Abstract
The cobalt- and iron-containing corrinoid iron–sulfur protein (CoFeSP) is functional in the acetyl-CoA (Ljungdahl–Wood) pathway of autotrophic carbon fixation in various bacteria and archaea, where it is essential for the biosynthesis of acetyl-CoA. CoFeSP acts in two methylation reactions: the transfer of a methyl group from methyltransferase (MeTr)-bound methyltetrahydrofolate to the cob(I)amide of CoFeSP and the transfer of the methyl group of methyl-cob(III)amide to the reduced Ni-Ni-[4Fe-4S] active site cluster A of acetyl-CoA synthase (ACS). We have solved the crystal structure of as-isolated CoFeSPCh from the CO-oxidizing hydrogenogenic bacterium Carboxydothermus hydrogenoformans at 1.9-Å resolution. The heterodimeric protein consists of two tightly interacting subunits with pseudo-twofold symmetry. The large CfsA subunit comprises three domains, of which the N-terminal domain binds the [4Fe-4S] cluster, the middle domain is a (βα)8-barrel, and the C-terminal domain shows an open fold and binds Coβ-aqua-(5,6-dimethylbenzimidazolylcobamide) in a “base-off” state without a protein ligand at the cobalt ion. The small CfsB subunit also displays a (βα)8-barrel fold and interacts with the upper side of the corrin macrocycle. Structure-based alignments show that both (βα)8-barrel domains are related to the MeTr in the acetyl-CoA pathway and to the folate domain of methionine synthase. We suggest that the C-terminal domain of the large subunit is the mobile element that allows the necessary interaction of CoFeSPCh with the active site of ACSCh and the methyltetrahydrofolate carrying MeTr. The conformation in the crystal structure shields the two open coordinations of cobalt and likely represents a resting state.
Keywords: acety-CoA pathway, Carboxydothermus hydrogenoformans, methyltransferase, cobalt
B12-dependent enzymes are widespread in nature and catalyze distinct reactions. They can be grouped in three major classes: the isomerases, reductive dehalogenases, and methyltransferases (MeTrs; ref. 1). The cobalamin-dependent MeTrs play important roles in the amino acid biosynthesis in many organisms, ranging from bacteria to humans, as well as in the one-carbon metabolism of bacteria and archaea. MeTrs typically catalyze the transfer of methyl groups between organic and inorganic donors and acceptors like S-adenosylmethionine, methyltetrahydrofolate, and cobalamins. A unique methyltransfer reaction is found within the acetyl-CoA (Ljungdahl–Wood) pathway of autotrophic carbon assimilation, which operates in the anaerobic acetogenic, sulfidogenic, methanogenic, and hydrogenogenic bacteria and archaea. In the final step of the pathway, acetyl-CoA is synthesized from a methyl cation, carbon monoxide (CO), and CoA, a reaction catalyzed by the Ni- and Fe-containing enzyme acetyl-CoA synthase (ACS; for a review, see ref. 2). Key to this reaction (Eq. 1) is the action of the Co- and Fe-containing corrinoid iron–sulfur protein (CoFeSP) that donates the methyl group from methyl-cob(III)amide to one of the two Ni ions in the active site cluster A of ACS (3). This is the only methyltransfer reaction known with metals as donors and acceptors of the methyl group:
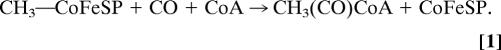 |
CoFeSP has been isolated and characterized from the acetogenic bacterium Moorella thermoacetica (4), the methanogenic archaeon Methanosarcina thermophila (5), and the hydrogenogenic bacterium Carboxydothermus hydrogenoformans (6).
C. hydrogenoformans can use CO as a sole source of energy and carbon under anaerobic chemolithoautotrophic conditions. Protons serve as terminal electron acceptor (7), and the methyl branch of the acetyl-CoA pathway is used for the assimilation of carbon (6). The oxidation of CO in this bacterium is catalyzed by two monofunctional NiFe-containing CODHs (CODHICh and CODHIICh) that harbor the [Ni-4Fe-5S] active site cluster C (7). A monomeric ACSCh, a 1:1 molar complex of ACSCh and CODHIIICh, and CoFeSPCh are expressed in functional form during the growth of C. hydrogenoformans using CO as the only substrate (6). The CoFeSPCh from C. hydrogenoformans is a heterodimeric protein composed of a large subunit (48.4 kDa, CfsA) and a small subunit (33.9 kDa, CfsB) and harbors one molecule of the cobalt-containing corrinoid cofactor and a single [4Fe-4S] cluster. The function of CoFeSPCh in C. hydrogenoformans is the transfer of a methyl group to the Ni-Ni-[4Fe-4S] active site cluster A of ACSCh or the ACSCh/CODHIIICh complex, where the synthesis of acetyl-CoA takes place (6).
The CoFeSPs from acetogens and methanogens are heterodimeric proteins sharing sequence identities of 35–53% with CoFeSPCh from C. hydrogenoformans. CoFeSPMt from M. thermoacetica was shown to contain two cofactors, 5-methoxybenzimidazolylcobamide (a variant of B12) and a [4Fe-4S]2+/1+ cluster, which gave the protein its name. For the cobamide cofactor, it was shown that neither the 5-methoxybenzimidazolyl group nor a nitrogen atom from a protein side chain is coordinating the cobalt (8). The low-potential [4Fe-4S]2+/1+ cluster is involved in the reductive reactivation of CoFeSPMt and is able to transfer electrons to cob(II)amide from NiFe-containing CODH, pyruvate:ferredoxin oxidoreductase or reduced ferredoxins (9). The physiological methyl donor for CoFeSPMt is the N5 atom of (6S)-methyltetrahydrofolate (CH3-H4folate). Methyltransfer to the cob(I)amide center of CoFeSPMt is facilitated by the MeTr (4), a homodimeric enzyme with two identical 28-kDa subunits with (βα)8-barrel fold (10).
Cobalamin-dependent methyl carriers like CoFeSP and methionine synthase share the difficult task of allowing specific and reversible interactions between methyl donors/acceptors and cobalamin. Both involve methyl group transfers between different modular domains in the case of methionine synthase and individual proteins in the case of CoFeSP, and most probably both systems use SN2 mechanisms (11). Biophysical characterizations of methionine synthase showed that the enzyme exists as an ensemble of conformations with equilibria that depend on the oxidation and ligation state of the cobalamin and the concentrations of methyl donors and acceptors (12).
Here, we report the 3D structure of the Co- and Fe-containing CoFeSPCh from C. hydrogenoformans as determined by x-ray crystallography to a resolution of 1.9 Å. Based on the structure of CoFeSPCh, we propose a model describing the function of this protein in the methyltransfer cycle from MeTr to ACS.
Results and Discussion
Description of the Overall Structure.
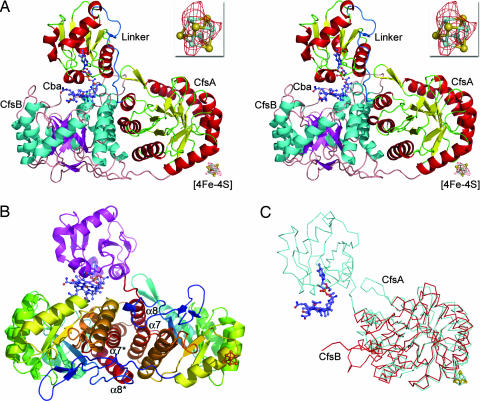
The crystal structure of CoFeSPCh from C. hydrogenoformans in the as-isolated oxidized state has been solved by multiple isomorphous replacement with anomalous scattering (MIRAS) phasing at a resolution of 1.9 Å. The asymmetric unit of the crystal contains the heterodimeric CoFeSPCh comprising the small (CfsB) and large (CfsA) subunits (Fig. 1A). The final model used for refinement contained residues 59–441 (complete 1–442) of CfsA, residues 3–309 (complete 1–309) of CfsB, Coβ-aqua-(5,6-dimethylbenzimidazolylcobamide), one [4Fe-4S] cluster, 16 iodide ions, and 564 water molecules. CfsB contains a single domain that folds into a (βα)8-barrel (Fig. 1 A and B). CfsA is composed of three domains. Its N-terminal domain (residues 1–58) contains the binding motif for the [4Fe-4S] cluster (motif Cys17X2Cys20X4Cys25X16Cys42, consensus CysX2CysX4CysX16Cys) and is disordered in the protein crystal. The middle domain (residues 59–316) is the dominating structural element of CfsA and displays a (βα)8-barrel. It is connected by a proline-rich linker of 18 residues to the C-terminal domain (334–441), which shows a Rossmann-like fold with a parallel four-stranded β-sheet with order 2134 that is flanked on both sides by two α-helices (Fig. 1A). An additional α-helix (cap-helix) is positioned approximately perpendicular to the β-sheet below the C-terminal end of the sheet (Figs. 1A and 2A).
Fig. 1.
Overall structure of CoFeSPCh from C. hydrogenoformans. (A) Stereo ribbon presentation of the CoFeSPCh structure. CfsA is shown with α-helices in red and β-sheets in yellow, and CfsB is shown with α-helices in cyan and β-sheets in magenta. Cba denotes the cobamide cofactor. The proline-rich linker connecting the middle and C-terminal domains of CfsA is blue and labeled as linker. Cofactors (corrinoid cofactor and [4Fe-4S] center) are presented as a ball-and-stick model (Co in wheat, C in light blue, O in red, N in dark blue, Fe in aquamarine, and S in dark yellow). (Inset) An anomalous difference Fourier map (red) around the [4Fe-4S] cluster, contoured at 3.0σ, is shown with the overall structure and magnified. (B) Ribbon presentation of CfsA and CfsB with the (βα)8-barrel domains in rainbow colors from the N termini in blue to the C termini in red. The C-terminal domain of CfsA is shown in magenta. Helices forming the four-helical bundle like arrangement in the subunit interface are labeled; a star denotes helices of CfsB. (C) Superimposition of CfsB (red) on CfsA (aquamarine). All pictures were prepared by using PyMol (26).
Fig. 2.
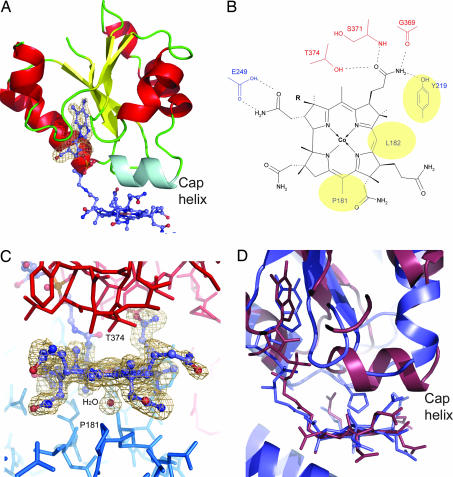
The corrinoid cofactor. (A) Ribbon representation of the C-terminal domain of CfsA with bound corrinoid cofactor. The cap-helix is aquamarine; otherwise, colors are as in Fig. 1A. An omit Fobs−Fcalc map for the ribose and dimethylbenzimidazol part of the cofactor is shown in light brown. (B) Schematic presentation of the corrin macrocycle and its interaction with the protein environment. The interacting residues are colored red if they belong to CfsA and blue if they are part of CfsB. Yellow ellipses mark residues with hydrophobic contacts to the β-side of the corrin macrocycle. (C) Stick presentation of amino acids near the corrinoid cofactor. Coloring of the cofactor as in Fig. 1A. An omit map excluding all atoms of the corrin macrocycle and the water ligand from the calculation of the phase set used for the Fobs−Fcalc synthesis is shown in blue; the contour level is 3σ. The ligand bound to the cobalt-β position is modeled as a water molecule shown as an orange sphere. (D) Superimposition of the cobalamin-binding domain of MetH (blue) and the C-terminal domain of CfsA (dark red).
Both subunits are closely associated by a side-to-side arrangement of the two (βα)8-barrels forming a four-helical bundle-like structure with a helix-to-helix angle of ≈45° at the interface (Fig. 1B), and the two barrels are rotated against each other by ≈130°. Dimerization covers ≈2,100 Å2 from each of the subunit surfaces and is mainly stabilized by hydrophobic interactions (74% based on the protein surface hydrophobicity; ref. 13). The two (βα)8-barrels share a very similar topology, including the insertion of a two-stranded antiparallel β-sheet covering the N-terminal opening of the barrel (Fig. 1C). A superimposition of the two (βα)8-barrel domains of CfsA and CfsB gives a distance matrix alignment (DALI) Z score of 29 and an rmsd value of 1.9 Å, the highest Z score and lowest rmsd value observed with the CoFeSP structure. A structure-based alignment shows that the barrel domains share a sequence identity of 21% and a similarity of 39% (Fig. 4, which is published as supporting information on the PNAS web site). A common origin for the two (βα)8-barrels is also in agreement with the observed pseudo-twofold symmetry of the heterodimeric structure (Fig. 1B), probably a relic of an ancestral homodimeric protein that evolved by gene duplication and by decoration of the gene coding for CfsA.
The Cofactors.
The [4Fe-4S] cluster was localized by using anomalous difference Fourier maps calculated from a data set collected at a wavelength of 1.734 Å (Fig. 1A and Table 1) and also shows a weak signal in 2Fobs−Fcalc maps. The [4Fe-4S] cluster refines with an approximately five times higher average B factor than the protein model (Table 2), which agrees with a large static disorder of the N-terminal domain in the crystal and its insufficient contribution to the measured reflection intensities. The [4Fe-4S] cluster is ≈8 Å away from the (βα)8-barrel domain of CfsA. The first amino acid of CfsA visible in the electron density (Pro-59) has a distance of 17 Å to the [4Fe-4S] cluster. No hydrophobic patch at the surface of the barrel domain of CfsA is observed near Pro-59 or the [4Fe-4S] cluster. In the crystal structure, the corrinoid cofactor is placed far away from the [4Fe-4S] cluster with a distance of ≈52 Å between the nearest Fe and Co, making a direct electron transfer between the two cofactors in the observed conformation unlikely.
Table 1.
Phasing statistics
| Wave length, Å | Space group | Unique/observed Reflections | Rs§ overall/last shell | Resolution overall/last shell, Å | Completeness, % overall/last shell | (I)/(σI) overall/last shell | Phasing power centric/acentric-Iso/-Ano | RCullis¶ acentric-Iso/-Ano | |
|---|---|---|---|---|---|---|---|---|---|
| Dano | 1.7340 | C2 | 39,116/73,264 | 0.056/0.184 | 20–2.54/2.60–2.54 | 98.4/92.1 | 9.5/4.0 | ||
| Native | 1.5418 | C2 | 75,235/185,280 | 0.098/0.236 | 20–2.04/2.10–2.04 | 99.0/90.9 | 8.6/3.1 | ||
| Thiom* | 1.5418 | C2 | 44,832/201,268 | 0.085/0.262 | 20–2.40/2.50–2.40 | 95.0/77.2 | 14.4/5.3 | 0.47/0.50/0.18 | 0.84/0.70 |
| Iodide, 0.3 M† | 1.5418 | C2 | 46,502/81,035 | 0.162/0.346 | 20–2.30/2.40–2.30 | 86.7/84.5 | 4.8/2.3 | 1.11/0.82/0.24 | 0.58/0.98 |
| Iodide, 1.0 M‡ | 1.5418 | C2 | 94,299/293,993 | 0.148/0.360 | 20–1.90/2.0–1.90 | 99.7/99.8 | 6.9/3.3 | 1.08/0.87/0.38 | 0.62/0.93 |
In the statistic, Friedel mates were treated as independent reflections.
*Heavy atom derivative: thiomersal (sodium 2-(ethylmercuriothio)benzoate) with four sites per asymmetric unit (a.u.).
†Heavy atom derivative: potassium iodide (300 mM) with one site per a.u.
‡Heavy atom derivative: potassium iodide (1 M) with seven sites per a.u.
§Rs = Σh Σi Ii(h) − 〈I(h)〉/Σh ΣiIi(h), where i are the independent observations of reflection h.
¶RCullis = Σh‖(FPH (h) − FP(h) − FHcalc(h))/Σh FPH(h) − FP(h).
Table 2.
Statistics on diffraction data and structure refinement
| Data set | CoFeSP |
|---|---|
| Total/unique reflections | 293,995/48,659 |
| Rs* | 0.149 (0.360) |
| Resolution, Å | 20–1.90 (2.0–1.9) |
| Completeness, % | 99.7 (99.8) |
| (I)/(σI) | 6.9 (3.3) |
| Model R/Rfree-factor, %† | 20.7/26.6 |
| rmsd from ideal geometry | |
| Bonds, Å | 0.017 |
| Angles, ° | 1.9 |
| Cruickshank DPI for coordinate error | 0.218 |
| Average temperature factors. Å2 | |
| CfsA (middle domain/C-terminal domain/[4Fe-4S]/corrinoid) | 13.2/17.6/72.9/14.6 |
| CfsB | 14.2 |
| Water | 18.9 |
| Ramachandran statistics, % | |
| Most favored/additional/generously allowed/disallowed regions | 86.0/13.8/0.3/0.0 |
In the refinement statistics, Friedel mates were merged.
*Rs = Σh Σi Ii(h) − 〈I(h)〉/ΣhΣiIi(h), where i are the independent observations of reflection h.
†The Rfree factor was calculated from 5% of the data, which were removed at random before the refinement was carried out.
It was shown for CoFeSPMt from M. thermoacetica that the isolated cobamide contains a 5-methoxybenzimidazolyl group (8, 14). The electron density for the bound cofactor in C. hydrogenoformans CoFeSPCh shows the presence of two short groups at positions 5 and 6, indicating the presence of a dimethylbenzimidazolyl group (Fig. 2A). The corrinoid cofactor is interacting with CfsB and the C-terminal domain of CfsA. The dimethylbenzimidazyolyl chain penetrates into the core of the C-terminal domain resulting in the “base-off” form of the cofactor. Its phosphate group is 3–4 Å away from the N-terminal end of the cap-helix, thus being well placed to interact with the helix-dipole. The corrin macrocycle is sandwiched between the C-terminal domain of CfsA and CfsB (Fig. 2 B and C). Because there is no direct interaction between these two structural elements, their relative orientation toward each other is indirectly established by their interaction with the corrin macrocycle. Neither dimethylbenzimidazol nor a protein side chain is bound to the cobalt ion in the structure. Based on a comparison of redox potentials between proteins with (5-methoxybenzimidazolyl)cobamide, free corrinoids, and CoFeSPMt from M. thermoacetica, it was suggested that the uncoordinated cob(I)amide is stabilized by approximately +100 mV compared with the “base-on” or “His-on” states (15). This “base-off/His-off” conformation increases the population of the cob(I)amide state and, in cases of unwanted oxidations of the cofactor, the more facile reduction of the cofactor. The corrin ring shows low ring flexure with a fold angle around the Co-C10 axis of 6.9° calculated from the two planes defined by the positions of C10, N22 and N21 and C10, N23, and N24. The lower face of the corrin ring interacts with the cap-helix of CfsA, which is in van der Waals distance. The side chain nearest to the cobalt ion is that of Thr-374, which is part of the cap-helix and has its hydroxyl group ≈3.6 Å away from the cobalt ion (Fig. 2C). The upper face of the macrocycle points to the βα-loops of CfsB, but the interaction is looser than at the lower face and allows the binding of a water molecule at the cobalt-β position (Co-O distance of 2.5 Å; Fig. 2C). The structure supports the assignment of the cofactor as Coβ-aqua-(5,6-dimethylbenzimidazolylcobamide), with the oxidation state of the cobalt ion being either +2 or +3. Atoms of the corrin macrocycle are positioned to make contacts with protein main- and side-chain atoms, whereas the ring circumference is solvent exposed at three of its four sides (Fig. 2 A and C). The presence of an axial water ligand bound to cobalt in both the +2 and +3 oxidation state is in agreement with recent spectroscopic data of CoFeSPMt (16).
Comparison of the CoFeSPCh Structure with That of Other Proteins.
To identify structures with high similarity to the subunits of CoFeSPCh, a DALI search (17) against the Protein Data Bank was carried out. Because of the high abundance of (βα)8-barrel-containing proteins, many structures were selected. The highest hit with both subunits of CoFeSPCh (DALI Z-score of 23) was observed with residues 354–649 of cobalamin-dependent methionine synthase (18). For a superimposition of 230 Cα-atoms of CfsB and methionine synthase, an rmsd value of 2.5 Å was obtained. Additionally, a structure-based alignment gives a sequence identity of 14% for the structurally corresponding parts (Fig. 4). Methionine synthase has a multimodular architecture and residues 354–649 constitute the methyltetrahydrofolate (CH3-H4folate)-binding domain, called the fol-domain (18), which is activating CH3-H4folate to donate its methyl-group to cob(I)amide bound to another module (residues 650–897; ref. 19). Of the six amino acids that bind CH3-H4folate in the complex structure of the fol-domain (18), all are conserved in MeTr, but only one (Asp-218 in MetH) is conserved in CfsA and CfsB and, correspondingly, binding of CH3-H4folate to CoFeSP has not been reported (Fig. 4). In addition to the obvious structural homology between the (βα)8-barrel structures, the C-terminal domain of CfsA and residues 650–897 of methionine synthase share a similar fold to bind the cobamide cofactor with an elongated dimethylbenzimidazolyl tail (Fig. 2D) and reveal an rmsd value of 3.4 Å for a superimposition of the core structure. However, the interaction between protein and corrin macrocycle in methionine synthase is due to βα-loops that allow the direct coordination of the cobalt ion by a histidine side chain (19), whereas in the case of CoFeSPCh, the cap-helix covers the lower side of the macrocycle, and a protein ligand to cobalt is not observed (Fig. 2D).
The second best hit, with a Z-score of 20, is observed for the MeTr from M. thermoacetica (10) that binds CH3-H4folate and is required for the methyltransfer reaction to the cob(I)amide of CoFeSPMt. CfsB and the MeTr show an rmsd value of 2.4 Å for 214 Cα-atoms and a sequence identity of 9% (Fig. 4). Both structural and sequence homologies indicate that the (i) methionine synthase and the MeTrs of the acetyl-CoA pathway are related, and (ii) proteins required for methyltransfer in the acetyl-CoA pathway itself are evolutionarily related. Furthermore, the link between the multimodular cobalamin-dependent methionine synthase and CoFeSPCh makes it likely that the more complex methionine synthase evolved from the methyltransfer system of the acetyl-CoA pathway.
Methyltransfer to and from CoFeSPCh.
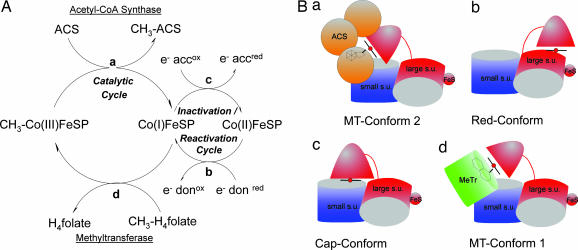
To gain further insight into the function of CoFeSPCh, the principal requirements of a complete catalytic cycle have to be analyzed (Fig. 3A). To complete a catalytic cycle, one conformation must allow methyltransfer from the MeTr to cob(I)amide (MT-Conform-1), and a second conformation is needed for methyltransfer from methyl-cob(III)amide to cluster A of ACS (MT-Conform-2). An additional conformation can help to avoid unspecific methylations by the methyl-cob(III)amide or reactions of the supernucleophile Co(I) (Cap-Conform). If cob(II)amide is generated through an oxidative side reaction, an additional conformation (Red-Conform) would be needed to allow reductive reactivation by electron transfer from the reduced [4Fe-4S] cluster to regenerate cob(I)amide.
Fig. 3.
Methyltransfer cycle of CoFeSPCh. (A) The methyltransfer and the inactivation/reactivation cycle of CoFeSPCh. (a–d) The different conformations and interactions of the protein during the catalytic cycle; “e donox” and “e donred” are used for the electron donor of the [4Fe-4S] cluster of CoFeSPCh in the oxidized and reduced state, and “e accox” and “e accred” are used for the unspecific electron acceptor of Cob(I) in the oxidized and reduced state. During the catalytic cycle, the protein cycles between CH3-Cob(III) and Cob(I). Methyltransfer reactions are from (d) CH3-H4folate to Cob(I) and (a) CH3-Cob(III) to the cluster A of ACSCh. (B) Hypothetical model for CoFeSPCh methyltransfer cycle. The model assumes the C-terminal domain of CfsA as the only mobile element and draws from the analogy to cobalamin-dependent methionine synthase. (a–d) The different states as used in A. For details, see Results and Discussion.
The likely position to add the methyl group to the cobalt ion is the cobalt-β position, which is positioned in the interface between CfsA and CfsB and is occupied by a water ligand in the structure. For CoFeSPMt, it was recently shown that a water ligand can also be found in the methylated state of the corrinoid cofactor (16), indicating structural changes that may allow a water molecule to bind to the cobalt-α position. It is evident from the crystal structure that any methyl group transfer to or from the cobamide cofactor depends on a conformation different from the one observed, because the cobalt-α and -β positions are shielded by the protein matrix. To allow for conformational changes, the protein structure would require flexible elements. The major structural elements in CoFeSPCh are the two (βα)8-barrel domains of CfsA and CfsB that show an extensive subunit interface, making rearrangements between these two elements unlikely. However, the structural unit that carries the cobamide cofactor as the reversible methyl acceptor/donor is the C-terminal domain of CfsA, which is only loosely connected to the rest of the protein. Because the C-terminal domain is connected by a 40-Å-long linker with the central (βα)8-barrel domain, it is free to adopt different conformations, which may include a 180° rotation or a translation by >30 Å. Its weak interactions with the rest of the protein make it likely that we have an ensemble of different conformations in solution, and that the conformation that was observed in the crystal structure is one of several different possible states. Changes in the oxidation state or the coordination of the cobalt ion can modulate the conformation of the corrin macrocycle, e.g., by changing the fold angle. Because the crucial interactions between the C-terminal domain and CfsB are mediated by the corrin macrocycle, changes at the cobalt ion could influence the populations in the ensemble of convertible conformational states of the protein, similar to what has been observed for cobalamin-dependent methionine synthase (12).
In the CoFeSPCh structure (Fig. 2C), the corrin ring is shielded on both faces by the protein matrix. The solved structure corresponds to CoFeSPCh in the as-isolated state, which is oxidized and contains either Co2+ or Co3+. The weak reducing conditions applied for crystallization (1 mM 2-mercaptoethanol) are not sufficient to reduce the [4Fe-4S] cluster or the cobalt ion to produce cob(I)amide. However, the observed conformation (Cap-Conform), with CfsB covering the upper face of cobamide, may also serve to protect cob(I)amide or the methylcob(III)amide forms from unspecific side reactions.
The Red-Conform must bring the two metal centers of CoFeSPCh close together to allow for an electron transfer between the reduced [4Fe-4S] cluster and cob(II)amide. Typically, a distance between the two cofactors of 8–14 Å would be anticipated (20). The cartoon of Fig. 3B postulates a specific complex in which the C-terminal cobamide domain of CfsA interacts with the central and N-terminal domains to bring together the [4Fe-4S] and cob(II)amide moieties.
MT-Conform-1 needs to enable the close interaction between cob(I)amide bound to CoFeSPCh and CH3-H4folate bound to MeTr. The structure of the fol-domain of methionine synthase shows CH3-H4folate bound near the C-terminal end of the central β-strands (18), and a similar position has been proposed for the binding of CH3-H4folate to MeTr (10). To correctly position the corrin-ring on top of CH3-H4folate, an arrangement between the C-terminal domain of CfsA and MeTr can be anticipated that resembles the complex between the C-terminal domain and CfsB (Fig. 3B).
The interaction between CoFeSPCh and ACSCh is probably the most intriguing, because it enables a methyltransfer between two metal sites (cobamide cofactor and Ni-Ni-[4Fe-4S] cluster A). Again, the flexibility of the C-terminal domain of CfsA is necessary to allow a close interaction between the active site of ACSCh and the methylated cob(III)amide cofactor in MT-Conform-2. A distance of ≈3–4 Å would be expected to allow a SN2-like methyltransfer between the donor and acceptor metal sites. Attempts to manually fit the C-terminal domain near cluster A of ACSCh failed to bring both metal sites closer than 8 Å because of clashes between the protein backbones. To reach the required short distance for methyltransfer, conformational changes at both proteins are likely to occur.
Materials and Methods
Crystallization and Data Collection.
Lithoautotrophic growth of C. hydrogenoformans on CO and purification of CoFeSPCh were carried out as detailed (7). Crystallization and soaking experiments were performed in an anaerobic glove box (Coy Laboratory Products, Grass Lake, MI) in an atmosphere of 95% nitrogen and 5% hydrogen and at a constant temperature of 15°C. The setup used hanging drops in the vapor diffusion method. The conditions used a reservoir solution with 20% PEG 3350/100 mM BaCl2/1 mM 2-mercaptoethanol/100 mM Bis·Tris (pH 7.0), and a protein-to-reservoir solution ratio of 1:1 was used for the crystallization drops. Crystals appeared after ≈3 weeks and belonged to space group C2 with approximate cell dimensions of 160×44×101 Å3, β = 118°. Heavy metal derivatization was conducted by soaking crystals for 20 min in crystallization solution containing 10 mM sodium 2-(ethylmercuriothio)benzoate. Soaks with potassium iodide were carried out by stepwise increasing the iodide concentration to either 300 mM or 1 M in the crystallization drop. Crystals were transferred to a cryo buffer (1:9 mixture of 2R,3R-butandiol and reservoir solution) and were frozen directly in the cold nitrogen stream, and diffraction data were collected at −180°C on a rotating copper anode x-ray generator (Nonius FR591, Bruker AXS, Karlsruhe, Germany) equipped with an image plate detector (mar345dtb, MAR-Research, Hamburg, Germany). Data sets at wavelengths of 1.734 and 1.7416 Å were collected at the European Synchrotron Radiation Facility (ESRF), beamline ID 14-2, to maximize the anomalous scattering contribution of iron.
Structure Solution and Refinement.
Isomorphous data were collected at −180°C. The initial heavy atom positions were located by using SHELXD, and MIRAS phasing was carried out by using SHARP (ref. 21; see Table 1 for details). Phases were modified by solvent flattening with SOLOMON (22). Model building and refinement were done with MAIN 2000 (23) and positional and temperature refinement in CNS (24). The final refinement statistics and stereochemical analyses using PROCHECK (25) are shown in Table 2. Omit maps were used to validate the position of cofactors and residues near the corrin macrocycle.
Supplementary Material
Acknowledgments
H.D., V.S., and O.M. acknowledge the Deutsche Forschungsgemeinschaft for funding [Grants DO 785/1-1/2 (to H.D.) and S.V. 10/1-1 (to V.S. and O.M.)]. H.D. acknowledges the Verband der Chemischen Industrie for funding.
Abbreviations
- ACS
acetyl-CoA synthase
- MeTr
methyltransferase.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2H9A).
References
- 1.Banerjee R, Ragsdale SW. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 2.Ragsdale SW, Kumar M. Chem Rev. 1996;96:2515–2539. doi: 10.1021/cr950058+. [DOI] [PubMed] [Google Scholar]
- 3.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 4.Hu SI, Pezacka E, Wood HG. J Biol Chem. 1984;259:8892–8897. [PubMed] [Google Scholar]
- 5.Maupin-Furlow J, Ferry JG. J Bacteriol. 1996;178:340–346. doi: 10.1128/jb.178.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svetlitchnyi V, Dobbek H, Meyer-Klaucke W, Meins T, Thiele B, Romer P, Huber R, Meyer O. Proc Natl Acad Sci USA. 2004;101:446–451. doi: 10.1073/pnas.0304262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svetlitchnyi V, Peschel C, Acker G, Meyer O. J Bacteriol. 2001;183:5134–5144. doi: 10.1128/JB.183.17.5134-5144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragsdale SW, Lindahl PA, Munck E. J Biol Chem. 1987;262:14289–14297. [PubMed] [Google Scholar]
- 9.Menon S, Ragsdale SW. Biochemistry. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- 10.Doukov T, Seravalli J, Stezowski JJ, Ragsdale SW. Structure Fold Des. 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig ML, Matthews RG. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 12.Bandarian V, Ludwig ML, Matthews RG. Proc Natl Acad Sci USA. 2003;100:8156–8163. doi: 10.1073/pnas.1133218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraczkiewicz R, Braun W. J Comp Chem. 1998;19:319–333. [Google Scholar]
- 14.Ljungdahl L, Irion E, Wood HG. Biochemistry. 1965;4:2771–2780. doi: 10.1021/bi00888a030. [DOI] [PubMed] [Google Scholar]
- 15.Harder SR, Lu WP, Feinberg BA, Ragsdale SW. Biochemistry. 1989;28:9080–9087. doi: 10.1021/bi00449a019. [DOI] [PubMed] [Google Scholar]
- 16.Stich TA, Seravalli J, Venkateshrao S, Spiro TG, Ragsdale SW, Brunold TC. J Am Chem Soc. 2006;128:5010–5020. doi: 10.1021/ja054690o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 18.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Proc Natl Acad Sci USA. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 20.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 21.La Fortelle E. d., Irwin JJ, Bricogne G. Cryst Comp. 1997;7:1–9. [Google Scholar]
- 22.Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. Collaborative Computational Project No. 4. [DOI] [PubMed] [Google Scholar]
- 23.Turk D. PhD thesis. München, Germany: Technische Universität München; 1992. [Google Scholar]
- 24.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.DeLano WL. San Carlos, CA: DeLano Scientific; 2002. PyMol. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.