Abstract
Combinatorial biosynthesis of type I polyketide synthases is a promising approach for the generation of new structural derivatives of polyketide-containing natural products. A target of this approach has been to change the extender units incorporated into a polyketide backbone to alter the structure and activity of the natural product. One limitation to these efforts is that only four extender units were known: malonyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA, and methoxymalonyl-acyl carrier protein (ACP). The chemical attributes of these extender units are quite similar, with the exception of the potential hydrogen bonding interactions by the oxygen of the methoxy moiety. Furthermore, the incorporated extender units are not easily modified by using simple chemical approaches when combinatorial biosynthesis is coupled to semisynthetic chemistry. We recently proposed the existence of two additional extender units, hydroxymalonyl-ACP and aminomalonyl-ACP, involved in the biosynthesis of zwittermicin A. These extender units offer unique possibilities for combinatorial biosynthesis and semisynthetic chemistry because of the introduction of free hydroxyl and amino moieties into a polyketide structure. Here, we present the biochemical and mass spectral evidence for the formation of these extender units. This evidence shows the formation of ACP-linked extender units for polyketide synthesis. Interestingly, aminomalonyl-ACP formation involves enzymology typically found in nonribosomal peptide synthesis.
Keywords: antibiotics, combinatorial biosynthesis
Natural products biosynthesized by type I polyketide synthases (PKSs) have an array of biological activities such as antimicrobial, anticancer, and immunosuppressant activities. Many of these products play important roles in the treatment of human, animal, and plant diseases. The diverse activities of these compounds are due to the extensive structural diversity available via type I PKS enzymology. Depending on the system, many variations are possible, including different starter units, different numbers and types of extender units, variations in the oxidation states and stereochemistry of the starter and extender units, and release of the final products in a linear or cyclic form (1). Although type I PKSs show extensive amino acid sequence similarity in their core enzymatic domains, thousands of structural outcomes are possible because of the inherent flexibility of these enzyme systems.
The flexibility of type I PKSs has been exploited for the generation of “unnatural” natural products through combinatorial biosynthesis. One example is to replace a catalytic domain from one type I PKS with an alternative domain from a different type I PKS, resulting in a hybrid enzyme that generates a hybrid product. This approach was shown to generate a library of nearly 60 “unnatural” erythromycin derivatives by exchanging catalytic domains from the erythromycin type I PKS with catalytic domains from other type I PKSs (2). Thus, combinatorial biosynthesis complements the more traditional approaches of using total or semisynthetic chemistry to generate structural diversity.
There is great interest in changing the extender unit(s) incorporated into a polyketide to vary the moiety that extends away from the backbone of the polyketide, which can have effects on its interaction with its biological target. Changes available by using this approach are somewhat limited because, until recently, there were only four proposed type I PKS extender units: malonyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA, and methoxymalonyl (MM)-acyl carrier protein (ACP) (1). These extender units result in the incorporation of acetyl, propionyl, butyryl, or methoxyacetyl moieties into the polyketide backbone, respectively. The latter offers hydrogen-bonding potential of the oxygen from the methoxy moiety. However, for all of these extender units, the moieties on the Cα lack simple chemical reactivity for further downstream modification by semisynthetic chemistry. Because of these limitations, there is an interest in identifying or generating new extender units with different chemical attributes to enhance structural diversification by combinatorial biosynthesis and increase the opportunities for downstream modification by semisynthetic chemistry.
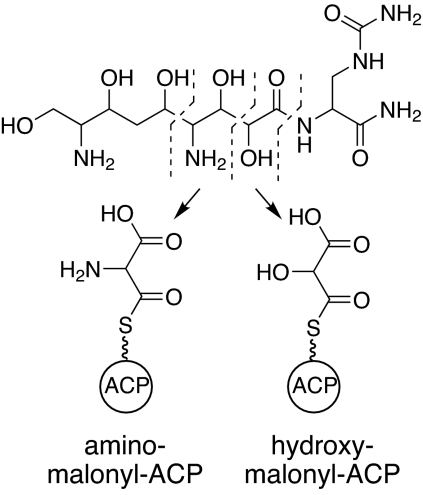
We recently proposed the existence of two previously undescribed type I PKSs extender units involved in the biosynthesis of the antibiotic zwittermicin A (ZMA) from Bacillus cereus (Fig. 1; ref. 3). These extender units, hydroxymalonyl (HM)-ACP and aminomalonyl (AM)-ACP, were proposed to form the glycolyl and ethanolamine units, respectively, found in ZMA (Fig. 1). The existence of these extender units not only provides insight into ZMA biosynthesis but also holds enormous potential for generating new structural derivatives of natural products through combinatorial biosynthesis. Their existence increases the number of extender units available for combinatorial biosynthesis by 50 percent and introduces previously undescribed functionality to a polyketide in the form of free hydroxyl or amino groups. Here, we present biochemical and mass spectral evidence for the formation of HM-ACP and AM-ACP, an important first step toward harnessing this biosynthetic potential to develop new bioactive molecules.
Fig. 1.
Chemical structures of ZMA (Upper) and the proposed ACP-linked type I PKS extender units (Lower). Dashed lines in ZMA structure delineate the ethanolamine and glycolyl moieties. These moieties are derived from aminomalonyl-ACP and hydroxymalonyl-ACP, respectively.
Results and Discussion
Bioinformatics Analysis and Protein Purification.
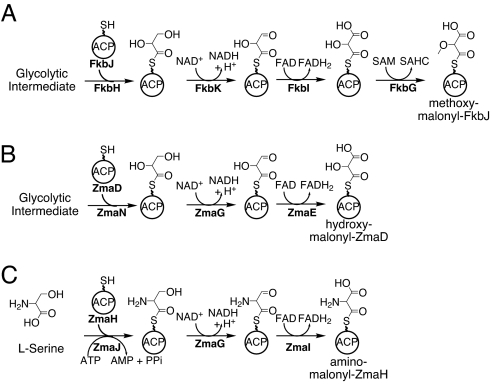
A set of five proteins is predicted to be involved in the formation of MM-ACP (Fig. 2A), an extender unit needed for the biosynthesis of FK520 (4). This proposal is based on bioinformatics analysis of the FK520 biosynthetic gene cluster and biosynthetic gene clusters for other methoxyacetyl-containing natural products (5–8). The proposed MM-ACP pathway is as follows (using FK520 protein nomenclature) (Fig. 2A): (i) FkbH binds to a glycolytic intermediate and dephosphorylates it while tethering it to an ACP, FkbJ, forming glyceryl-ACP; (ii) FkbK catalyzes the oxidation of the glyceryl-ACP to 2-hydroxy-3-oxopropionyl-ACP (or 2,3,3-trihydroxypropionyl-ACP; ref. 9); (iii) FkbI converts the FkbK product to HM-ACP; and (iv) FkbG catalyzes the O-methylation to form MM-ACP. Floss et al. (7) subsequently showed that during ansamitosin biosynthesis, O-methylation occurs before incorporation by the PKS; therefore, MM-ACP is the in vivo extender unit, not HM-ACP. Whereas genetic experiments support the hypothesis that these enzymes are involved in the incorporation of a MM-ACP extender unit (7, 10, 11) and a crystal structure of FkbI has been determined (9), biochemical evidence for MM-ACP has yet to be provided.
Fig. 2.
Proposed pathways for ACP-linked PKS extender units. (A) MM-ACP formation during FK520 biosynthesis. (B) HM-ACP formation during ZMA biosynthesis. (C) AM-ACP formation during ZMA biosynthesis. The squiggle denotes the 4′ Ppant prosthetic groups of the ACPs. For correlating previous Orf names (3) to those shown above: ZmaD, Orf3; ZmaE, Orf1; ZmaG, Orf4; ZmaH, Orf5; ZmaI, Orf6; ZmaJ, Orf7.
We analyzed a part of the ZMA biosynthetic pathway and identified five genes coding for homologs of the MM-ACP pathway from FK520 (3). Sequencing of the ZMA biosynthetic gene cluster has been completed, and we have identified a gene, zmaN, coding for a homolog of FkbH. Thus, the ZMA biosynthetic gene cluster codes for one homolog of both FkbH and FkbK and two homologs of FkbJ and FkbI. An FkbG homolog is not coded within the biosynthetic gene cluster (data not shown). The presence of two homologs of the ACP that is proposed to carry the methoxymalonyl moiety for the MM-ACP extender unit suggested the existence of two ACP-linked type I PKS extender units for ZMA production.
One of these extender units, HM-ACP, would be generated in a similar manner to that proposed for MM-ACP, but the lack of an FkbG homolog results in the formation of HM-ACP (Fig. 2B). The second pathway was proposed to involve an enzyme showing sequence similarity to adenylation domains of nonribosomal peptide synthetases (NRPSs) (12); this enzyme, ZmaJ, contains an amino acid specificity code for the activation of l-serine (l-Ser) (3). We proposed that ZmaJ tethers l-Ser to ZmaH, and the seryl-ACP is then oxidized to AM-ACP (Fig. 2C). There is only one homolog of FkbK, the enzyme that catalyzes the first oxidation of glyceryl-ACP, suggesting ZmaG catalyzes the first oxidation step in both pathways (Fig. 2 B and C).
To test our hypotheses, we took a biochemical and mass spectral approach to investigate whether HM-ACP and AM-ACP formation could be reconstituted in vitro. For this analysis, each protein was overproduced in Escherichia coli as an N-terminally histidine-tagged protein and purified to near homogeneity by using nickel-chelate chromatography (Fig. 7, which is published as supporting information on the PNAS web site). Using these purified proteins, each pathway was tested for in vitro reconstitution.
Formation of HM-ACP.
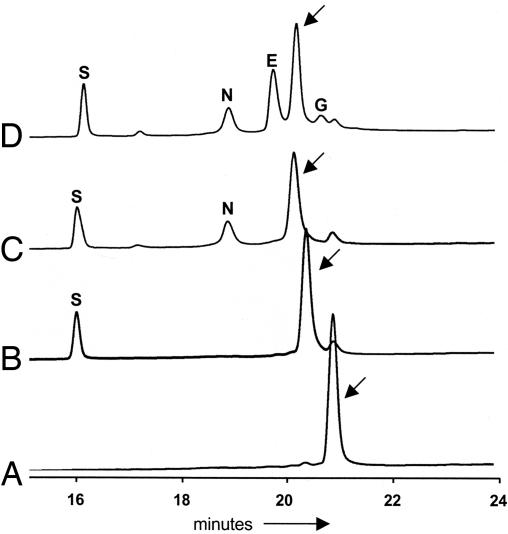
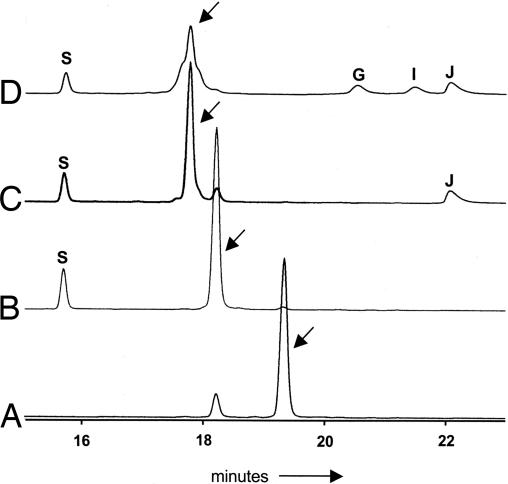
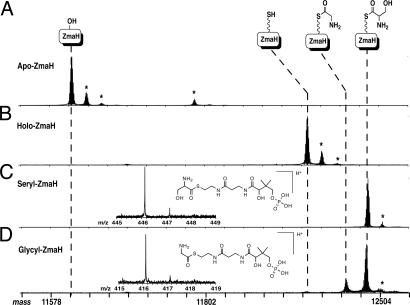
The proposed pathway for HM-ACP formation involves tethering a substrate to the 4′-phosphopantetheinyl (4′ Ppant) prosthetic group of the ACP ZmaD, followed by modification of the tethered intermediate (Fig. 2B). Using HPLC and MALDI-TOF MS analysis, we determined that the ZmaD purified from E. coli lacked the required prosthetic group (Fig. 3 and Table 1). To generate holo-ZmaD, in vitro phosphopantetheinylation was performed by using the 4′ Ppant transferase Sfp as described in ref. 13. HPLC and MALDI-TOF MS analysis determined that Sfp converted most of the apo-ZmaD to holo-ZmaD (Fig. 3 and Table 1).
Fig. 3.
HPLC analysis of ZmaD. Representative HPLC traces of reaction mixtures containing apo-ZmaD (A); apo-ZmaD, Sfp (B); apo-ZmaD, Sfp, ZmaN (C); apo-ZmaD, Sfp, ZmaN, ZmaG, ZmaE (D). Protein elution was monitored at 220 nm. Arrows identify the peak associated with ZmaD derivatives, which were collected and analyzed by MS. The letters above absorbance peaks identify the elution of a protein from the reaction mixture: S, Sfp; N, ZmaN; E, ZmaE; G, ZmaG.
Table 1.
MS analysis of purified apo- and holo-proteins
| Protein* | MALDI-MS |
ESI-FT-ICR-MS |
4′ Ppant elimination |
||||
|---|---|---|---|---|---|---|---|
| Theo. [M + H]+ | Exper. [M + H]+ | Theo. avg mass | Exp. avg mass | Moiety | Theo. [M + H]+ | Exp. [M + H]+ | |
| ZmaD | |||||||
| apo-ZmaD | 12,239 | 12,239 | 12,238.7 | 12,238.3 | |||
| holo-ZmaD | 12,579 | 12,579 | 12,579.0 | 12,579.3 | |||
| glyceryl-ZmaD | 12,667 | 12,668† | 12,667.1 | 12,667.4† | Glyceryl | 447.120 | 447.121 |
| 12,670‡ | |||||||
| HM-ZmaD | 12,681 | ND | 12,681.1 | 12,681.3 | HM | 461.099 | 461.099 |
| glycolyl-ZmaD | 12,637 | 12,636 | 12,637.1 | 12,637.4 | Glycolyl | 417.109 | 417.110 |
| ZmaH | |||||||
| apo-ZmaH | 11,609 | 11,605 | 11,609.1 | 11,608.8 | |||
| holo-ZmaH | 11,949 | 11,946 | 11,949.4 | 11,949.1 | |||
| seryl-ZmaH | 12,037 | 12,038§ | 12,036.5 | 12,036.8 | Seryl | 446.136 | 446.136 |
| 12,036¶ | |||||||
| AM-ZmaH | 12,051 | ND | |||||
| glycyl-ZmaH | 12,007 | 12,007 | 12,006.5 | 12,006.8 | Glycyl | 416.125 | 416.124 |
Masses are in daltons. Theo., theoretical; Exp., experimental; ND, not detected.
*The mass of ZmaD and ZmaH derivatives is calculated after removal of the first methionine.
†The mass of glyceryl-ZmaD detected when 3-PG was the starting substrate.
‡The mass of glyceryl-ZmaD detected when glyceraldehyde-3-phosphate was the starting substrate.
§The mass of seryl-ZmaH detected in the ZmaJ, holo-ZmaH reaction.
¶The mass of seryl-ZmaH detected in the ZmaJ, holo-ZmaH, ZmaG, and ZmaI reaction.
FkbH is proposed to recognize a glycolytic intermediate, dephosphorylate it, and tether it to its partner ACP to form glyceryl-ACP (Fig. 2A). The most likely substrate for FkbH is 1,3-bisphosphoglycerate (1,3-bPG) (4, 14). With that assumption, we investigated whether ZmaN would catalyze the formation of glyceryl-ZmaD in the presence of 1,3-bPG. The formation of glyceryl-ZmaD likely would be detected by a change in the elution time of holo-ZmaD from the HPLC and a change in its mass as detected by MALDI-TOF MS.
Although there is no commercial source of 1,3-bPG, enzymatic synthesis is possible by using 3-phosphoglycerate (3-PG), ATP, and 3-PG phosphokinase (PK). Incubation of these components with ZmaN and holo-ZmaD did result in a shift in holo-ZmaD elution time; however, this shift did not require the addition of 3-PGPK (Fig. 3). This result suggested either the substrate for ZmaN is 3-PG, or the reaction mixtures were contaminated with an unidentified source of 3-PGPK and the substrate was the resulting 1,3-bPG. Consistent with the latter hypothesis, the rate of holo-ZmaD modification was enhanced when the 3-PGPK was added to the reaction (1.44 nmol·min−1 with 3-PGPK, 0.08 nmol·min−1 without 3-PGPK). Furthermore, the equivalent change in elution time of holo-ZmaD was observed when GAPDH was used to convert GAPDH to 1,3-bPG (data not shown). Based on these observations, we conclude that 1,3-bPG is the substrate for ZmaN-catalyzed modification of holo-ZmaD.
To investigate whether the modified holo-ZmaD was glyceryl-ZmaD, the purified ZmaD derivatives from the reactions containing the contaminating kinase and the GAPDH were analyzed by MALDI-TOF MS. The mass of holo-ZmaD had shifted to be consistent with the formation of glyceryl-ZmaD (Table 1). Importantly, holo-ZmaH, discussed below, was not a substrate for ZmaN-catalyzed formation of glyceryl-ACP (data not shown), suggesting ZmaN specifically recognizes holo-ZmaD.
A mass consistent with 3-phosphoglyceryl-ZmaD was not detected, suggesting the phosphatase activity of ZmaN occurs before the acylation of holo-ZmaD. The 3-phosphoryl group may be extraneous because it plays no obvious role in the HM-ACP pathway. Its presence is likely because 1,3-bPG is the most readily available glyceryl primary metabolite containing an activated acid, thereby requiring the removal of the 3-phosphoryl group before downstream reactions. ZmaN must coordinate this phosphatase activity with its acyltransferase activity. ZmaN and its homologs contain the DXDX(T/V) motif of the phosphatase members of the haloacid dehalogenase superfamily of hydrolases (14, 15). During catalysis, these enzymes remove the phosphate from the substrate first by forming a phosphoaspartyl intermediate, then by hydrolyzing the phosphate from the enzyme. Structural analyses of members of this enzyme superfamily suggest the phosphoaspartyl formation is associated with a conformation change of the enzyme (16, 17). The phosphoaspartyl intermediate in ZmaN catalysis may alter the conformation of ZmaN to enhance its interactions with holo-ZmaD once the 3-phosphoryl group is removed from the substrate, thereby coordinating the phosphatase and acyltransferase activities. Additional studies on the mechanism of ZmaN are needed to test this hypothesis.
ZmaG and ZmaE are predicted to catalyze the oxidation of glyceryl-ZmaD to HM-ZmaD (Fig. 2B). These enzymes were incubated independently or together with their appropriate coenzymes along with glyceryl-ZmaD. A subtle change in elution time of glyceryl-ZmaD was observed (Fig. 3); this change required both ZmaG and ZmaE. Independently, neither enzyme was found to modify the glyceryl-ZmaD intermediate (data not shown). Replacement of ZmaE with ZmaI also resulted in a change in elution time and mass (data not shown), suggesting either FkbI homolog may be involved in HM-ACP formation.
Analysis of the purified ZmaD-tethered product from the ZmaG/ZmaE reaction by MALDI-TOF MS detected a mass consistent with the decarboxylated form of HM-ZmaD, glycolyl-ZmaD (Table 1). This decarboxylation could be the result of the instability of the product under our assay conditions or to the sample preparation and ionization process for MALDI-TOF MS. However, the detection of glycolyl-ZmaD is indicative of the formation of HM-ZmaD.
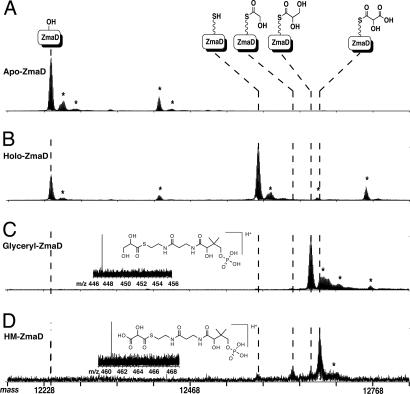
To more directly identify the ZmaD product of each reaction and to investigate whether an alternative MS technique could detect the HM-ZmaD final product, we used electrospray ionization-Fourier transform ion cyclotron resonance (ESI-FT-ICR)-MS to analyze the ZmaD products. The intact mass spectra collected showed mass shifts consistent with the formation of holo-ZmaD (+340 Da), glyceryl-ZmaD (+428 Da), HM-ZmaD (+442 Da), and glycolyl-ZmaD (+398 Da) (Fig. 4). MS/MS data localized the active site to the sequence G Y V N S, where the S is the site of 4′ Ppant (data not shown), and mass shifts were confirmed through measurement of the 4′ Ppant elimination product (Fig. 4 Insets). Importantly, ESI-FT-ICR-MS enabled detection and mass spectral analysis of HM-ZmaD.
Fig. 4.
ESI-FT-ICR-MS spectra of the intermediates in HM-ZmaD formation. The top of the figure depicts ZmaD intermediates of interest [from left to right: apo-, holo- (+340 Da), glycolyl- (+398 Da), glyceryl- (+428 Da), and HM-ZmaD (+442 Da)] and alignment to the representative peaks in the mass spectra as indicated by vertical dashed lines. Shown are the loading and corresponding mass shifts (1,015–1,070 m/z, +12 ions converted to mass scale) of apo-ZmaD (A), holo-ZmaD (B), glyceryl-ZmaD (C), and HM-ZmaD (D); asterisks indicate signals arising from artifactual adduction: sodium (+22 Da), potassium (+38 Da), phosphate (+98 Da), and oxidation of Met/Cys residues (+16 Da). (Insets) Mass spectra and structures of 4′ Ppant elimination product.
These data are consistent with the successful in vitro reconstitution of HM-ACP formation and present direct evidence for the existence of ACP-linked type I PKS extender units. These data also suggest that the substrate for both HM-ACP and MM-ACP formation is 1,3-bPG.
Formation of AM-ACP.
We previously proposed that the extender unit AM-ACP accounts for the ethanolamine unit in ZMA (ref. 3; Fig. 1). ZmaJ, a homolog of adenylation domains of NRPSs, is proposed to recognize l-Ser and tether it onto the ACP homolog ZmaH. The seryl moiety subsequently is oxidized in two steps to AM-ACP, analogous to the conversion of glyceryl-ACP to HM-ACP (Fig. 2).
Analysis of the amino acid substrate specificity of ZmaJ by standard ATP/PPi exchange assays by using l-Ser and structurally related amino acids (l-2,3-diaminopropionate, l-Thr, Gly, and l-Cys) determined ZmaJ activated only l-Ser (data not shown). Determination of the kinetic parameters of ZmaJ for l-Ser activation yielded values expected for adenylation domains of NRPSs (Km = 1.8 ± 0.2 mM; kcat = 42 ± 2 min−1) (e.g., ref. 12).
To test whether ZmaJ tethers l-Ser to ZmaH, holo-ZmaH was needed. By following the same protocol for the conversion of apo-ZmaD to holo-ZmaD, it was determined that a majority of ZmaH is purified from E. coli in its apo form, and apo-ZmaH is efficiently converted to holo-ZmaH by Sfp (Fig. 5 and Table 1). Incubation of holo-ZmaH with ZmaJ, l-Ser, and ATP resulted in a change in the holo-ZmaH elution time from the HPLC, and the mass of the purified product was consistent with the formation of seryl-ZmaH (Table 1). Importantly, ZmaJ was not able to aminoacylate holo-ZmaD (data not shown), highlighting the specificity of ZmaJ for holo-ZmaH.
Fig. 5.
HPLC analysis of ZmaH. Representative HPLC traces of reaction mixtures containing apo-ZmaH (A); apo-ZmaH, Sfp (B); apo-ZmaH, Sfp, ZmaJ (C); and apo-ZmaH, Sfp, ZmaJ, ZmaG, ZmaI (D). Each reaction also contained the required cofactors and substrates. Protein elution was monitored at 220 nm. Arrows identify the peak associated with ZmaH derivatives, which were collected and analyzed by MS. The letters above peaks identify the elution of a protein in the reaction mixture: S, Sfp; G, ZmaG; I, ZmaI; J, ZmaJ.
To test for AM-ZmaH formation, ZmaG and ZmaI were added independently or together with seryl-ZmaH. Only when ZmaG and ZmaI were added together, along with NAD+ and FAD, was a change in the seryl-ZmaH elution profile observed, as indicated by a broadening of the seryl-ZmaH peak (Fig. 5). ZmaE, the homolog of ZmaI from the HM-ZmaD pathway, could not replace ZmaI (data not shown). These data suggest ZmaI is specific for AM-ZmaH formation, whereas ZmaE is involved in HM-ZmaD formation. Analysis of the ZmaG/ZmaI-modified seryl-ZmaH derivative by MALDI-TOF MS was consistent with the formation of glycyl-ZmaH, the product expected if AM-ZmaH becomes decarboxylated, along with unreacted seryl-ZmaH (Table 1). As discussed below, longer incubation of the reaction resulted in a nearly complete conversion of seryl-ZmaH to glycyl-ZmaH.
The observed decarboxylation product could be due to the instability of the AM-ZmaH or to the decarboxylation caused by the MALDI-TOF MS analysis as discussed above with HM-ZmaD. To identify more directly the ZmaH-tethered products of each reaction, ESI-FT-ICR-MS analysis was performed on ZmaH purified from each reaction mixture. The intact mass spectra collected showed mass shifts consistent with the formation of holo-ZmaH (+340 Da), seryl-ZmaH (+427 Da), and glycyl-ZmaH (+397 Da) (Fig. 6). MS/MS data localized the active site to the sequence G L V N S, where the S is the site of 4′ Ppant (data not shown), and mass shifts were confirmed through measurement of the 4′ Ppant elimination product (Fig. 6 Inset). To address the lack of observable AM-ZmaH and lack of conversion of all of the seryl-ZmaH, the seryl-ZmaH was incubated for 5 or 40 min with ZmaG and ZmaI. After 40 min, the seryl-ZmaH was almost completely converted to glycyl-ZmaH (Fig. 8, which is published as supporting information on the PNAS web site). AM-ZmaH, however, was not detected.
Fig. 6.
ESI-FT-ICR-MS spectra of the intermediates in AM-ZmaH formation. The top of the figure depicts ZmaH intermediates of interest [from left to right: apo-, holo- (+340 Da), glycyl- (+397 Da), seryl-ZmaH (+427 Da)], and alignment to the representative peaks in the mass spectra are as indicated by vertical dashed lines. Shown are the loading and corresponding mass shifts (825–865 m/z, +14 ions converted to mass scale) of apo-ZmaH (A), holo-ZmaH (B), seryl-ZmaH (C), and glycyl-ZmaH (D); asterisks indicate signals arising from artifactual adduction: sodium (+22 Da), potassium (+38 Da), phosphate (+98 Da), and oxidation of Met/Cys residues (+16 Da). (Insets) Mass spectra and structures of 4′ Ppant elimination product.
The finding of glycyl-ZmaH by using both MS approaches suggests assay conditions cause the decarboxylation of AM-ZmaH. This finding is not surprising based on the high level of spontaneous decarboxylation that occurs with aminomalonate (18). Under acidic conditions, this decarboxylation is very rapid. Our HPLC assays conditions include 0.1% TFA in the solvents to function as an ion-pairing agent to enhance the resolution of proteins eluting from the HPLC. The presence of TFA in the solvents reduces the pH of the solvents to 2.0 and would likely expedite the decarboxylation of AM-ACP to glyceryl-ACP. Attempts were made to adjust the HPLC conditions to separate the proteins in the reaction at neutral pH, but these attempts were not successful. Analysis of reaction mixtures directly by MS without prior HPLC separation also detected a mass consistent with glycyl-ZmaH (data not shown). Regardless, the presence of glycyl-ZmaH is indicative of AM-ZmaH formation.
The data presented here are consistent with the formation of the AM-ACP extender unit. Interestingly, the enzyme that catalyzes the first oxidation of both glyceryl-ZmaD and seryl-ZmaH has no selectivity between the two intermediates; however, the second oxidation is catalyzed by a pathway-specific dehydrogenase. The formation of AM-ACP is an example of an amino acid being converted to a PKS extender unit.
Conclusions
In summary, we have presented biochemical and mass spectral evidence for the existence of two type I PKS extender units, HM-ACP and AM-ACP. This empirical evidence shows the formation of ACP-linked type I PKS extender units. Additionally, we establish that 1,3-bPG is the likely in vivo precursor for the formation of HM-ACP and MM-ACP. Interestingly, the AM-ACP pathway uses NRPS enzymology (an adenylation and carrier protein pair) for the formation of a type I PKS extender unit. We propose that AM-ACP will function as an NRPS extender unit in other enzyme systems. For example, the antibiotic GE23077 is a cyclic nonribosomal peptide that contains an aminomalonate residue (19). Therefore, AM-ACP has the potential to function as a type I PKS or an NRPS extender unit.
Defining the existence and mechanism of formation of HM-ACP and AM-ACP is of particular interest for combinatorial biosynthesis of natural products that are synthesized by type I PKS enzymology. The ability to incorporate glycolyl or ethanolamine units into a polyketide backbone introduces significantly different chemical attributes to the molecule. Hydroxyl or amino groups increase the potential for hydrogen bonding between a polyketide-containing natural product and its biological target, resulting in altered interactions that may enhance a desired biological activity. These hydroxyl and amino groups also offer functional groups for downstream chemical modifications. The challenges ahead to harness the full combinatorial biosynthetic potential of these extender units include investigations into the substrate flexibility of acyltransferase (20) and ketosynthase (21) domains of type I PKSs to recognize and incorporate HM-ACP and AM-ACP extender units. However, by defining the existence of HM-ACP and AM-ACP and the enzymatic requirements to make these precursors available, we have taken an important step toward this goal.
Materials and Methods
Cloning of Biosynthetic Genes.
Standard PCR-based cloning was used to introduce zmaD, zmaE, zmaG, zmaH, zmaI, zmaJ, and zmaN into the E. coli overexpression vector pET28b (Novagen, Madison, WI). All expression vector clones result in the production of a protein with an N-terminal histidine tag. All clones were verified by sequencing at the University of Wisconsin Biotechnology Sequencing Center (Madison, WI).
Heterologous Overproduction of Proteins.
All expression constructs were introduced into E. coli BL21 (λDE3). For overproduction of ZmaD, ZmaG, ZmaH, ZmaJ, and ZmaN, cells were grown at 25°C in LB medium containing 50 μg/ml kanamycin. At an OD600 of 0.4–0.6, the temperature was reduced to 15°C, and cells were grown for 1.5–2.5 h. IPTG then was added to 60 μM, and the cells were grown an additional 16 h at 15°C. For overproduction of ZmaE and ZmaI, cells were grown at 25°C for 24 h in LB medium containing 50 μg/ml kanamycin without IPTG addition. Cells were harvested by centrifugation.
Purification of Proteins.
Cells were resuspended in histidine-tag purification buffer [20 mM Tris, pH 8/300 mM NaCl/10% (vol/vol) glycerol; for ZmaG: 20 mM Tris, pH 8/300 mM NaCl/10% (wt/vol) sucrose]. Cells were broken by sonication, and cell debris was removed by centrifugation. Imidazole was added to the cell-free extract to 5 mM, and the mixture was incubated with 1–2 ml of Ni-NTA agarose resin (Qiagen, Valencia, CA) for 1–2 h at 4°C with gentle rocking. The resin was collected by centrifugation and loaded into a column. The resin was washed with histidine-tag buffer containing 5 mM imidazole, and stepwise elutions were performed with buffer containing increasing imidazole concentrations (20, 40, 60, 100, and 250 mM). Fractions containing purified protein based on SDS/PAGE/Coomassie blue staining were pooled and dialyzed at 4°C in dialysis buffer [50 mM Tris pH 8/100 mM NaCl/10% (vol/vol) glycerol; for ZmaG: 50 mM Tris, pH 8/100 mM NaCl/10% (wt/vol) sucrose; for ZmaI: 50 mM Tris, pH 8/300 mM NaCl/10% (vol/vol) glycerol]. ZmaE was dialyzed further in high salt buffer [50 mM Tris, pH 8/300 mM NaCl/10% (vol/vol) glycerol], and ZmaI was dialyzed in high salt buffer containing 100 μM FAD for 5 h and then in high salt buffer lacking FAD. All proteins were concentrated, flash frozen with liquid nitrogen, and stored at −80°C. Protein concentrations were determined by the calculated molar extinction coefficients (ZmaD, 2,560 M−1cm−1; ZmaE, 44,410 M−1cm−1; ZmaG, 21,180 M−1cm−1; ZmaH, 2,560 M−1cm−1; ZmaI, 44,770 M−1cm−1; ZmaJ, 46760 M−1cm−1; and ZmaN, 41,070 M−1cm−1).
Phosphopantetheinylation of ZmaD and ZmaH.
Sfp from Bacillus subtilis was used for apo- to holo-ACP conversion as described in ref. 13. Each reaction mixture contained 12.5 μM apo-ACP, 75 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM Tris(2-carboxyethylphosphine) (TCEP), 50 or 500 μM CoA, and 1 μM Sfp and was incubated at 22°C for 1 h.
Formation of Glyceryl-ZmaD.
Reactions with 3-PG (130 or 260 μl) contained 10 μM holo-ZmaD, 58 mM Tris (pH 7.5), 8 mM MgCl2, 0.8 mM TCEP, 385 μM CoA, 0.8 μM Sfp, 250 μM D(-)3-PG (Sigma, St. Louis, MO), 5 mM ATP, and 1 μM ZmaN and were run for 40 min at 22°C. Control reactions were run without ZmaN, ATP, or with 2 units of 3-PGPK (Sigma). A time course comparing reactions with and without 3-PGPK contained 50 μM 3-PG, and the reactions were run for 0.5, 2.0, 5.0, and 10 min. Reactions containing glyceraldehyde-3-phosphate (150 μl final volume) contained 12.5 μM holo-ZmaD, 75 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM TCEP, 100 mM NaCl, 500 μM CoA, 1 μM Sfp, 250 μM dl-glyceraldehyde-3-phosphate (Sigma), 1 unit of GAPDH (Sigma), and 1 μM ZmaN were run for 40 min at 22°C. Control reactions without GAPDH, dl-glyceraldehyde-3-phosphate, Sfp, or ZmaN were run, as well as control reactions in which holo-ZmaH replaced holo-ZmaD.
Formation of HM-ACP.
Reaction mixtures (130 or 260 μl) contained the following components: 10 μM glyceryl-ZmaD, 58 mM Tris (pH 7.5), 8 mM MgCl2, 0.8 mM TCEP, 385 μM CoA, 0.8 μM Sfp, 250 μM D(−)3-PG, 5 mM ATP, 1 μM ZmaN, 1 μM ZmaG, 200 μM NAD+, 1 μM ZmaE, and 100 μM FAD. The reaction mixture was incubated at 22°C for 1 h before injection onto an HPLC as discussed below. Reactions were repeated without ZmaG, ZmaE, or replacing ZmaE with 1 μM ZmaI.
Formation of Seryl-ZmaH.
Reaction mixtures (130 or 260 μl) contained 10 μM holo-ZmaH, 58 mM Tris (pH 7.5), 8 mM MgCl2, 0.8 mM TCEP, 385 μM CoA, 0.8 μM Sfp, 250 μM l-serine, 5 mM ATP, and 1 μM ZmaJ were incubated at 22°C for 1 h. To test whether seryl-ZmaD could be formed, 10 μM holo-ZmaD replaced the 10 μM holo-ZmaH in a 130-μl reaction mixture.
Formation of AM-ZmaH.
Reaction mixtures (130 or 260 μl) contained the following components: 10 μM ZmaD, 58 mM Tris (pH 7.5), 8 mM MgCl2, 0.8 mM TCEP, 385 μM CoA, 0.8 μM Sfp, 250 μM l-serine, 5 mM ATP, 1 μM ZmaJ, 1 μM ZmaG, 200 μM NAD+, 1 μM ZmaI, and 100 μM FAD were run for 5 or 40 min at 22°C. Reactions were repeated without ZmaG, ZmaI, or replacing ZmaI with 1 μM ZmaE.
HPLC Analysis of ZmaD and ZmaH Reaction Products.
HPLC analysis of enzymatic reaction products was performed with a Vydac (Hesperia, CA) C18 peptide column (250 × 4.6 mm). One hundred to two hundred microliters of apo- or holo-ZmaD or 130–200 μl of acylated ZmaD reactions were injected, and products were separated by using a 20–80% acetonitrile/0.1% TFA gradient over 20 min at a flow rate of 1 ml/min; elution was monitored at A220. One hundred to two hundred microliters of apo- or holo-ZmaH or 130–200 μl of aminoacylated ZmaH reaction products was injected, and products were separated by using the same method as above.
MALDI-TOF MS Analysis of ZmaD and ZmaH Reaction Products.
The enzymatic reaction products were collected as they eluted from the HPLC, flash-frozen with CO2(s)/ethanol (95%), and lyophilized overnight. Lyophilized samples were resuspended in double distilled H2O and added to 1 μl of the sinipinic acid matrix (10 mg/ml in 50% acetonitrile/0.05% TFA). MALDI-TOF MS analysis was performed by using a Voyager Biospectrometry Workstation (DE Pro; Applied Biosystems, Foster City, CA) in linear mode. Calibration was performed by using apomyoglobin, bovine insulin, and cytochrome C (Sigma).
ATP-PPi Exchange Assays for ZmaJ.
ZmaJ used in these assays was purified by an additional anion exchange chromatography step, which had no effect on seryl-ZmaH formation. ATP-PPi exchange assays were performed as described in refs. 12 and 22. Reactions for amino acid specificity contained 70 nM ZmaJ and 1 mM of the following amino acids: l-Ser, l-2,3-diaminopropionate, l-Thr, Gly, and l-Cys. To determine the kinetic parameters of ZmaJ activation of l-Ser, 100-μl reactions containing 70 nM ZmaJ, 3.5 mM ATP, and varying concentrations of l-Ser (0.35–10 mM), in triplicate, were incubated for 10 min before stopping the reactions. The reactions were in the linear range for enzyme concentration and <10% substrate-to-product conversion.
Analysis of Samples by ESI-FT-ICR-MS.
Mass analysis was performed on a custom-built 8.5 T Quadrupole-Enhanced Fourier Transform Ion Cyclotron Resonance Mass Spectrometer of the Marshall design (23). In general, a sample of interest was introduced to the mass spectrometer by using a NanoMate 100 for automated nanospray (Advion Biosciences, Ithaca, NY). Targeted species were externally accumulated in an octopole ion trap and either transferred to the ICR cell, isolated by stored waveform inverse Fourier transform (24), and fragmented by infrared multiphoton dissociation (25) or fragmented in the trap (OCAD) (23) and then transferred to the ICR cell for mass measurement. Collected data were analyzed by using THRASH (26) and/or manually interpreted, producing sets of intact mass data and fragment ion peak lists, which were uploaded onto the ProSightPTM (27) web server for analysis in single protein mode. All ESI-FT-ICR-MS experiments for glyceryl-ZmaD and HM-ZmaD used 3-PG as the starting substrate.
Supplementary Material
Acknowledgments
We thank Dr. M. R. Rondon for critical reading of the manuscript; Dr. H. Floss for discussions; Drs. S. van Lanen, P. Dorrestein, and B. Shen for helpful discussions and sharing of unpublished data; and Dr. J. Ravel and The Institute for Genomic Research (Rockville, MD) for the zmaN sequence. Y.A.C. was supported by a Chemical–Biology Interface National Institutes of Health (NIH) Training Grant and a William H. Peterson Predoctoral Fellowship. M.T.B. was supported on an NIH Institutional National Research Service Award 5T32 GM 07283 in Cellular and Molecular Biology. This work was funded, in part, by NIH Grants AI065850 (to M.G.T.) and GM067725 (N.L.K.).
Abbreviations
- ACP
acyl carrier protein
- AM
aminomalonyl
- ESI
electrospray ionization
- FT
Fourier transform
- ICR
ion cyclotron resonance
- HM
hydroxymalonyl
- 3-PG
3-phosphoglycerate
- 3-PGPK
3-PG phosphokinase
- 1,3-bPG
1,3-bisphosphoglycerate
- MM
methoxymalonyl
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- 4′ Ppant
4′-phosphopantetheinyl
- TCEP
Tris(2-carboxyethylphosphine)
- ZMA
zwittermicin A.
Note Added in Proof:
While this work was under review, a paper describing the analysis of an FkbH homolog was published (28).
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence for zmaN reported in this paper has been deposited in the GenBank database (accession no. DQ830808).
References
- 1.Rawlings BJ. Nat Prod Rep. 2001;18:231–281. doi: 10.1039/b100191o. [DOI] [PubMed] [Google Scholar]
- 2.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Ashley G. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emmert EAB, Klimowicz AK, Thomas MG, Handelsman J. Appl Environ Microbiol. 2004;70:104–113. doi: 10.1128/AEM.70.1.104-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K, Chung L, Revill WP, Katz L, Reeves CD. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 5.Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, Lewis M, Hutchinson CR. FEMS Microbiol Lett. 2003;218:223–230. doi: 10.1016/S0378-1097(02)01148-5. [DOI] [PubMed] [Google Scholar]
- 6.Rascher A, Hu Z, Buchanan GO, Reid R, Hutchinson CR. Appl Environ Microbiol. 2005;71:4862–4871. doi: 10.1128/AEM.71.8.4862-4871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll BJ, Moss SJ, Bai L, Kato Y, Toelzer S, Yu T-W, Floss HG. J Am Chem Soc. 2002;124:4176–4177. doi: 10.1021/ja0124764. [DOI] [PubMed] [Google Scholar]
- 8.Ligon J, Hill S, Beck J, Zirkle R, Molnar I, Zawodny J, Money S, Schupp T. Gene. 2002;285:257–267. doi: 10.1016/s0378-1119(02)00396-7. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe K, Khosla C, Stroud RM, Tsai S-C. J Mol Biol. 2003;334:435–444. doi: 10.1016/j.jmb.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Kato Y, Bai L, Xue Q, Revill WP, Yu T-W, Floss HG. J Am Chem Soc. 2002;124:5268–5269. doi: 10.1021/ja0127483. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez E, Ward S, Fu H, Revill WP, McDaniel R, Katz L. Appl Microbiol Biotechnol. 2004;66:85–91. doi: 10.1007/s00253-004-1658-7. [DOI] [PubMed] [Google Scholar]
- 12.Stachelhaus T, Marahiel MA. J Biol Chem. 1995;270:6163–6169. doi: 10.1074/jbc.270.11.6163. [DOI] [PubMed] [Google Scholar]
- 13.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 14.Walton LJ, Corre C, Challis GL. J Ind Microbiol Biotechnol. 2006;33:105–120. doi: 10.1007/s10295-005-0026-7. [DOI] [PubMed] [Google Scholar]
- 15.Collet J-F, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. J Biol Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 16.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. Proc Natl Acad Sci USA. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Kim R, Jancarik J, Yokota H, Kim SH. Structure (London) 2001;9:65–71. doi: 10.1016/s0969-2126(00)00558-x. [DOI] [PubMed] [Google Scholar]
- 18.Matthew M, Neuberger A. Biochem J. 1963;87:601–612. doi: 10.1042/bj0870601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciciliato I, Corti E, Sarubbi E, Stefanelli S, Gastaldo L, Montanini N, Kurz M, Losi D, Marinelli F, Selva E. J Antibiot. 2004;57:210–217. doi: 10.7164/antibiotics.57.210. [DOI] [PubMed] [Google Scholar]
- 20.Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, Konig A, Marsden AF, Galloway IS, Staunton J, Leadlay PF. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe K, Wang CC, Boddy CN, Cane DE, Khosla C. J Biol Chem. 2003;278:42020–42026. doi: 10.1074/jbc.M305339200. [DOI] [PubMed] [Google Scholar]
- 22.Thomas MG, Burkart MD, Walsh CT. Chem Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 23.Patrie SM, Charlebois JP, Whipple D, Kelleher NL, Hendrickson CL, Quinn JP, Marshall AG, Mukhopadhyay B. J Am Soc Mass Spectrom. 2004;15:1099–1108. doi: 10.1016/j.jasms.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Wang TC, Ricca TL, Marshall AG. Anal Chem. 1986;58:2935–2938. doi: 10.1021/ac00127a009. [DOI] [PubMed] [Google Scholar]
- 25.Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 26.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GK, Kim YB, Forbes AJ, Meng F, McCarthy R, Kelleher NL. Anal Chem. 2003;75:4081–4086. doi: 10.1021/ac0341721. [DOI] [PubMed] [Google Scholar]
- 28.Dorrestein PC, Van Lanen SG, Li W, Zhao C, Deng Z, Shen B, Kelleher NL. J Am Chem Soc. 2006;128:10386–10387. doi: 10.1021/ja0639362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.