′(t) varies annually according to a cosine function β′(t) = β(1 + δcos(2π t)), where β is the base transmission rate and δ specifies the magnitude of seasonal variation. For the deterministic model, δ is fixed. For the stochastic model, it is randomly varied by up to ±25% every 6 months by using uniformly distributed random numbers. The average duration of infection/infectiousness is given by γ. The population is assumed to be well mixed and have a constant size with the birth and death rates both equal to μ.
Thus, the system is described by the five ordinary differential equations:
Supplementary Material
Acknowledgments
Financial support for this work was provided by The Wellcome Trust, Japan Society for the Promotion of Science, and the Military Infectious Diseases Research Program of the U.S. Department of Defense.
Abbreviation
- DENV
dengue virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Grenfell BT, Pybus OG, Gog JR, Wood JLN, Daly JM, Mumford JA, Holmes EC. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 2.WHO. State of the Art of New Vaccines: Research and Development Initiative for Vaccine Research. Geneva: WHO; 2003. [Google Scholar]
- 3.Halstead SB. In: Dengue and Dengue Hemorrhagic Fever. Gubler DJ, Kuno G, editors. New York: CABI; 1997. pp. 23–44. [Google Scholar]
- 4.Rothman AL. J Clin Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada A, Kawaoka Y. Rev Med Virol. 2003;12:387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- 6.Lobigs M, Pavy M, Hall R. Vaccine. 2003;21:1572–1579. doi: 10.1016/s0264-410x(02)00743-0. [DOI] [PubMed] [Google Scholar]
- 7.Mady BJ, Erbe DV, Kurane I, Fanger MW, Ennis FA. J Immunol. 1991;147:3139–3144. [PubMed] [Google Scholar]
- 8.Ferguson NM, Anderson RM, Gupta S. Proc Natl Acad Sci USA. 1999;96:790–194. doi: 10.1073/pnas.96.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings DAT, Schwartz IB, Billings L, Shaw B, Burke DS. Proc Natl Acad Sci USA. 2005;102:15259–15264. doi: 10.1073/pnas.0507320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi I, Sasaki A, Boots M. Proc R Soc London Ser B; 2003. pp. 2241–2247. [Google Scholar]
- 11.Cummings DAT, Irizarry RA, Huang NE, Endy TP, Nisalak A, Ungchusak K, Burke DS. Nature. 2003;427:344–347. doi: 10.1038/nature02225. [DOI] [PubMed] [Google Scholar]
- 12.Chareonsook O, Foy HM, Teeraratkul A, Silarug N. Epidemiol Infect. 1999;122:161–166. doi: 10.1017/s0950268898001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nisalak A, Endy TP, Nimmannitya S, Kalayanarooj S, Thisayakorn U, Scott RM, Burke DS, Hoke CH, Innis BL, Vaughn DW. Am J Trop Med Hyg. 2003;68:191–202. [PubMed] [Google Scholar]
- 14.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatansen S, Salitul V, Phanthumachinda B, Halstead SB. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 15.Endy TP, Chunsuttiwat S, Nisalak A, Libraty DH, Green S, Rothman AL, Vaughn DW, Ennis FA. Am J Epidemiol. 2002;156:32–39. doi: 10.1093/aje/kwf006. [DOI] [PubMed] [Google Scholar]
- 16.Kliks SC, Nimmanitya S, Nisalak A, Burke DS. Am J Trop Med Hyg. 1988;38:411–419. doi: 10.4269/ajtmh.1988.38.411. [DOI] [PubMed] [Google Scholar]
- 17.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayokorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 18.Roehrig JT. In: Dengue and Dengue Hemorrhagic Fever. Gubler DJ, Kuno G, editors. New York: CABI; 1997. pp. 199–220. [Google Scholar]
- 19.Twiddy SS, Farrar JJ, Chau NV, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Virology. 2002;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- 20.Holmes EC, Twiddy SS. Infect Genet Evol. 2003;3:19–28. doi: 10.1016/s1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 21.Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Mol Biol Evol. 2003;20:1650–1658. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- 22.Foster JE, Bennett SN, Carrington CVF, Vaughan H, McMillan WO. Virology. 2004;324:48–59. doi: 10.1016/j.virol.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Thu HM, Lowry K, Jian L, Hlaing T, Holmes EC, Aaskov J. Virology. 2005;336:163–172. doi: 10.1016/j.virol.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 24.A-Nuegoonpipat A, Berlioz-Arthuad A, Chow V, Endy T, Lowry K Mai le Q, Ninh TU, Pyke A, Reid M, Reynes JM, et al. Virology. 2004;329:505–512. doi: 10.1016/j.virol.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Sittisombut N, Sistayanarain A, Cardosa MJ, Salminen M, Damrongdachakul S, Kalayanarooj S, Rojanasuphot S, Supawadee J, Maneekarn M. Am J Trop Hyg Med. 1997;57:100–108. doi: 10.4269/ajtmh.1997.57.100. [DOI] [PubMed] [Google Scholar]
- 26.Thu HM, Lowery K Myint TT, Shwe TN, Han AM, Khin KK, Thant KZ, Aaskov J. Emerg Infect Dis. 2004;10:593–597. doi: 10.3201/eid1004.030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Mammen MP, Jr, Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, Nimmannitya S, Kalayandarooj S, Holmes EC. J Virol. 2005;79:15123–15130. doi: 10.1128/JVI.79.24.15123-15130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Mammen MP, Jr, Chinnawirotpisan P, Klungthong C, Rodpradit P, Nisalak A, Nimmannitya S, Kalayanarooj S, Vaughn DW, Holmes EC. J Gen Virol. 87:873–883. doi: 10.1099/vir.0.81486-0. [DOI] [PubMed] [Google Scholar]
- 29.Anderson RM, May RM. Infectious Diseases of Humans. Dynamics and Control. Oxford: Oxford Univ Press; 1991. pp. 122–143. [Google Scholar]
- 30.Keeling MJ, Rohani P, Grenfell BT. Physica D. 2001;148:317–335. [Google Scholar]
- 31.Kamo M, Sasaki A. Physica D. 2002;165:228–241. [Google Scholar]
- 32.Greenman J, Kamo M, Boots M. Physica D. 2004;190:136–151. [Google Scholar]
- 33.Jordan DW, Smith P. Nonlinear Ordinary Differential Equations. Oxford: Oxford Univ Press; 1999. [Google Scholar]
- 34.Aron JL, Schwartz IB. J Theor Biol. 1984;110:665–679. doi: 10.1016/s0022-5193(84)80150-2. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz IB, Smith HL. J Math Biol. 1983;18:233–253. doi: 10.1007/BF00276090. [DOI] [PubMed] [Google Scholar]
- 36.Nisbet RM, Gurney WSC. Modelling Fluctuating Populations. New York: Wiley; 1982. pp. 48–51. [Google Scholar]
- 37.Dushoff J, Plotkin JB, Levin SA, Earn DJD. Proc Natl Acad Sci USA. 2005;101:16915–16916. doi: 10.1073/pnas.0407293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuno G. In: Dengue and Dengue Hemorrhagic Fever. Gubler DJ, Kuno G, editors. New York: CABI; 1997. pp. 61–88. [Google Scholar]
- 39.Ferguson NM, Donnelly CA, Anderson RM. Philos Trans R Soc London B; 1999. pp. 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferguson NM, Galvani AP, Bush RM. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 41.Waris M. J Infect Dis. 1991;163:464–469. doi: 10.1093/infdis/163.3.464. [DOI] [PubMed] [Google Scholar]
- 42.Klungthong S, Zhang C, Mammen MP, Jr, Ubol S, Holmes EC. Virology. 2004;329:168–179. doi: 10.1016/j.virol.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 2003. Ver 4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.