Abstract
Granulocyte colony-stimulating factor (GCSF) administration has been linked to the development of monosomy 7 in severe congenital neutropenia and aplastic anemia. We assessed the effect of pharmacologic doses of GCSF on monosomy 7 cells to determine whether this chromosomal abnormality developed de novo or arose as a result of favored expansion of a preexisting clone. Fluorescence in situ hybridization (FISH) of chromosome 7 was used to identify small populations of aneuploid cells. When bone marrow mononuclear cells from patients with monosomy 7 were cultured with 400 ng/ml GCSF, all samples showed significant increases in the proportion of monosomy 7 cells. In contrast, bone marrow from karyotypically normal aplastic anemia, myelodysplastic syndrome, or healthy individuals did not show an increase in monosomy 7 cells in culture. In bone marrow CD34 cells of patients with myelodysplastic syndrome and monosomy 7, GCSF receptor (GCSFR) protein was increased. Although no mutation was found in genomic GCSFR DNA, CD34 cells showed increased expression of the GCSFR class IV mRNA isoform, which is defective in signaling cellular differentiation. GCSFR signal transduction via the Jak/Stat system was abnormal in monosomy 7 CD34 cells, with increased phosphorylated signal transducer and activation of transcription protein, STAT1-P, and increased STAT5-P relative to STAT3-P. Our results suggest that pharmacologic doses of GCSF increase the proportion of preexisting monosomy 7 cells. The abnormal response of monosomy 7 cells to GCSF would be explained by the expansion of undifferentiated monosomy 7 clones expressing the class IV GCSFR, which is defective in signaling cell maturation.
Keywords: hematopoiesis, myelodysplasia, clonal evolution, leukemia, marrow failure
Treatment of aplastic anemia (AA) with immunosuppressive drugs such as antithymocyte globulin and cyclosporine usually improves blood counts, but a minority of patients develop recurrent or refractory cytopenias, or frank leukemia, associated with the expansion in the marrow of dysplastic and cytogenetically abnormal clones, a pathophysiology termed “clonal evolution” (1, 2). Of the cytogenetic abnormalities, monosomy 7 is the most common and has the worst prognosis (3, 4). Some studies of AA in children and adults, and observations in children with congenital neutropenia (Kostmann's syndrome), have suggested a correlation between development of monosomy 7 and long-term use of high doses of granulocyte colony-stimulating factor (GCSF) (5–7). It is uncertain whether GCSF is directly responsible for cytogenetic abnormalities because other linked factors such as unresponsiveness to immunosuppressive therapy, persistent neutropenia, and concurrent infections might also be implicated (8). Some investigators report a comparable incidence of monosomy 7 in patients who have not been treated with GCSF (2, 9).
Using the sensitive quantitative method fluorescence in situ hybridization (FISH), we previously found small numbers of monosomy 7 cells (among other chromosomal abnormalities) in AA patients sampled at the time of presentation of their disease and before treatment (10). Routine cytogenetic analysis of AA bone marrow samples is often unsuccessful because many patients have too few dividing bone marrow cells to permit analysis of 20 metaphase cells. However, FISH makes possible the examination of 400 or more interphase cells because the technique does not require cells to be in metaphase. In one study of marrow-failure patients, only 28% of chromosomal abnormalities detected by FISH were observed by conventional cytogenetics (10). GCSF levels are generally an order of magnitude higher in severe AA than in neutropenia of other origins (11, 12). An increased prevalence of monosomy 7 among untreated cases could, therefore, be explained by high endogenous levels of GCSF in AA.
Monosomy 7 in AA could be a consequence of GCSF-induced errors in the cell cycle, DNA replication and repair, or chromosome segregation errors provoked by increasing cell replication. Alternatively, GCSF might favor the expansion of a preexisting clone. In support of the latter hypothesis, dominant proliferative signaling from a mutant GCSF receptor (GCSFR) has been reported in children with severe congenital neutropenia (SCN) who require prolonged GCSF treatment for survival (13). To distinguish between these two possibilities, we used FISH for chromosome 7 to examine cells from patients with AA, myelodysplastic syndrome (MDS) with normal cytogenetics, MDS with cytogenetically documented monosomy 7, and normal controls, in order to assess the effect of GCSF on bone marrow cells grown in short-term culture. We also cultured cytogenetically normal samples obtained at presentation of pancytopenia from patients who later developed monosomy 7 with GCSF. Lastly, we sought evidence of abnormal GCSF signal transduction in monosomy 7 cells.
Results
Study Population.
Twenty-eight de novo MDS patients with monosomy 7 (24 positive by FISH and conventional cytogenetics and 4 MDS patients positive by FISH alone), 4 additional patients with a history of AA who developed monosomy 7, and 26 individuals with normal karyotype by cytogenetics and FISH (6 MDS patients, 10 AA patients, and 10 healthy controls) were studied. Four AA patients who initially had normal cytogenetics, but who later developed monosomy 7 were also studied. Data for these subjects are provided in Table 6, which is published as supporting information on the PNAS web site.
GCSF Promotes Expansion of Preexisting Monosomy 7 Clones.
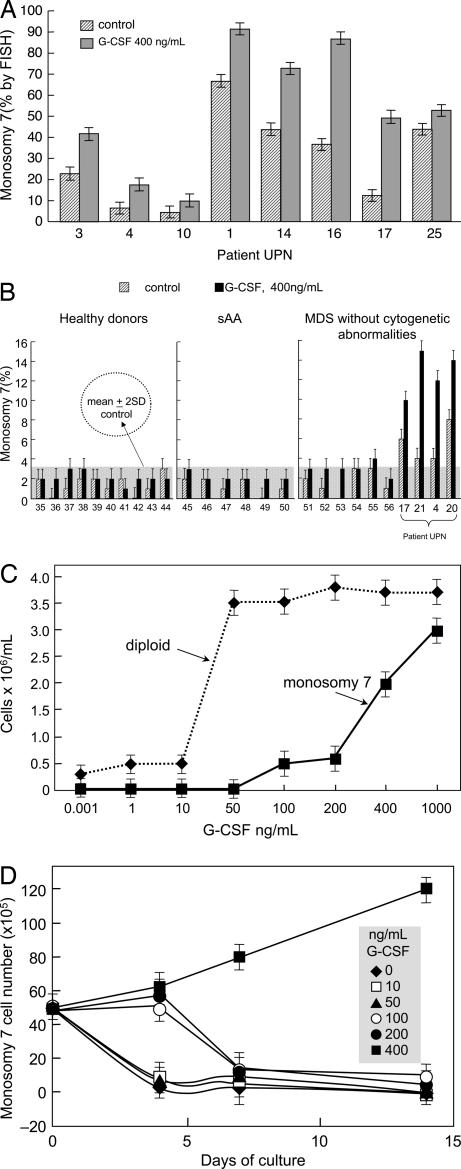
When bone marrow mononuclear cells (BMMNC) from eight patients with monosomy 7 (as determined by cytogenetics) were cultured in liquid media for 7 days in the presence or absence of pharmacological doses of GCSF (400 ng/ml), all cytokine-treated samples showed increased proportions of cells with monosomy 7 (Fig. 1A) (P < 0.01). Similarly, samples from four patients with MDS and monosomy 7 identified only by FISH (presumably below the threshold of detection by cytogenetics) showed expansion of monosomy 7 cell numbers after culture (Fig. 1B). When MDS BMMNC from five additional patients with monosomy 7 by cytogenetics were cultured in semisolid media for 2 weeks and individual colonies were picked for FISH analysis, a similar increase was seen in the number and proportion of hematopoietic colonies with monosomy 7, primarily among myeloid progeny (Table 1). Colonies were homogeneous (i.e., they were composed of either all diploid cells or all monosomy 7 cells). GCSF-stimulated cultures of monosomy 7 cells showed large numbers of undifferentiated cells by microscopy (data not shown). In contrast, BMMNC from 10 AA and 6 MDS patients with normal chromosome 7 numbers by FISH had no increase in monosomy 7 cells after culture with GCSF (Fig. 1B; P < 0.01). Four patients with AA had BMMNC stored at the time of first clinical presentation; FISH had disclosed a detectable monosomy 7 clone of cells in all of the samples, despite normal conventional cytogenetics, and we were able to expand this clone in vitro with GCSF (Table 2). The proportion and number of monosomy 7 cells were unaltered in vitro by incubation with other cytokines, IL-3, or granulocyte-macrophage colony-stimulating factor, even at high concentrations (data not shown).
Fig. 1.
GCSF increases the proportion of monosomy 7 cells in patients with a preexisting monosomy 7 clone. BMMNC were placed in culture with and without pharmacologic doses (400 ng/ml) of GCSF for 7 days, as described in Materials and Methods. (A) All 24 patients who demonstrated monosomy 7 by conventional cytogenetics showed increased proportions and numbers of cells with monosomy 7. Results are shown for eight patients. (B) None of the 10 normal controls or 6 AA patients without clinical monosomy 7 showed a significant increase in the proportion of cells with monosomy 7 in tissue culture as a result of exposure to high GCSF doses, whereas MDS patients with monosomy 7 detectable by FISH all showed expansion of this clone after GCSF exposure. (C) A dose–response curve for BMMNC performed on three patients with monosomy 7 showing decreased sensitivity to low doses of GCSF when cultured short-term (4 days) compared with diploid cells. (D) When the cells described in C were subjected to longer culture times (14 days), monosomy 7 cells declined at lower GCSF concentrations.
Table 1.
Expansion of monosomy 7 CD34 cells exposed to in vitro pharmacologic concentrations of GCSF
| Patient no. | % monosomy 7 CD34 | % CFU-GM monosomy 7 |
% CFU-E monosomy 7 |
||
|---|---|---|---|---|---|
| −GCSF | +GCSF | −GCSF | +GCSF | ||
| 24 | 25 | 35 | 75 | 5 | 13 |
| 21 | 9 | 12 | 35 | 0 | 7 |
| 7 | 3 | 0 | 8 | 2 | 12 |
| 22 | 6 | 2 | 8 | 0 | 5 |
| 13 | 8 | 10 | 25 | 2 | 12 |
CD34 cells were obtained by flow cytometry, and FISH was performed by using a centrometric probe for chromosome 7. CD34 cells were plated in semisolid media with and without pharmacologic concentrations of GCSF. Colonies derived from granulocyte-macrophage hematopoietic progenitors (CFU-GM) and erythroid hematopoietic progenitors (CFU-E) were isolated, and FISH was performed on individual colonies. Colonies were homogeneous, containing only monosomy 7 cells or diploid cells. Results are shown as a percentage of colonies in which FISH showed monosomy 7. The monosomy 7 clone expanded in samples containing pharmacologic concentrations of GCSF, mostly in myeloid colonies.
Table 2.
Serial analysis of BMMNC from AA patients with small monosomy 7 clones exposed in vitro to GCSF
| Patient no. | −18 mo NT, % | −18 mo + GCSF, % | −12 mo NT, % | −12 mo+ GCSF, % | −6 mo NT, % | −6 mo + GCSF, % | 0 mo NT, % |
|---|---|---|---|---|---|---|---|
| 29 | 5 | 7 | 4 | 7 | 10 | 30 | 60 |
| 30 | 6 | 10 | 7 | 14 | 12 | 45 | 46 |
| 31 | 12 | 23 | 10 | 16 | 18 | 35 | 38 |
| 32 | ND | ND | ND | ND | 24 | 41 | 53 |
Frozen samples from AA patients were obtained before the development of any clinical cytogenetic abnormality BMMNC cultured with GCSF. Results are shown as percentage of monosomy 7 cells. Time 0 is when monosomy 7 was clinically first evident on routine cytogenetics. NT, not treated with GCSF; ND, not done.
Decreased numbers of monosomy 7 cells relative to diploid cells were evident in cultures after 4 days at low GCSF concentrations, but significantly more monosomy 7 cells were seen growing at very high GCSF concentrations, in the range of those present in the circulation of patients with marrow failure (11, 14) (Fig. 1C). Inferior survival of monosomy 7 cells after 14 days of culture with GCSF was evident at low concentrations of GCSF (Fig. 1D).
GCSFR Expression Is Increased on CD34 Cells from Monosomy 7 Patients and Preferentially in Monosomy 7 Cells.
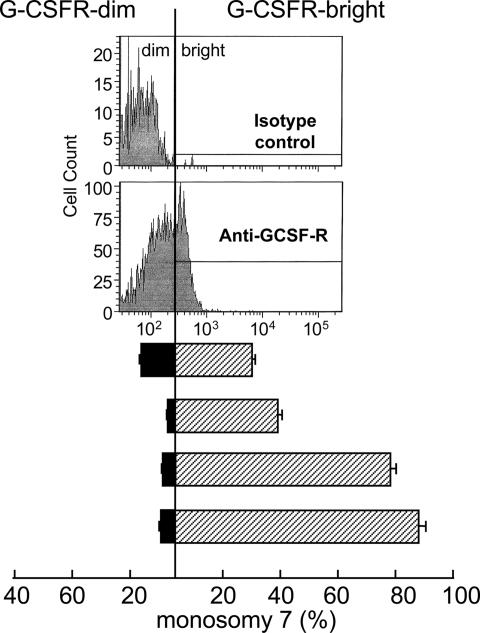
We next examined GCSFR expression on cells from monosomy 7 patients by using flow cytometry. Because hematopoietic cells increase expression of GCSFR as they mature, we specifically studied flow-cytometrically purified bone marrow CD34 cells. When CD34 cells were sorted on the basis of GCSFR expression, the percentage of cells with monosomy 7 assessed by FISH was significantly increased in the fraction of cells with higher expression of the receptor (Fig. 2).
Fig. 2.
GCSFR protein is increased on monosomy 7 CD34 cells. When CD34 cells from monosomy 7 patients were sorted into GCSFR-negative and -positive fractions by flow cytometry and FISH was performed, the monosomy 7 cells were predominantly in the GCSFR-positive population.
CD34 Cells Expressing GCSFR Isoform IV Are Increased in Monosomy 7 Patients.
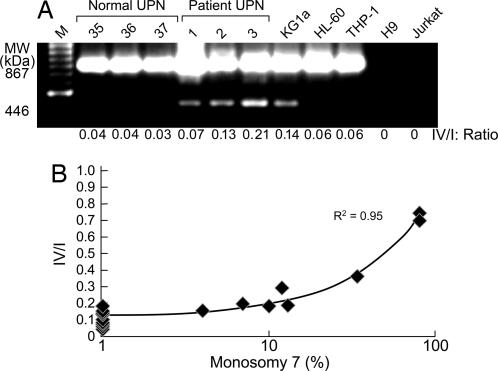
Mutations in the genomic DNA encoding the GCSFR, as have been described in SCN (15), or alternative splicing of the mRNA, as occurs in leukemia (16), could account for altered sensitivity to GCSF, especially if the genetic alteration were to remove the distal cytoplasmic domain responsible for myeloid lineage differentiation. The nucleotide sequences of the GCSFR of two patients and of a monosomy 7 cell line showed several polymorphisms but no mutations (see Table 7, which is published as supporting information on the PNAS web site). We then examined GCSFR mRNA, assaying specifically for the differentiation-defective isoform IV, which lacks 87 aa at the C terminus as a result of alternative splicing. Overexpression of this isoform has been described in some patients with leukemia (17) and has been implicated in disordered myeloid maturation. Monosomy 7 CD34 cell mRNA was extracted and amplified. PCR products were electrophoresed, and band intensities were quantified and compared with those of CD34 cells from 10 normal donors and 6 patients with MDS with normal cytogenetics. Analysis of relative amounts of GCSFR isoforms in six monosomy 7 patients with MDS showed an increased ratio of isoform IV relative to isoform I in the monosomy 7 cell population (see example in Fig. 3A) but not in four patients with MDS and normal cytogenetics or in six healthy controls. To confirm these findings, real-time PCR was performed on lymphocyte-depleted BMMNC from an additional seven patients with monosomy 7 MDS patients, eight healthy controls, and seven MDS patients with other cytogenetic abnormalities or normal cytogenetics. The numbers of monosomy 7 cells correlated with increased relative expression of isoform IV (Fig. 3B).
Fig. 3.
Bone marrow CD34 cells from patients with monosomy 7 express abnormal amounts of the shortened isoform IV of GCSFR. RNA from CD34 cells from monosomy 7 patients was subjected to DNase treatment (Invitrogen, Carlsbad, CA) followed by oligo(dT)-primed RT-PCR using the SuperScript First Strand synthesis system for RT-PCR (Invitrogen). GCSFR isoform amplification was performed as described in ref. 17. (A) Samples from three healthy volunteers, three patients, and cell lines KGi1, HL60, and THP-1, showing increased expression of shortened isoform IV. (B) When real-time PCR was performed on BMMNC, as described in Materials and Methods, an increased ratio of GCSFR-IV/I was observed that was proportional to the number of monosomy 7 cells in the bone marrow sample.
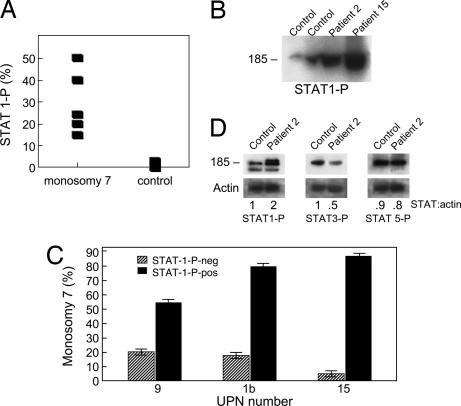
STAT1-P and STAT5-P Are Up-Regulated in CD34 Cells of Patients with Monosomy 7.
In experimental mouse models and in children with SCN, abnormal GCSFRs have been associated with defects in internalization, resulting in membrane GCSFR overexpression and a hyperproliferative cellular response (18). Transfected murine cells overexpressing class IV and SCN cells expressing the truncated GCSFR show constitutive STAT1 phosphorylation and an increased ratio of STAT5-P/STAT3-P. To determine whether STAT expression was similarly disordered in monosomy 7 MDS, we measured STAT1-P expression in CD34 cells from patients with significant proportions of monosomy 7 cells in the bone marrow, as determined by flow cytometry (Fig. 4A). Immunoblot verified the overexpression of STAT1-P in monosomy 7 patients (Fig. 4B). Similarly, when BMMNC were sorted by flow cytometry into CD34-positive fractions containing STAT1-P-positive and -negative fractions, the STAT1-P-positive fraction contained most of the monosomy 7 cells (Fig. 4C). Next, we stimulated BMMNC from monosomy 7 patients and compared levels of STAT1-P, STAT3-P, and STAT5-P with those of normal controls. GCSF-stimulated BMMNC demonstrated increased STAT1-P, decreased STAT3-P, and increased STAT5-P compared with normal by immunoblot (Fig. 4D), and confirmed on flow cytometry studies (Table 3).
Fig. 4.
STAT P expression is abnormal in monosomy 7. Unstimulated BMMNC were stained with CD34-PE and anti-STAT1-P mAbs. (A) When CD34 cells from monosomy 7 cells not subjected to GCSF stimulation were stained with STAT1-P-FITC mAb, there was increased STAT1-P expression in patients compared with controls. (B) Constitutive expression of STAT1-P in monosomy 7 patients is shown on immunoblot. (C) When CD34 cells were sorted by flow cytometry and FISH was performed, there was a preponderance of monosomy 7 cells in the STAT1-P+ group, indicating constitutive expression of this protein in monosomy 7 cells. (D) When BMMNC from two healthy donors, four MDS patients without monosomy 7, and three patients with monosomy 7 were stimulated with GCSF (400 ng/ml) for 30 min, immunoblotting showed increased expression of STAT1-P and decreased STAT3-P (example shown).
Table 3.
Monosomy 7 BMMNC constitutively express STAT1-P and have an increased STAT5/STAT3 ratio
| Patient no. | % monosomy 7 | % STAT1-P expression | STAT5-P/STAT3-P ratio |
|---|---|---|---|
| 4 | 9 | 14 | 6.6 |
| 2 | 65 | 30 | 20 |
| 15 | 68 | 24 | 26 |
| 13 | 7 | 4 | 2.6 |
| MDS* mean (n = 5) | 2 | 2 | 0.8 |
| Normal (n = 10) | 0 | 1 | 0.12 |
CD34 cells from monosomy 7 patients were stained for STAT1-P and STAT5-P and examined by flow cytometry. The cells demonstrated constitutive expression of STAT1-P and an increased STAT3/STAT5 ratio consistent with abnormalities in GCSFR signal transduction. Patients with small numbers of monosomy 7 cells showed STAT-P and isoform values similar to those for normal controls.
*Denotes MDS with normal cytogenetics.
Discussion
Here, we show that GCSF does not induce the karyotypic abnormality of monosomy 7 in patients with bone marrow failure; rather, high concentrations of GCSF favor the expansion of preexisting monosomy 7 clones. This conclusion is based on our ability to demonstrate that the survival and proliferation of monosomy 7 cells were inferior to those of diploid cells at very low GCSF concentrations. These results suggest that the GCSFR, which mediates proliferation, maturation, and survival of myeloid progenitor cells, was abnormal in monosomy 7. Although genomic DNA GCSFR was normal in monosomy 7 cells, there was an increase in the class IV GCSFR isoform (which lacks the membrane-distal region of the receptor responsible for differentiation). The class IV GCSFR isoform, like the GCSFR in SCN, lacks the same 87 C-terminal amino acids that are present in the full-length receptor; however, the class IV isoform differs from the GCSFR in SCN because this segment is replaced by a shorter, novel 34-aa sequence. The class IV isoform is the product of alternative splicing, which is responsible for tissue-specific variation in the receptor. Myeloid blasts with normal genomic GCSFR DNA also express an increased amount of isoform IV, again resulting from alternative splicing of the mRNA (17). In murine CD34 cells transfected with the isoform IV gene, relatively small increases in the ratio of isoform IV relative to full-length isoform I cause substantial defects in maturation without affecting proliferation (which is controlled by the membrane-proximal cytoplasmic domain of the receptor) (19). Our data are consistent with those of other investigators who report that the class IV isoform results in reduced, rather than increased, proliferative signaling in BAF3 and 32D cells at GCSF concentrations <100 ng/ml (20), with decreased survival at lower concentrations of GCSF and increased proliferation at concentrations >200 ng/ml (21). In monosomy 7, increased class IV GCSFR isoform expression was comparable to, or greater than, that measured in leukemia and in the murine model (17, 21). The pattern of STAT activation we observed in monosomy 7 was similar to that described in SCN (22–24), consistent with functional dominance of the class IV over the class I isoforms. The apparent functional dominance of the class IV isoform in monosomy 7 cells in leukemic blasts and in transfected murine cells could be explained if, like the truncated form of GCSFR seen in SCN (18), the class IV isoform failed to normally internalize following GCSF binding. Both the leukemic class IV isoform and the truncated SCN GCSFR lack the dileucine residue required for normal internalization (21), and defective internalization of GCSFR in response to GCSF binding is well described in SCN (18).
Monosomy 7 cells with up-regulated membrane GCSFR would be expected to have a survival advantage compared with diploid cells because forced cell surface expression of the mutated GCSFR of SCN (25) or of isoform IV (26) confers resistance to apoptosis by up-regulation of Akt (27, 28). If GCSF only facilitates proliferation of a stable, preexisting monosomy 7 clone, a decline in monosomy 7 cells should occur at lower GCSF levels. In support of this hypothesis, two of our patients showed substantial decreases in the number of monosomy 7 bone marrow cells (one to a fully normal karyotype) following clinical responses to immunosuppressive therapy (unpublished data); others have reported similar declines in monosomy 7 after successful treatment (29) or on withdrawal of GCSF administration (3, 30). Previous studies demonstrated a dosage effect in gene transcription in trisomy 8 and monosomy 7 (31, 32). Multiple genes on chromosome 7 regulate myeloid proliferation. CDP/cut or CCAAT displacement is a transcription factor, located on the long arm of chromosome 7, which indirectly down-regulates membrane GCSFR expression by negatively regulating C/EBP-ε (33, 34). Down-regulation of CDP/cut would be expected to be accompanied by up-regulation of C/EBP-ε, which in turn increases GCSFR expression (35).
In conclusion, monosomy 7 cells are abnormally sensitive to high concentrations of GCSF. Our findings link a GCSF-mediated advantage for monosomy 7 cells to an overexpressed isoform of the GCSFR. The abnormalities of proliferation and differentiation closely mimic those of SCN, which has defects in the membrane distal portion of the GCSFR. Although this study reveals similarities between SCN and monosomy 7, the mechanism of GCSFR class IV overrepresentation in monosomy 7 remains unknown. Our data strongly suggest a mechanism in which GCSF expands an existing monosomy 7 clone; however, we cannot exclude the possibility that GCSF facilitates genomic instability with longer exposure. Nevertheless, our findings should be reassuring to physicians who administer this agent to healthy donors of blood products and to patients not experiencing bone marrow failure.
Materials and Methods
Patients.
Bone marrow was obtained from 35 patients with MDS (as defined in ref. 36): 28 with monosomy 7, 10 with AA with normal cytogenetics, and 10 healthy adult volunteers. All individuals gave written informed consent, in accordance with protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute.
Cell Preparation.
BMMNC were aspirated from the posterior iliac crest into syringes containing media supplemented 1:10 with heparin (O'Neill and Feldman, St. Louis, MO) and prepared by density gradient centrifugation using lymphocyte separation medium (Organon, Durham, NC). Duplicate aliquots of freshly isolated BMMNC were placed in Myelocult (StemCell Technologies, Vancouver, Canada) in the presence of growth factors (37). In some instances, cells were plated in methylcellulose with similar growth factor support. At 4, 7, and 14 days later, colonies were counted, individual colonies were picked, cells were lysed, and each colony's nuclei were subjected to FISH for chromosome 7.
FISH.
FISH was performed as described in ref. 37, using probes for chromosome 7 (Vysis, Downers Grove, IL). Percent positive staining was based on counting of 400 cells assessed. Three different observers, who were blinded with respect to sample identity, scored three different sets of slides. Scores were averaged, and the mean of the three was recorded.
Sequence Analysis of the GCSFR Coding Regions.
Cryopreserved BMMNC samples from patients confirmed by FISH to contain monosomy 7 in >50% of the cell populations were thawed. One million cells were subjected to lymphocyte depletion by using Dynabeads M-450 CD3 (Pan T) (Dynal, Oslo, Norway) in accordance with the manufacturer's instructions. Depletion yielded 4 × 105 cells, which were used to isolate genomic DNA by using the QIAmp DNA blood mini kit (Qiagen, Valencia, CA). To analyze for the presence of mutations, PCR was performed with the primers listed in Table 4. Because of the limited number of cells, nested PCR was performed to generate sufficient quantities of template for sequencing. Initially, a region encompassing exons 4–6 was amplified using outer primers F1-1/R1-1, followed by second-round PCR using inner primers F1-2/R1-2. Similarly, exons 7–10 were amplified using F2-1/R2-1 and F2-2/R2–2; exons 11 and 12 were amplified using F3-1/R3-1 and F3-2/R3-2; and, finally, exons 13–17 were amplified using primers F4-1/R4-1 and F4-2/R4-2. PCR amplification conditions using the DNA thermal cycler 480 (Perkin-Elmer, Foster City, CA) were as follows: 1 cycle at 95°C for 1 min; followed by 35 cycles of 95°C for 30 s, 52°C for 50 s, and 72°C for 1 min, with a 10-s increase per cycle; and ending with 1 cycle of 72°C for 5 min. Second-round PCR was then performed. Secondary PCR samples were electrophoresed on 1% agarose gels and stained with ethidium bromide to assess the purity and size of the DNA fragments; samples were subsequently purified by using the QIA Quick PCR purification kit (Qiagen). We amplified and then directly sequenced all exons of the GCSF receptor by using the BigDye Terminator cycle sequencing kit (Version 3.1; Applied Biosystems, Foster City, CA) and the ABI Prism 3100 genetic analyzer (Applied Biosystems). The oligonucleotide primers used for sequencing are listed in Table 5.
Table 4.
Primers for real-time PCR
| Gene | Designation | Sequence (5′ → 3′) |
|---|---|---|
| ABL | Forward primer | GATACGAAGGGAGGGTGTACCA |
| Reverse primer | CTCGGCCAGGGTGTTGAA | |
| TaqMan probe | TGCTTCTGATGGCAAGCTCTACGTCTCCT | |
| GCSFR1 | Forward primer | GGATCCGGGTCCATGGG |
| Reverse primer | TTAAGAGGCAGGCCCAAGAAG | |
| TaqMan probe | AACCCCAGGAAGCCCTA | |
| GCSFR4 | Forward primer | CAGCCCCAATCCCAGTCT |
| Reverse primer | GGAGCATGATCTGGTCCTTAAAGT | |
| TaqMan probe | TCAGGCTGGGCCTCC |
Table 5.
Primer sequences for GCSFR sequencing
| Regions | PCR primers | Sequence primers |
|---|---|---|
| I | F1-1 (8512): CTT GAA GGG TGA GAC AGG AAG GTG | FS1-A (8552): GGA GTA CAA AGG CCT AGA TTA TAG C |
| R1-1 (10991): CTT CTG TGC CTC TGT CTC TAG GTC | FS1-B (10295): CAG ACA AGG TGA CAA CAG AGG AAC | |
| 8529–10970 | F1-2 (8529): GAA GGT GAG GCC TCT GAG TCT AGG | RS1-A (9024): GAA TAC AGG CGT GAG CCA ACG TGC |
| R1-2 (10970): G TCT TCC AGT GTC TTC TTG TCT GTC | RS1-B (10948): GT CGC TAG TTC TGT GTT TCC CTC TCC | |
| II | F2-1 (11477): CAT AGA GAT CGA ACC ACA GAA ACG | FS2-A (11527): GAA CCA CAG ACA GGG AGA CAA GAG |
| R2-1 (10034): GAA CTC CTG ACC TCG GAT GAT CTG | FS2-B (12560): CTC CGT TTG TAT GAT CCA TCC ACC | |
| 11499–13000 | F2-2 (11499): CGT GTC ATT GGC AAC GTG GAC AGA TG | RS2-A (12332): CTC CCA GAC CTG TTG GAG TCC TAA G |
| R2-2 (13000): CCT CCC AAA GTG CTG GGA TTA CAG | RS2-B (12950): GGA CTA GAT TT AAC CCA GGC AGT C | |
| III | F3-1 (14307): CTA CTT TAC AGA TGA GGA AAC TGA G | FS3-A (14381): CAG GAT TTG AAC CCA GGC TTC TGG |
| R3-1 (15308): CTG TAG GGA TCC AGT GTA AGC CAG | ||
| RS3-A (15234): GTT CCC TAT ACT TCT GAT TGC TGG | ||
| 14332–15263 | F3-2 (14332): GCT AAG ATG TTG ACC TAC CAA AGG | |
| R3-2 (15263): CAG GGC TGG AAG TAT GGT AGG AAG | ||
| F4-1 (15917): GAT GGA ATC TGG ATT AAA TCC CTG G | FS4-A (15997): GTG TCT GGG AAG CCA CAA GAA GTC | |
| IV | R4-1 (18330): GAA GAA AGG ACC AAC TCT GAG AAG | FS4-B (16561): GGA CTG ACT TTG AAT CCC CTG GTC |
| FS4-C (17362): CAC CAG TAA CAA CAG TTG AAT GTC | ||
| 15962–18314 | F4-2 (15962): GAG TCC CCT TGG ACT CTT TTC TCA C | |
| R4-2 (18314): CTG AG AAG TTT CCC CCT GAC TGC TG | RS4-A (16553): CTC TCA AAA TCA GCA TCC TTT GGG | |
| RS4-B (17146): GCT TCA GAA GGT GTC CCT TCA CTG | ||
| RS4-C (18276): AAA TAA CAG AGA CTC AGG TAC ACC |
Measurement of GCSFR Isoforms.
For initial semiquantitation of GCSFR isoforms, RNA samples were extracted from column-purified CD34 cells and subjected to DNase treatment (Invitrogen, Carlsbad, CA) followed by oligo(dT)-primed RT-PCR using the SuperScript first strand synthesis system for RT-PCR (Invitrogen). GCSFR isoform amplification was performed as described in ref. 17. PCR products were electrophoresed on 1% agarose gels and stained with ethidium bromide to assess the purity and size of the DNA fragments and were subsequently imaged for densitometry calculations. Gel images were captured with the ChemiImager 5500 system (Alpha Innotech, San Leandro, CA), and quantification of band intensities was determined by using AlphaEaseFC software (Alpha Innotech).
For real-time PCR measurement of the GCSFR-I and GCSFR-IV isoforms, RNA was treated with DNase I (Invitrogen) to eliminate genomic DNA, and random hexamer-primed cDNA was synthesized by using the Advantage RT-for-PCR kit (Clontech, Mountain View, CA). TaqMan probe-and-primer reagents were designed by using the Custom TaqMan Gene Expression Assays File Builder software (Applied Biosystems). ABL expression, measured by using 300-nM primers and a 200-nM probe, was used as the endogenous cDNA quantity control for all samples (38). Expression of the GCSFR-I and -IV isoforms was assessed by quantitative real-time RT-PCR using the ABI PRISM 7900 sequence detection system (Applied Biosystems). All reactions were performed in triplicate on 10 μl volume under standard conditions with 40 cycles of amplification. Primers and probes for real-time PCR are listed in Table 4.
Flow Cytometry.
Intracellular staining for STAT1-P expression was performed with the Pharmingen (San Diego, CA) intracellular staining kit (14). First, double-color surface staining was performed with phycoerythrin (PE)-conjugated anti-CD34 mAb, and cells were then permeabilized by using a saponin-based method (Pharmingen) and stained with FITC-conjugated −STAT1-P mAbs (Pharmingen). When appropriate, CD34 cells were sorted with the Altra flow cytometry sorter (Beckman Coulter, Miami, FL) into STAT1-P-positive and -negative fractions for FISH. Staining of bone marrow aspirate smears, which were subsequently analyzed for the presence of monosomy 7 by using FISH, was performed as described in ref. 37.
Immunoblotting.
Minces were used for preparation of the extracts, as described in ref. 39. Proteins (10 μg per lane) were resolved in 12% Tris-glycine SDS gel (Invitrogen) electrophoresis at 125 V. Resolved proteins from the gel were transferred to PVDF membranes (Invitrogen). To evaluate equal loading of the lanes, membranes were reblotted with anti-actin polyclonal Ab. Densitometry analysis of the bands of interest was performed with ImageQuant analysis software (Amersham Biosciences, Piscataway, NJ). The anti-actin Ab and the HRP-conjugated Abs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistics.
Statistical significance was calculated by using the Mann–Whitney test. All quoted P values are from two-sided tests, with values <0.05 considered significant.
Acknowledgments
We thank Dr. Ivo Touw for helpful comments and advice.
Abbreviations
- AA
aplastic anemia
- BMMNC
bone marrow mononuclear cells
- GCSF
granulocyte colony-stimulating factor
- GCSFR
GCSF receptor
- MDS
myelodysplastic syndrome
- SCN
severe congenital neutropenia
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Young NS. Blood. 1992;79:1385–1392. [PubMed] [Google Scholar]
- 2.Rosenfeld R, Follman D, Nunez O, Young N. J Am Med Assoc. 2003;289:1130–1135. doi: 10.1001/jama.289.9.1130. [DOI] [PubMed] [Google Scholar]
- 3.Maciejewski J, Risitano A, Sloand EM, Nunez O, Young NS. Blood. 2002;99:3129–3135. doi: 10.1182/blood.v99.9.3129. [DOI] [PubMed] [Google Scholar]
- 4.Hashino S, Imamura M, Tanaka J, Kobajashi S, Musashi M, Kasai M, Asaka M. Ann Hematol. 1996;72:337–339. doi: 10.1007/s002770050183. [DOI] [PubMed] [Google Scholar]
- 5.Kojima S, Ohara A, Tsuchida M, Kudoh T, Hanada R, Okimoto Y, Kaneko T, Takano T, Ikuta K, Tsukimoto I. Blood. 2002;100:786–790. doi: 10.1182/blood.v100.3.786. [DOI] [PubMed] [Google Scholar]
- 6.Germeshausen M, Schulze H, Kratz C, Wilkens L, Repp R, Shannon K, Welte K, Ballmaier M. Leukemia. 2005;19:611–617. doi: 10.1038/sj.leu.2403663. [DOI] [PubMed] [Google Scholar]
- 7.Touw IP. Baillieres Clin Haematol. 1997;10:577–587. doi: 10.1016/s0950-3536(97)80027-3. [DOI] [PubMed] [Google Scholar]
- 8.Young NS. Blood. 2002;100:741. [Google Scholar]
- 9.Locasciulli A, Arcese W, Locatelli F, Di Bona E, Bacigalupo A. Lancet. 2001;357:43–44. doi: 10.1016/s0140-6736(00)03574-1. [DOI] [PubMed] [Google Scholar]
- 10.Kearns WG, Sutton JF, Maciejewski JP, Young NS, Liu JM. Am J Hematol. 2004;76:220–224. doi: 10.1002/ajh.20101. [DOI] [PubMed] [Google Scholar]
- 11.Omori F, Okamura S, Shimoda K, Otsuka T, Harada M, Niho Y. Biotherapy. 1992;4:147–153. doi: 10.1007/BF02171759. [DOI] [PubMed] [Google Scholar]
- 12.Kavgaci H, Ozdemir F, Aydin F, Yavuz A, Yavuz M. J Exp Clin Cancer Res. 2002;21:475–479. [PubMed] [Google Scholar]
- 13.Touw IP, Dong F. Leuk Res. 1996;20:629–631. doi: 10.1016/0145-2126(96)00017-3. [DOI] [PubMed] [Google Scholar]
- 14.Watari K, Asano S, Shirafuji N, Kodo H, Ozawa K, Takaku F, Kamachi S. Blood. 1989;73:117–122. [PubMed] [Google Scholar]
- 15.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 16.Dong F, van Paassen M, van Buitenen C, Hoefsloot LH, Lowenberg B, Touw IP. Blood. 1995;85:902–911. [PubMed] [Google Scholar]
- 17.White SM, Ball ED, Ehmann WC, Rao AS, Tweardy DJ. Leukemia. 1998;12:899–906. doi: 10.1038/sj.leu.2401062. [DOI] [PubMed] [Google Scholar]
- 18.Ward AC, van Aesch YM, Schelen AM, Touw IP. Blood. 1999;93:447–458. [PubMed] [Google Scholar]
- 19.White SM, Alarcon MH, Tweardy DJ. Blood. 2000;95:3335–3340. [PubMed] [Google Scholar]
- 20.Dong F, van Buitenen C, Pouwels K, Hoefsloot LH, Lowenberg B, Touw IP. Mol Cell Biol. 1993;13:7774–7781. doi: 10.1128/mcb.13.12.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White SM, Alarcon MH, Tweardy DJ. Blood. 2000;95:3335–3340. [PubMed] [Google Scholar]
- 22.de Koning JP, Dong F, Smith L, Schelen AM, Barge RM, van der Plas DC, Hoefsloot LH, Lowenberg B, Touw IP. Blood. 1996;87:1335–1342. [PubMed] [Google Scholar]
- 23.Dong F, Liu X, de Koning JP, Touw IP, Henninghausen L, Larner A, Grimley PM. J Immunol. 1998;161:6503–6509. [PubMed] [Google Scholar]
- 24.Dong F, Qiu Y, Yi T, Touw IP, Larner AC. J Immunol. 2001;167:6447–6452. doi: 10.4049/jimmunol.167.11.6447. [DOI] [PubMed] [Google Scholar]
- 25.Zhu QS, Robinson LJ, Roginskaya V, Corey SJ. Blood. 2004;103:3305–3312. doi: 10.1182/blood-2003-06-1861. [DOI] [PubMed] [Google Scholar]
- 26.Hunter MG, Avalos BR. Blood. 2000;95:2132–2137. [PubMed] [Google Scholar]
- 27.Hermans MH, Antonissen C, Ward AC, Mayen AE, Ploemacher RE, Touw IP. J Exp Med. 1999;189:683–692. doi: 10.1084/jem.189.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aarts LH, Roovers O, Ward AC, Touw IP. Blood. 2004;103:571–579. doi: 10.1182/blood-2003-07-2250. [DOI] [PubMed] [Google Scholar]
- 29.Socie G, Rosenfeld S, Frickhofen N, Gluckman E, Tichelli A. Semin Hematol. 2000;37:91–101. [PubMed] [Google Scholar]
- 30.Nishimura M, Yamada T, Andoh T, Tao T, Emoto M, Ohji T, Matsuda K, Kameda N, Satoh Y, Matsutani A, et al. Int J Hematol. 1998;68:203–211. doi: 10.1016/s0925-5710(98)00047-4. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Zeng W, Miyazato A, Billings E, Maciejewski JP, Kajigaya S, Sloand EM, Young NS. Blood. 2004;104:4210–4218. doi: 10.1182/blood-2004-01-0103. [DOI] [PubMed] [Google Scholar]
- 32.Schoch C, Kohlmann A, Dugas M, Kern W, Hiddemann W, Schnittger S, Haferlach T. Leukemia. 2005;19:1224–1228. doi: 10.1038/sj.leu.2403810. [DOI] [PubMed] [Google Scholar]
- 33.Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N. Proc Natl Acad Sci USA. 2001;98:8000–8005. doi: 10.1073/pnas.141229598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu K, Kitabayashi I, Kamada N, Abe T, Maseki N, Suzukawa K, Ohki M. Blood. 2000;96:288–296. [PubMed] [Google Scholar]
- 36.Bennett JM. Int J Hematol. 2000;72:131–133. [PubMed] [Google Scholar]
- 37.Sloand EM, Kim S, Fuhrer M, Risitano AM, Nakamura R, Maciejewski JP, Barrett AJ, Young NS. Blood. 2002;100:4427–4432. doi: 10.1182/blood-2002-01-0096. [DOI] [PubMed] [Google Scholar]
- 38.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, et al. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 39.Solomou EE, Keyvanfar K, Young NS. Blood. 2006;107:3983–3991. doi: 10.1182/blood-2005-10-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]