Abstract
SLAM (signaling lymphocyte activation molecule)-associated protein (SAP) is a Src homology 2 (SH2) domain-containing adaptor expressed in T cells and natural killer cells. Its essential role in immune responses is underscored by the recent finding that mutations in SAP result in a rare but fatal X-linked lymphoproliferative disease (XLP). Although SAP is known to associate with SLAM-family receptors, the exact molecular mechanism by which SAP regulates lymphocyte signaling remains elusive. We here report that in T cells, SAP associates with the PAK-interacting exchange factor (PIX), a guanine nucleotide exchange factor (GEF) specific for Rac/Cdc42 GTPases. Moreover, SAP, PIX, and an activated form of Cdc42 form a complex in mammalian cells. We demonstrate that the SAP-PIX interaction is specific and is mediated by the C-terminal region of the SAP SH2 domain and the PIX SH3 domain. We further show that SAP is required for the recruitment of PIX to the SLAM-family receptors. Interestingly, overexpression of SAP, but not its homolog EAT-2, leads to a synergistic activation of nuclear factor of activating T cells (NFAT) in combination with a calcium signal in T cells. This SAP-mediated activation appears to be receptor-dependent and can be blocked by a dominant negative form of PIX. Taken together, our data strongly suggest that, in addition to the known SAP-interacting kinase Fyn, PIX may be another key player in SAP-mediated T cell activation.
Keywords: nuclear factor of activating T cells (NFAT), Cdc42, 2B4, Fyn
X-linked lymphoproliferative disease (XLP) is an inherited severe or fatal immune dysfunction generally triggered by infection with Epstein–Barr virus (EBV) (1). XLP patients often develop a lethal fulminant infectious mononucleosis after EBV infection. Some patients also develop progressive dysgammaglobulinemia, malignant lymphomas, or autoimmunity such as vasculitis (2–4). Several immune cell types have been shown to exhibit functional alterations in XLP, including CD4+ T cells, CD8+ T cells, natural killer cells, and B cells (3). In 1998, three groups identified the gene aberrant in XLP, named as SAP/SH2D1A/DSHP (5–7). The human SLAM (signaling lymphocyte activation molecule)-associated protein (SAP) is comprised of 128 amino acids, of which residues 6–102 contribute to an N-terminal SH2 domain. Many experiments have shown that the SH2 domain of SAP preferentially recognize specific tyrosine-based motifs containing TxYxxV/I that are present within the cytoplasmic tails of SLAM and its related receptors, including 2B4, CD84, Ly9, NTB-A, and CRACC (1, 8, 9). Because XLP patients lack functional SAP, it has been speculated that the phenotypes of XLP likely reflect perturbed signaling downstream of one or more of these receptors, ultimately resulting in abnormal immune responses.
Given the severe clinical manifestations in XLP patients, recent efforts have been focused on elucidating the signaling mechanism by which SAP regulates in T and natural killer cells. It has been shown that SAP recruits a Src kinase Fyn to the SLAM-related receptors via the Fyn SH3 domain, and such recruitment and downstream signaling may be responsible for the SLAM receptor mediated T cell activation (10–13). We report here a specific interaction between SAP and the PAK-interacting exchange factor (PIX), a guanine nucleotide exchange factor (GEF) for Rac/Cdc42 small GTPases (14). We show that SAP interacts with PIX directly and that the interaction involves a second binding surface in the SH2 domain of SAP and the SH3 domain of PIX. Such interaction can facilitate the PIX recruitment to the SAP coupling receptors. We further show that SAP, PIX, and an activated form of Cdc42 can form a complex. Interestingly, overexpression of SAP in T cells together with a calcium signal results in a synergistic activation of nuclear factor of activating T cells (NFAT). Such activation is inhibited specifically by the SH3 domain of PIX, but not Fyn, indicating that the SAP–PIX interaction may have a specific role in T cell activation. This report describes the physical and potential functional interaction between SAP and a Rac/Cdc42 GEF, which may provide insight into the pathophysiology of XLP.
Results
SAP Associates with PIX in T Cells.
We used a yeast two-hybrid system to screen a human lymphoid cDNA library with human SAP. The positive clones identified included SLAM and other SLAM-related molecules, consistent with the previous finding and providing validity of our screening. Interestingly, we have identified multiple clones containing various lengths of the cDNA sequence corresponding to βPIX, also known as p85 PIX. The SAP–βPIX interaction was further supported by the identification of αPIX (p90 PIX) in the same screening.
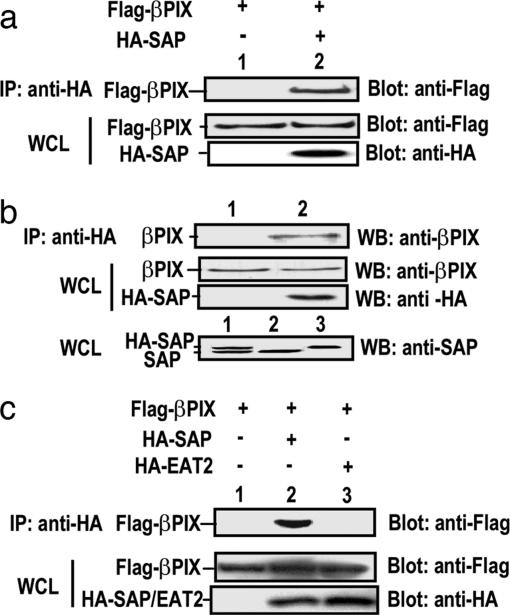
To examine the in vivo interacting and physiological relevance between SAP and PIX, we transfected 293T cells with a FLAG-βPIX in combination with a vector or an HA-SAP. As shown in Fig. 1a, HA-SAP co-immunoprecipitated with FLAG-βPIX, thus confirming the interaction in mammalian cells. Similarly, we have shown that SAP also interacted with αPIX (data not shown). To avoid the overexpression caveat and to further examine their physiological association in T cells, we generated a stable Jurkat cell transfectant expressing HA-SAP, because the anti-SAP antibody blocked the SAP-PIX interaction, and the anti-PIX antibodies failed to immunoprecipitate the endogenous PIX efficiently (data not shown). HA-SAP was expressed at a level comparable with endogenous SAP in the transfectants (Fig. 1b). Using an anti-HA antibody, we could readily detect the SAP–PIX complex, demonstrating that SAP and PIX interact in T cells at a physiological level. It has been recently shown that EAT-2, another SH2 domain-containing SAP homolog, is expressed in lymphocytes and also binds to the SLAM-family receptors (15). In contrast to SAP, EAT-2 failed to interact with PIX (Fig. 1c). Taken together, our data demonstrated a specific interaction between SAP and PIX in T cells.
Fig. 1.
SAP associates with PIX in T cells. (a) SAP and βPIX interacts in mammalian 293T cells. Cells were transiently transfected with FLAG-βPIX in combination with either control vector (lane 1) or HA-SAP (lane 2). Cell lysates were immunoprecipitated with anti-HA antibody and the immune complex were analyzed with anti-FLAG (Upper). Expressions of FLAG-βPIX and HA-SAP in the whole lysates were shown in Lower. (b) Association of SAP with endogenous βPIX. Lysates from HA-SAP Jurkat cells were immunoprecipitated with mouse IgG (lane 1) or anti-HA antibody (lane 2), and the immune complexes were analyzed with βPIX-specific antibody (Top). Expressions of endogenous βPIX and HA-SAP in the whole lysates were determined by Western blotting with βPIX-specific or anti-HA antibodies, respectively (Middle). (Bottom) Lysates of stable HA-SAP Jurkat cells (lane 1), Jurkat cells (lane 2), and Jurkat cells transiently transfected with HA-SAP (lane 3) were immunoblotted for anti-SAP antiserum. The addition of HA epitope resulted in a mobility shift of SAP. (c) No interaction between EAT-2 and βPIX. 293T cells were transiently transfected with FLAG-βPIX in combination with either control vector (lane 1), HA-SAP (lane 2), or HA-EAT-2 (lane 3) and assayed as described in a.
The C Terminus of the SAP SH2 Domain Is Required for Its Binding to PIX.
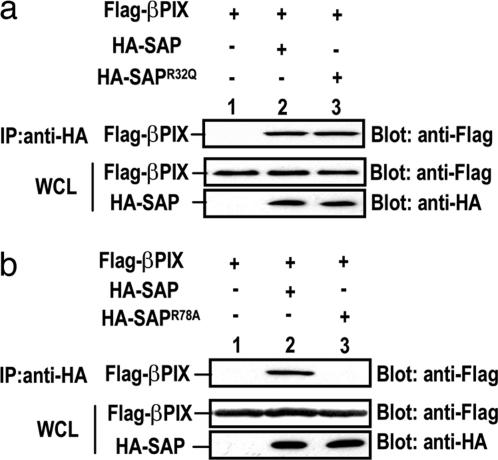
SAP is composed almost exclusively of an SH2 domain that mediates its interaction with the SLAM family of receptors. The conserved residue, Arg-32, within the SAP SH2 domain is required for its binding to the TxYxxV/I motifs in these receptors (10, 13, 16). Unlike the conventional SH2 domain and phosphotyrosine interaction, SAP is capable of binding to the nonphosphorylated form of the receptors (17). To address whether the interaction between SAP and PIX depends on the receptor-binding pocket in the SAP SH2 domain, we generated a point mutant, in which the Arg-32 was changed to a glutamine or a lysine residue. We showed that neither mutation of Arg-32 impaired the SAP's ability to bind to PIX (Fig. 2a, and data not shown), suggesting that PIX does not occupy the receptor-binding region within SAP.
Fig. 2.
The C terminus of the SAP SH2 domain is required for its binding to PIX. (a) Receptor-binding site in the SAP SH2 domain is not required for its binding to PIX. 293T cells were transiently transfected with FLAG-βPIX in combination with control vector (lane 1), HA-SAP (lane 2), or HA-SAPR32Q (lane 3) and assayed as described in Fig. 1a. (b) Mutation of Arg-78 abolishes binding of SAP to βPIX. 293T cells were transiently transfected with FLAG-βPIX in combination with either control vector (lane 1), HA-SAP (lane 2), or HA-SAPR78A (lane 3) and assayed as described in Fig. 1a.
To date, it has been well demonstrated that SAP recruits tyrosine kinase Fyn to the SLAM-family receptors in T and natural killer cells (10, 13). It has been demonstrated that SAP interacts with Fyn via a second binding surface within the SAP SH2 domain and the Fyn SH3 domain. The unique Fyn-binding region in SAP is centered on Arg-78, which is opposite to the phosphotyrosine-binding pocket of the SH2 domain. This feature presumably enables a single SAP molecule to bind both SLAM and Fyn simultaneously. Interestingly, we found that HA-SAPR78A completely failed to interact with βPIX (Fig. 2b), indicating that Arg-78, and likewise the Fyn-binding region within SAP, is also required for binding of βPIX. In fact, our biochemical experiments showed that PIX and Fyn competed with each other to bind to SAP, and PIX appeared to have higher binding affinity to SAP than Fyn in mammalian cells (data not shown).
SAP-Dependent Binding of βPIX to the SLAM-Family Receptor 2B4.
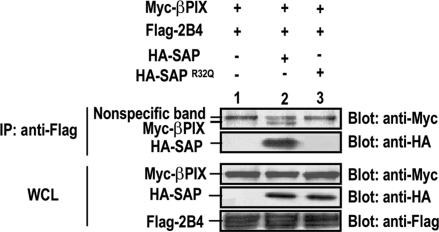
Recent biochemical and functional data have provided evidence that SAP couples with Fyn to mediate activation of the SLAM-family receptors (18). However, given the pleiotropic immune dysfunctional phenotype found in SAP-deficient mice and XLP patients, SAP likely recruits other signaling molecules to the receptors to participate in downstream signaling cascades. We therefore tested whether the SLAM-family receptors, SAP, and PIX may form a complex together. We generated an extracellular FLAG-tagged 2B4 molecule and coexpressed it with myc-βPIX in the presence of either HA-SAP or HA-SAPR32Q. As demonstrated in Fig. 3, myc-βPIX did not bind to FLAG-2B4 when coexpressed. However, myc-βPIX became readily detectable in the anti-FLAG-2B4 immunoprecipitate in the presence of HA-SAP. In contrast, HA-SAPR32Q, although capable of associating with βPIX, failed to bind to FLAG-2B4 or to mediate the PIX recruitment to 2B4 (Fig. 3 and data not shown). Similarly, myc-βPIX was absent in the anti-FLAG-2B4 immunoprecipitate when the PIX-binding mutant HA-SAPR78A was coexpressed (data not shown). Taken together, our data demonstrated that SAP is able to recruit PIX to the SLAM-related receptors, implicating a potential role of PIX in mediating the biological functions of these receptors.
Fig. 3.
SAP-dependent binding of βPIX to the SLAM-family receptor 2B4. 293T cells were transiently transfected with FLAG-2B4 and Myc-βPIX in combination with control vector (lane 1), HA-SAP (lane 2), or HA-SAPR32Q (lane 3). Cell lysates were immunoprecipitated with anti-FLAG antibody and the immune complex were analyzed by Western blotting with anti-Myc or anti-HA antibodies (Upper). Expressions of each protein in the whole lysates are shown in Lower.
SAP Interacts with the PIX SH3 Domain and Forms a Complex with an Activated Form of Cdc42.
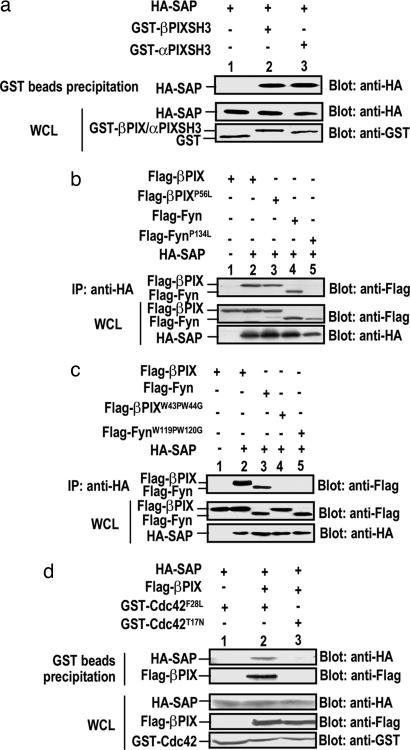
Given the recent finding that the Fyn SH3 domain interacts with the SAP SH2 domain (10, 13), we postulate that the SH3 domain present in the N terminus of PIX may also mediate its association with SAP. As shown in Fig. 4a, GST-PIX-SH3 was sufficient for binding to HA-SAP. Moreover, an SH3 domain truncated form of βPIX was incapable of binding to SAP (data not shown), indicating that the SH3 domain of PIX is necessary and sufficient to associate with SAP. We further investigated whether any conserved residues in the PIX SH3 domain are critical for its binding to SAP. Previous studies have revealed a proline residue, Pro-134, in the Fyn SH3 domain that is essential for its binding to SAP (13). Intriguingly, a point mutation in the corresponding proline residue (P56) in the βPIX SH3 domain did not impair its interaction with SAP (Fig. 4b). It has been shown that the conserved tryptophan residues 43 and 44 in PIX SH3 domain are critical for the PAK–PIX interaction (14). We attempted to assess whether these two conserved residues in the PIX or the Fyn SH3 domain are important in their interaction with SAP. We transiently transfected HA-SAP with FLAG-PIX, FLAG-Fyn, FLAG-PIXW43P/W44G, or FLAG-FynW119P/W120G. Interestingly, those mutations in both PIX and Fyn abolished their interaction with SAP (Fig. 4c). Taken together, these data suggest different SAP-binding mechanisms between PIX and Fyn.
Fig. 4.
SAP interacts with the PIX SH3 domain and forms a complex with an activated form of Cdc42. (a) PIX SH3 domains are sufficient for interacting with SAP. 293T cells were transiently transfected with HA-SAP in combination with GST alone vector (lane 1), GST-βPIX-SH3 (lane 2), or GST-αPIX-SH3 (lane 3). Cell lysates were incubated with glutathione beads, and absorbed proteins were analyzed by Western blotting with anti-HA antibody (Upper). Expressions of each protein in the whole lysates are shown in Lower. (b) The corresponding SH3 domain point mutation that abrogates SAP-Fyn association does not impair PIX's interaction with SAP. 293T cells were transiently transfected with FLAG-βPIX (lanes 1 and 2), FLAG-βPIXP56L (lane 3), FLAG-Fyn (lane 4), or FLAG-FynP134L (lane 5) in combination with either control vector (lane 1) or HA-SAP (lanes 2–5). Lysates were immunoprecipitated with anti-HA antibody, and the immune complexes were analyzed by Western blotting with anti-FLAG antibody (Upper). Expressions of each protein in the whole lysates are shown in Lower. (c) The conserved tryptophan mutations in both PIX and Fyn abolish their interaction with SAP. 293T cells were transiently transfected with FLAG-βPIX (lanes 1 and 2), FLAG-Fyn (lane 3), FLAG-βPIXW43P/W44G (lane 4), or FLAG-FynW119P/W120G (lane 5) in combination with either control vector (lane 1) or HA-SAP (lanes 2–5) and assayed as described in b. (d) SAP, PIX, and an activated form of Cdc42 form a complex. 293T cells were transiently transfected with HA-SAP, in combination with either control vector (lane 1) or FLAG-βPIX (lanes 2 and 3), and with either GST-Cdc42F28L (lanes 1 and 2) or GST-Cdc42T17N (lane 3). Lysates were incubated with glutathione beads, and absorbed proteins were analyzed by Western blotting with anti-HA or anti-FLAG antibodies, respectively (Upper). Expressions of each protein in the whole lysates are shown in Lower.
PIX has been shown to interact with PAK and an activated form of Cdc42 via its SH3 and DH domains, respectively (14). We therefore tested whether PAK and SAP compete with each other for PIX binding. We found that with increasing amounts of PAK, the SAP–PIX interaction diminished (data not shown). In addition, we showed that binding of SAP to PIX did not appear to interfere with the ability of PIX to interact with Cdc42. More importantly, SAP, PIX, and an activated Cdc42, but not a dominant negative Cdc42, could form a complex (Fig. 4d), suggesting that SAP may use PIX to regulate downstream Cdc42-mediated pathways.
Overexpression of SAP Leads to Synergistic Induction of NFAT Activity with a Calcium Signal in T Cells.
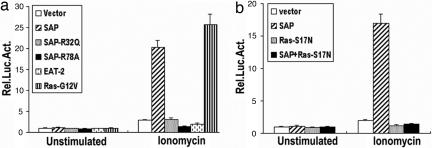
The aberrant immune responses in SAP-deficient mice have strongly suggested that SAP regulates multiple, and possibly distinct, signal transducing pathways in T cells. Recent studies have in fact revealed that SAP deficiency also resulted in TCR-mediated functional defects (19–21). We used a reporter constructs containing a key promoter element NFAT to explore potential function of SAP and PIX in T cell activation. We overexpressed a control vector, wild-type SAP, SAPR32Q, SAPR78A, or EAT-2 and examined their effects on the NFAT activity in Jurkat T cells. Phorbal myristate acetate (PMA) and ionomycin, which stimulate the calcineurin and the Ras pathways respectively, gives rise to a synergistic NFAT activity in T cells. Surprisingly, overexpression of wild-type SAP resulted in a 15- to 20-fold increase in NFAT activity in the presence of ionomycin, but not PMA (Fig. 5a and data not shown). In contrast, overexpression of either SAP mutant, SAPR32Q or SAPR78A, or EAT-2 completely failed to elicit such an effect, suggesting that the SAP effect is rather specific and that the stimulation may depend on the SAP binding to the upstream receptors as well as the downstream effectors such as PIX or Fyn. The synergistic effect of SAP with ionomycin on NFAT activity was reminiscent to that of activated form of Ras (RasG12V) (Fig. 5a). In addition, the effect of SAP with ionomycin on NFAT activation was completely abolished by the dominant negative RasR17N (Fig. 5b), suggesting that Ras or Ras-related pathway may be involved in SAP function in T cells.
Fig. 5.
Overexpression of SAP leads to synergistic activation of NFAT with ionomycin in T cells. (a) Similar to v-Ras, SAP and ionomycin synergistically activate NFAT. TAg Jurkat cells were cotransfected with NFAT-Luc together with empty vector, wild-type SAP, SAPR32Q, SAPR78A, EAT-2, or RasG12V. Cells were either unstimulated or stimulated with 1 μM ionomycin and were assayed for luciferase activity. The results are shown as the fold induction of luciferase activity as compared with the activity in unstimulated cells transfected with NFAT-Luc and empty vector. (b) Ras is required for the synergistic effect of SAP on ionomycin-induced NFAT activity. TAg Jurkat cells were cotransfected with NFAT-Luc together with empty vector, SAP, RasS17N, or SAP plus RasS17N. Western blot analysis showed comparable levels of SAP (data not shown). Cells were either unstimulated or stimulated with 1 μM ionomycin and were assayed for luciferase activity as described in a. Luciferase activity was determined in triplicate in each experimental condition. The data are representative of at least two independent experiments. Error bars represent standard errors of the means.
The SH3 Domain of PIX Inhibits SAP-Mediated NFAT Activation.
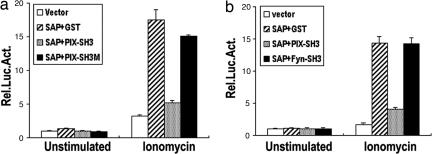
Because PIX and Fyn interact with SAP in T cells, it is likely that either or both of them may mediate the SAP effect. However, we failed to observe any significant changes in NFAT activation in the presence of ionomycin when either PIX or Fyn was overexpressed (data not shown). It is likely that either PIX or Fyn may need SAP for their recruitment to the receptors to exert their function. We therefore tested whether the SH3 domains of PIX or Fyn, which is sufficient for SAP binding, may interfere with the SAP-mediated function. As shown in Fig. 6a, the SH3 domain of PIX dramatically inhibited the SAP-mediated NFAT activation in the presence of ionomycin. In contrast, the PIX SH3 domain mutation (PIX-SH3 W43P/W44G), which failed to bind to SAP, had no effect on the SAP-stimulated effect. Interestingly, the SH3 domain of Fyn, when expressed at a level comparable with PIX SH3 domain, did not have any significant inhibitory effect on the SAP-induced NFAT activation (Fig. 6b). These data suggest that SAP may use PIX, rather than Fyn, to mediate such function in T cells.
Fig. 6.
The SH3 domain of PIX, but not Fyn, inhibits SAP-mediated NFAT activation. TAg Jurkat cells were cotransfected with of NFAT-Luc together with empty vector, SAP plus vector, SAP plus PIX-SH3, or SAP plus PIX-SH3M (a); or empty vector, SAP plus vector, SAP plus PIX-SH3, or SAP plus Fyn-SH3 (b). Western blot analysis showed comparable levels of SAP, PIX, or Fyn in the transfectants (data not shown). Cells were either unstimulated or stimulated with 1 μM ionomycin and were assayed for luciferase activity as described in Fig. 5a. Luciferase activity was determined in triplicate in each experimental condition. The data are representative of at least two independent experiments. Error bars represent standard errors of the means.
Discussion
Recent genetic and biochemical evidence demonstrates that the SAP plays an essential role in coupling a group of immune receptors, including SLAM and 2B4, to mediate proper immune activation (11, 22–25). Although a good deal of information has been accumulated regarding the SAP recruitment of Fyn to SLAM-family receptors and the importance of the SAP-Fyn interaction in immune cell functions (12, 18, 26, 27), little is known about the mechanism by which SAP initiates various signaling processes. Here, we identified another important family of signaling molecules including αPIX and βPIX, which interacted with SAP in T cells. We provided evidence suggesting that SAP–PIX complex may be involved in receptor signaling transduction leading to T cell activation. Given the complexity of XLP pathophysiology and the multiple immune defects found in SAP-deficient animals, we hypothesize that through binding to different partners including Fyn, PIX, and likely others, SAP and its coupling receptors may selectively turn on or regulate various signaling pathways in T and natural killer cells.
PIX was originally identified as a GEF for Rac and Cdc42 small GTPases and as a binding partner for PAK in fibroblasts (14). However, instead of possessing GEF activity, it binds preferentially to the activated form of Cdc42 (28, 29). Consistently, we also found that SAP, PIX, and an activated form of Cdc42 were able to form a complex in vivo. In addition, SAP competes with PAK to interact with PIX and Cdc42. It is noteworthy that Cbl also binds to the SH3 domain of PIX, and such interaction prevents Cbl from associating with EGF receptors (28). However, our data demonstrate a stable complex among 2B4, SAP, and βPIX. In addition, we did not observe any significant increase in the association between R78A, a PIX-binding SAP mutant, and 2B4 when compared with wild-type SAP (data not shown). Therefore, it does not appear that the SAP–PIX association leads to sequestration of SAP from the receptors; rather, SAP likely serves as an adaptor to recruit PIX, and possibly an activated Cdc42, to the SLAM-related receptors. Furthermore, the C-terminal regions of PIX proteins are also capable of interacting with GIT1 and GIT2, which associate with many focal adhesion complex proteins such as focal adhesion kinase and paxillin (30). Therefore, our identification of the SAP–PIX interaction raises a possibility that the SLAM receptors may be brought to the site of integrin interaction with the extracellular matrix, or they may be directly involved in cell adhesion and cytoskeletal rearrangement, a process vital for immune cell activation.
Our identification of SAP–PIX interaction brings to three the total number of proteins capable of binding to the PIX SH3 domain: PAK, Cbl, and SAP (14, 28). A typical SH3 domain interacts with a conserved PxxP motif in their ligands (31, 32). Interestingly, neither of these PIX-binding partners contains such a conventional proline-rich SH3-interacting motif, nor do they share any significant homology within their PIX binding regions. Recent reports describing the NMR and the crystal structure of the βPIX SH3 domain have revealed an atypical SH3/peptide binding surface with a unique large, planar hydrophobic binding pocket (33). A similar binding surface has been described in a Grb2-related signaling adaptor Gads, in which its SH3 domain interacts with SLP-76 with high affinity (34). The crystal structure of the Gads SH3 domain/SLP-76 peptide complex has revealed that, instead of a canonical PxxP motif, it prefers to bind a motif containing RxxK. Interestingly, SAP also contains an RxxK motif, of which the R78 mutation abolished its interaction with the PIX SH3 domain. It will be interesting to ascertain the exact SH3 binding sequence within SAP and further examine whether it also binds to the PIX SH3 domain with unusually high affinity. The unique binding surface within the PIX SH3 domain also predicts that it may allow rather promiscuous binding to other partners (33). In fact, neither Cbl nor PAK contains a putative RxxK motif, but they compete with each other for binding to the PIX SH3 domain (28). It is noteworthy that phosphorylation of the PAK peptide significantly reduced its binding affinity for the PIX SH3 domain, thus preventing PAK's recruitment to the PIX–GIT1 complex (35). Although our preliminary analyses did not reveal any significant change in the spectrum of complexes with cellular stimulation (data not shown), it is conceivable that certain stimuli may lead to distinct or sequential binding partners, triggering different signaling processes and downstream biological consequences.
Our attempt to elucidate the molecular mechanism of SAP–PIX in T cells led to a surprising finding that SAP overexpression resulted in a dramatic synergistic activation of an IL-2 promoter element NFAT with a calcium signal. To date, only an ectopical expression of an activated form of Ras can trigger such characteristic induction, suggesting that SAP may activate, or be associated with, the Ras-like pathways. Similar to an activated form of Ras, overexpression of SAP did not further augment TCR-induced NFAT activation (data not shown). This SAP effect does not appear to be nonspecific, because the loss-of-binding mutations to either the SLAM receptors or PIX abolished such ability to synergize with ionomycin. Moreover, EAT-2, although capable of binding to the SLAM-family receptors, completely failed to exhibit such an effect. Interestingly, nontransformed T cells from XLP patient exhibited diminished MAPK activation, a biochemical event downstream of Ras (19). Moreover, these SAP-deficient T cells have an enhanced level of calcium signaling after TCR stimulation. Because activation of calcium and Ras pathway synergistically turn on T cells, our data implicating SAP in Ras activation may predict a compensatory enhancement of calcium pathway due to SAP deficiency. It remains intriguing how overexpression of SAP, a Rac/Cdc42 GEF-binding protein, may lead to Ras activation in T cells, because overexpression of an activated form of Cdc42 failed to synergize with ionomycin in activating NFAT (data not shown). It would be interesting to further explore any potential interplay between Rac/Cdc42 and Ras pathways.
Although SAP is believed to serve as a master molecular switch for the SLAM-family receptors, a number of reports also demonstrate specific defects in TCR-mediated activation. In particular, one study has shown that nontransformed T cells from XLP patients were defective in several TCR-induced activation events, such as IL-2 production and CD25 up-regulation (19). It has been hypothesized that SAP may directly participate in TCR-mediated activation through an as-yet-unknown mechanism. However, we found that neither the R32Q nor the R78A mutant of SAP acted as a dominant negative to inhibit TCR-induced NFAT activation, suggesting that it may not function directly downstream of TCR. Because many of the SLAM members exhibit homophilic interactions (36, 37), another plausible scenario is that these SAP-associated receptors may activate certain signaling events that influence the TCR activation. Supporting this hypothesis, one study has demonstrated that SLAM/SAP engagement was critical for TCR-induced PKC-θ recruitment and subsequent downstream cytokine production (11).
Given the vast amount of convincing evidence that implicates Fyn in SAP-mediated function, it is still unclear how SAP selectively interacts with PIX and Fyn during T cell activation. The inability of the Fyn SH3 domain to inhibit the SAP-mediated NFAT induction suggests that PIX may be another primary target of SAP in the T cell signaling pathways. Most importantly, because both SAP R78A mutant reconstituted cells and knockin mice have been used to prove the role of Fyn in SAP-mediated immune responses (11–13), we now have to bear in mind that this mutation also abolishes SAP binding to another important signaling adaptor PIX. Therefore, some of the defective phenotypes associated with this SAP mutant may also be a consequence of an aberrant interaction between SAP and PIX.
Materials and Methods
Antibodies.
Anti-HA and anti-Myc monoclonal antibodies were obtained from Roche (Indianapolis, IN); anti-FLAG monoclonal antibody was obtained from Sigma (St. Louis, MO); and polyclonal rabbit antibody against βPIX was obtained from Chemicon (Temecula, CA); anti-GST monoclonal antibody, anti-mouse IgG horseradish peroxidase (HRP), and anti-rabbit IgG HRP were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-SAP antiserum was a gift from K. M. Nichols (Children's Hospital of Philadelphia, Philadelphia, PA).
Cell Lines and Transfectants.
Human 293T cells and TAg Jurkat cells were cultured in DMEM and RPMI medium 1640, respectively. HA-SAP Jurkat cells was generated by retroviral infection. Briefly, a retroviral vector containing human SAP cDNA with a C-terminus HA tag, pCI-PG8, and pCI-VSV were cotransfected into 293T cells. Viral supernatants were collected and used to infect Jurkat T cells (ATCC TIB-152). Cells were then selected, cloned, and screened for SAP expression.
DNA Constructs.
Human SAP, mutant SAP, EAT-2, and mutant Ras cDNAs were cloned in a mammalian expression vector pCDEF3 with a C-terminal HA tag. Human βPIX, mutant βPIX, Fyn, and mutant Fyn cDNAs were cloned in a pCDEF3 vector with an N-terminal FLAG tag. Human 2B4 cDNA with an extracellular FLAG tag (9) was subcloned in pCDEF3 vector. Human Cdc42, βPIX SH3 domain, αPIX SH3 domain, and Fyn SH3 domain were cloned in pCDEF3 with a GST tag. All mutant cDNAs were generated by site-directed mutagenesis. The pCDEF3 vector and NFAT luciferase reporter construct were gifts from Arthur Weiss (University of California, San Francisco, CA).
Yeast Two-Hybrid Screening.
The full-length human SAP cDNA was cloned into pGBKT7 vector and was used to screen a pACT2-human lymphoid cDNA library in a yeast two-hybrid system (Clontech, Mountain View, CA). β-galactosidase activities were measured by using o-nitrophenyl-galactoside as a substrate. Clones activating the β-galactosidase reporter gene were sequenced and analyzed.
Transfections, Immunoprecipitations, and Western Blotting.
293T cells were cotransfected with 3 μg of each indicated plasmids by using calcium phosphate. Cells were cultured for 24 h after transfection, and cells were harvested and lysed in lysis buffer as previously described (9). Lysates were subjected to immunoprecipitation with the indicated antibodies. For Western blotting, the immune complexes were resolved by SDS/PAGE and transferred, and membranes were probed with the indicated primary antibodies followed by a HRP-conjugated secondary antibody and then assayed by ECL (Pierce, Rockford, IL).
NFAT Luciferase Reporter Assay.
TAg Jurkat cells were transfected with 1 μg of NFAT Luciferase plasmid together with 2 μg of indicated plasmids by SuperFect transfection (Qiagen, Valencia, CA). Cells were cultured for 40 h after transfection, aliquoted into a 96-well plate (Corning, Acton, MA) at 1 × 105 cells per 100 μl of RPMI growth medium per well. Cells were unstimulated or stimulated at 37°C in the presence of 1 μM ionomycin. After 8 h stimulation, cells were lysed in 25 μl of 5X reporter lysis buffer (Promega, San Luis Obispo, CA). Luciferase activity was assayed with a Lumat LB 9507 luminometer (Berthold Technologies, Oak Ridge, TN).
Acknowledgments
We thank Bing-e Xu and Rui Jin for experimental design and manuscript preparation, Arthur Weiss for pCDEF3 vector and NFAT-Luc constructs, and Kim E. Nichols for anti-SAP antiserum. This work was supported by National Natural Science Foundation of China Grant 30430630, National Outstanding Youth Fund Grant 30225022, Key Program of Basic Research Grant STC 05JC14087, and the Cancer Council of New South Wales, Australia.
Abbreviations
- XLP
X-linked lymphoproliferative disease
- SLAM
signaling lymphocyte activation molecule
- SAP
SLAM-associated protein
- PAK
p21-activated kinase
- PIX
PAK-interacting exchange factor
- NFAT
nuclear factor of activated T cells
- SH2
Src homology 2
- GEF
guanine nucleotide exchange factor.
Footnotes
Conflict of interest statement: A.W. is a consultant of Shanghai Genomics.
References
- 1.Engel P, Eck MJ, Terhorst C. Nat Rev Immunol. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar HB, Sharifi R, Gilmour KC, Thrasher AJ. Br J Haematol. 2002;119:585–595. doi: 10.1046/j.1365-2141.2002.03851.x. [DOI] [PubMed] [Google Scholar]
- 3.Seemayer TA, Gross TG, Egeler RM, Pirruccello SJ, Davis JR, Kelly CM, Okano M, Lanyi A, Sumegi J. Pediatr Res. 1995;38:471–478. doi: 10.1203/00006450-199510000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Sumegi J, Huang D, Lanyi A, Davis JD, Seemayer TA, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo DT, et al. Blood. 2000;96:3118–3125. [PubMed] [Google Scholar]
- 5.Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, et al. Proc Natl Acad Sci USA. 1998;95:13765–13770. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, et al. Nat Genet. 1998;20:129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 7.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, et al. Nature. 1998;395:462–469. [PubMed] [Google Scholar]
- 8.Ma CS, Nichols KE, Tangye SG. Annu Rev Immunol. doi: 10.1146/annurev.immunol.25.022106.141651. in press. [DOI] [PubMed] [Google Scholar]
- 9.Tangye SG, Lazetic S, Woollatt E, Sutherland GR, Lanier LL, Phillips JH. J Immunol. 1999;162:6981–6985. [PubMed] [Google Scholar]
- 10.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 11.Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, Ohno S, Yanagi Y, Veillette A. Immunity. 2004;21:707–717. doi: 10.1016/j.immuni.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A. Nat Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 14.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 15.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, et al. EMBO J. 2001;20:5840–5852. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang PM, Li C, Morra M, Lillywhite J, Muhandiram DR, Gertler F, Terhorst C, Kay LE, Pawson T, Forman-Kay JD, Li SC. EMBO J. 2002;21:314–323. doi: 10.1093/emboj/21.3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, Eck MJ. Mol Cell. 1999;4:555–561. doi: 10.1016/s1097-2765(00)80206-3. [DOI] [PubMed] [Google Scholar]
- 18.Bloch-Queyrat C, Fondaneche MC, Chen R, Yin L, Relouzat F, Veillette A, Fischer A, Latour S. J Exp Med. 2005;202:181–192. doi: 10.1084/jem.20050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanzone S, Zeyda M, Saemann MD, Soncini M, Holter W, Fritsch G, Knapp W, Candotti F, Stulnig TM, Parolini O. J Biol Chem. 2003;278:29593–29599. doi: 10.1074/jbc.M300565200. [DOI] [PubMed] [Google Scholar]
- 20.Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, et al. Nat Immunol. 2001;2:410–414. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
- 21.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, et al. Proc Natl Acad Sci USA. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A. Nat Immunol. 2001;2:681–690. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 23.Bottino C, Falco M, Parolini S, Marcenaro E, Augugliaro R, Sivori S, Landi E, Biassoni R, Notarangelo LD, Moretta L, Moretta A. J Exp Med. 2001;194:235–246. doi: 10.1084/jem.194.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumaresan PR, Lai WC, Chuang SS, Bennett M, Mathew PA. Mol Immunol. 2002;39:1–8. doi: 10.1016/s0161-5890(02)00094-9. [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Relouzat F, Roncagalli R, Aoukaty A, Tan R, Latour S, Veillette A. Mol Cell Biol. 2004;24:5144–5156. doi: 10.1128/MCB.24.12.5144-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benoit L, Wang X, Pabst HF, Dutz J, Tan R. J Immunol. 2000;165:3549–3553. doi: 10.4049/jimmunol.165.7.3549. [DOI] [PubMed] [Google Scholar]
- 27.Latour S, Veillette A. Semin Immunol. 2004;16:409–419. doi: 10.1016/j.smim.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Wu WJ, Tu S, Cerione RA. Cell. 2003;114:715–725. doi: 10.1016/s0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- 29.Feng Q, Albeck JG, Cerione RA, Yang W. J Biol Chem. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- 30.Zhao ZS, Manser E, Loo TH, Lim L. Mol Cell Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 32.Lim WA, Richards FM, Fox RO. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Liu X, Sun F, Gao J, Zhou H, Gao GF, Bartlam M, Rao Z. Biochem Biophys Res Commun. 2006;339:407–414. doi: 10.1016/j.bbrc.2005.10.212. [DOI] [PubMed] [Google Scholar]
- 34.Harkiolaki M, Lewitzky M, Gilbert RJ, Jones EY, Bourette RP, Mouchiroud G, Sondermann H, Moarefi I, Feller SM. EMBO J. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mott HR, Nietlispach D, Evetts KA, Owen D. Biochemistry. 2005;44:10977–10983. doi: 10.1021/bi050374a. [DOI] [PubMed] [Google Scholar]
- 36.Mavaddat N, Mason DW, Atkinson PD, Evans EJ, Gilbert RJ, Stuart DI, Fennelly JA, Barclay AN, Davis SJ, Brown MH. J Biol Chem. 2000;275:28100–28109. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- 37.Martin M, Romero X, de la Fuente MA, Tovar V, Zapater N, Esplugues E, Pizcueta P, Bosch J, Engel P. J Immunol. 2001;167:3668–3676. doi: 10.4049/jimmunol.167.7.3668. [DOI] [PubMed] [Google Scholar]