Abstract
Rho-associated coiled-coil protein kinase 1 (ROCK-1) is a direct cleavage substrate of activated caspase-3, which is associated with heart failure. In the course of human heart failure, we found marked cleavage of ROCK-1 resulting in a 130-kDa subspecies, which was absent in normal hearts and in an equivalent cohort of patients with left ventricular assist devices. Murine cardiomyocytes treated with doxorubicin led to enhanced ROCK-1 cleavage and apoptosis, all of which was blocked by a caspase-3 inhibitor. In addition, a bitransgenic mouse model of severe cardiomyopathy, which overexpresses Gq protein and hematopoietic progenitor kinase-/germinal center kinase-like kinase, revealed the robust accumulation of the 130-kDa ROCK-1 cleaved fragment. This constitutively active ROCK-1 subspecies, when expressed in cardiomyocytes, led to caspase-3 activation, indicating a positive feed-forward regulatory loop. ROCK-1-dependent caspase-3 activation was coupled with the activation of PTEN and the subsequent inhibition of protein kinase B (Akt) activity, all of which was attenuated by siRNA directed against ROCK-1 expression. Similarly, ROCK-1-null mice (Rock-1−/−) showed a marked reduction in myocyte apoptosis associated with pressure overload. These data suggest an obligatory role for ROCK-1 cleavage in promoting apoptotic signals in myocardial hypertrophy and/or failure.
Keywords: heart failure, left ventricle assist device, phosphatase and tensin homolog deleted on chromosome ten
Heart failure is an eventual outcome for diverse cardiovascular disorders and the leading cause of combined morbidity and mortality in the United States and other developed industrial nations (1). Diverse signal transduction pathways, G proteins and protein kinases among them, likely contribute to heart failure, and the identification of essential control points have both fundamental and translational importance (2, 3). Recent findings suggest a role for the activation of the apoptotic cascade in heart failure, which may involve the activation of proteolytic caspase-3 and cardiomyocyte loss (1, 4). Although the level of apoptosis detected in the failing heart are variable (4–6), a low prevalence of apoptosis is sufficient to cause cardiac contractile depression (7). Accounting for the most conservative rate of cardiomyocyte death, the normal heart would lose most of its mass in a few decades, but the senile and failing heart lose myocytes in a matter of several months to a few years (8). This dilemma raised the issue of the imbalance between the continual loss of cardiomyocytes and the long interval for the chronic progression in heart failure. There are critical deficiencies in the available information regarding the relationship between apoptosis in the failing heart and depressed contractile function. Other mechanisms might contribute to heart failure besides cell loss. For example, we showed that activated caspase-3 mediated the cleavage of serum response factor (SRF). Cleaved SRF became a dominant negative factor that down-regulated SRF target genes, many of which are contractile proteins (9). Therefore, the identification of other caspase-specific substrates may yield new insights into apoptosis-related cardiac depression.
Cellular membrane blebbing, a hallmark of cellular apoptosis, has led to the identification of Rho-associated coiled-coil protein kinase 1 (ROCK-1), one of only two isoforms that gets constitutively activated by caspase-3 cleavage in fibroblasts (10–12). ROCK is a downstream mediator of RhoA and is believed to play a critical role in mediating the effects of RhoA on stress fiber formation, smooth muscle contraction, cell adhesion, membrane ruffling, and cell motility (13, 14). The Rho/ROCK pathway is an attractive candidate in the pathogenesis of heart failure, with a potential role in both ventricular remodeling and impaired peripheral vascular resistance (15, 16). We asked whether endogenous ROCK-1 was cleaved in heart disease, and if so, what the functional consequences were. Myocardial ROCK-1 was found to be a natural substrate for caspase-3 cleavage in human heart failure and in genetic cardiomyopathy mouse models. Here, we observed truncated ROCK-1-facilitated caspase-3 activation, which led to a feed-forward loop promoting apoptosis. Loss-of-function studies in cultured cardiomyocytes with ROCK-1 siRNA significantly decreased cardiac apoptosis induced by ceramide. In ROCK-1-null mouse mutants (ROCK-1−/−), we observed reduced apoptosis in response to pressure overload. Furthermore, cleaved ROCK-1 also led to the activation of PTEN (phosphatase and tensin homolog deleted on chromosome ten) and subsequent inhibition of the protein kinase B (Akt) survival pathway. Caspase-3-mediated ROCK-1 cleavage is a functionally important activator of the apoptotic pathway associated with cardiac overload and/or failure.
Results
Demographics.
Table 1 shows the demographics of the human population studied, in which all 23 patients were in the New York Heart Association functional class III or IV with a left ventricle ejection fraction of <20%. Ten patients underwent placement of left ventricle assist device (LVAD) support for progressive cardiac deterioration (despite maximal i.v. inotropic support) and impending end-organ failure. The mean duration of LVAD support was 41 ± 10 days (range 14–91 days).
Table 1.
Demographics of human heart failure patients
| Patient without LVAD | Age/sex | Diagnosis | Patient with LVAD | Age/sex | Diagnosis | Implant duration, days |
|---|---|---|---|---|---|---|
| 1 | 63/M | ICM | 1 | 49/M | ICM | 21 |
| 2 | 60/M | ICM | 2 | 66/M | DCM | 90 |
| 3 | 69/M | ICM | 3 | 61/M | DCM | 35 |
| 4 | 54/M | ICM | 4 | 48/F | DCM | 15 |
| 5 | 56/F | DCM | 5 | 60/F | CDM | 14 |
| 6 | 54/M | DCM | 6 | 57/M | DCM | 21 |
| 7 | 69/M | DCM | 7 | 44/F | DCM | 91 |
| 8 | 41/M | DCM | 8 | 70/M | ICM | 14 |
| 9 | 53/F | DCM | 9 | 44/M | DCM | 61 |
| 10 | 67/M | ICM | 10 | 42/M | DCM | 43 |
| 11 | 54/M | ICM | ||||
| 12 | 64/M | ICM | ||||
| 13 | 52/F | HCM |
ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; M, male; F, female.
ROCK-1 Cleaved in Human Failing Hearts and Rescued by Ventricular Mechanical Support.
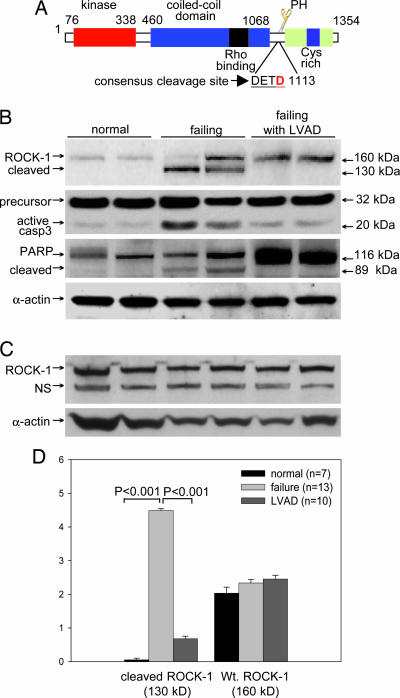
Because caspase-3 was activated in human heart failure (9, 17), and the consensus recognition sequence DETD (at amino acid position 1113) for caspase-3 cleavage was a conserved sequence in ROCK-1 mammalian species (Fig. 1A), we asked whether ROCK-1 cleavage occurred in failing human hearts. Ventricular myocardial samples were collected from 13 patients with end-stage heart failure at the time of transplant, 7 patients who died of noncardiac causes (control group), and 10 patients with end-stage heart failure who were being maintained on LVAD support awaiting transplantation (Fig. 1B). Protein blot analysis detected a ROCK-1 fragment of 130 kDa in all failing hearts without LVAD support, corresponding to the predicted ROCK-1 cleavage product, in addition to the 160-kDa native protein. In contrast, normal hearts and failing hearts with LVAD support contained only the intact 160-kDa protein. As Chang et al. (9) and Narula et al. (17) reported, a significant increase in caspase-3 activity was observed in nontreated failing hearts when compared with either the normal hearts or the failing hearts treated with LVAD. The cleavage of poly(ADP-ribose) polymerase (PARP), a known caspase-3 substrate, was also observed (Fig. 1B). Thus, increased caspase-3 activity in failing hearts was accompanied by the cleavage of ROCK-1 and the reciprocal appearance of a ROCK-1 subspecies that corresponded to the predicated cleavage product. To further confirm that the 130-kDa fragment was cleaved ROCK-1, we used a second antibody specific against the C-terminal end of ROCK-1 (epitope maps between residues 1300–1354). Only the 160-kDa full-length ROCK-1 was detected in the hearts of all patient groups (Fig. 1C). Densitometry analyses of ROCK-1 cleavage pooled from several Western blots for all of the 13 heart failure patients, 10 LVAD patients, and 7 normal controls were summarized in Fig. 1D. Statistical analysis showed a significant (P < 0.001) difference between failing heart and normal heart samples for the presence of the cleaved subspecies. Reduced amounts of ROCK-1 cleavage were apparent in patients placed on LVAD (P < 0.001), whereas ROCK-1 cleavage in the LVAD group did not appear differently from the control cohorts (Fig. 1D). No significant differences were observed among all groups of patients for levels of the native full-length ROCK-1 protein. Moreover, we did not observe any differences in ROCK-1 cleavage with regard to the etiology of heart failure.
Fig. 1.
ROCK-1 cleaved in human failing hearts. (A) A schematic diagram of ROCK-1 structural domains. The caspase-3 consensus cleavage sequence (DETD, amino acids 1110–1113) is conserved among ROCK-1 genes in mammals. Cleavage of ROCK-1 removes the inhibitory C terminus, generating a 130-kDa fragment with constitutive kinase activity. (B) Representative Western blots of human myocardial samples. ROCK-1 protein levels were assessed from myocardial samples taken from patients with end-stage heart failure with and/or without LVAD treatment and from normal hearts. An increase in caspase-3 activity was observed in human failing hearts, which returned to control levels after LVAD treatment. Cleavage of PARP, a known caspase-3 substrate during apoptosis, is also shown as an internal control. Even protein loading for PAGE was shown by a blot probed with a specific antibody against α-actin. (C) A specific antibody directed against the C-terminal end of ROCK-1 detected only full-length ROCK-1 in all patient groups. NS, nonspecific. (D) Densitometric analysis of cleaved ROCK-1 and full-length ROCK-1 levels were taken from two to three replicates that covered 13 heart failure patients, 10 LVAD patients, and 10 noncardiac disease control patients. The statistical analysis showed a significant difference (P < 0.001) in comparison between failing myocardium samples and normal myocardium samples in the cleaved ROCK-1 subspecies (130 kDa).
ROCK-1 Was Cleaved by Caspase-3 in Apoptotic Cardiomyocytes and Genetic Models of Murine Heart Failure.
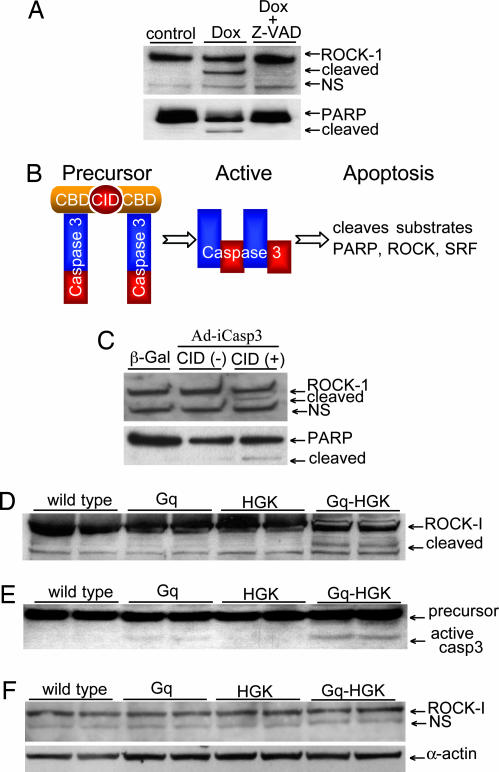
Neonatal rat cardiomyocytes were subjected to doxorubicin treatment, a potent apoptotic trigger, at 10 μM for 20 h. The cleaved 130-kDa ROCK-1 subspecies appeared in doxorubicin-treated myocytes, which also paralleled the cleavage of PARP (Fig. 2A). Cleavage of ROCK-1 and PARP was blocked by a caspase-3 inhibitor, Z-VAD-fmk. Next, caspase-3 activity was induced by introducing dimerizer-dependent caspase-3 via adenovirus, Ad-G/iCasp 3 (Fig. 2B) (18). The adenovirus expresses a caspase-3 precursor fusion gene linked in-frame with a modified FK506-binding domain and chemical inducer binding domain, the binding site for a chemical inducer of dimerization (CID). Adding CID brings together the modified caspase-3 precursors, leading to its activation and rapid cellular apoptosis. ROCK-1 remained intact after Ad-G/iCasp 3 infection of cardiomyocytes, but after the addition of CID, ROCK-1 cleavage proceeded with the appearance of the 130-kDa subspecies, along with PARP cleavage. Thus, activated caspase-3 induced by forced dimerization caused ROCK-1 cleavage (Fig. 2C).
Fig. 2.
ROCK-1 cleaved by caspase-3 in apoptotic cardiomyocytes and in transgenic heart failure mouse models. (A) A Western blot of whole-cell lysates from controls and doxorubicin-treated (Dox) neonatal rat cardiomyocytes revealed enhanced ROCK-1 and PARP cleavage. Caspase-3 inhibitor Z-VAD-fmk blocked caspase-3 cleavage. (B) A schematic diagram of a dimerizer inducible activation system for caspase-3. An adenovirus expressed a caspase-3 precursor fused to a modified FK506-binding domain, and a chemical inducer binding domain. Dimerization of caspase-3 precursors via CID forces caspase-3 activation and rapid cellular apoptosis. (C) A Western blot of whole-cell lysates from neonatal rat cardiomyocytes showed intact ROCK-1 after caspase-3 adenovirus infection. The addition of CID led to ROCK-1 cleavage shown by the appearance of the 130-kDa species. Control adenovirus express β-gal. (D and E). To assess ROCK-1 cleavage in vivo, hearts from Gq, HGK, and bigenic Gq-HGK mice were assessed by Western blot analysis for ROCK-1 and caspase-3 activity. A 130-kDa fragment of ROCK-1 was observed only in hearts from bitransgenic mice paralleled with a significant increase in caspase-3 activity. (F) The same heart specimens assessed by a specific antibody against C-terminal ROCK-1 revealed only a full-length species of ROCK-1.
To determine whether ROCK-1 cleavage occurred in the intact heart, we took advantage of transgenic mouse lines that display cardiomyopathies. One line expresses Gq, a pivotal G protein for diverse hypertrophic signals in the heart with a predisposition to apoptosis (19). Hematopoietic progenitor kinase-/germinal center kinase-like kinase (HGK, MAP4K4), an upstream mediator of cytokine signaling, imposes little or no baseline phenotype in the heart but induces florid apoptotic cardiomyopathy in concert with Gq (M.X. and M.D.S., unpublished results). Hearts from αMHC-Gq, αMHC-HGK, and bigenic αMHC-Gq-HGK transgenic mice were analyzed by using protein immunoblots. As indicated in Fig. 2 D and E, the clipped 130-kDa ROCK-1 fragment was observed in hearts from bigenic mice, which coincided with a significant increase in caspase-3 activity. Cleaved ROCK-1 fragments were not detected in mice inheriting overexpression of only HGK or Gq transgenes, although a small increase in caspase-3 activity was measurable in Gq hearts that was probably insufficient to induce ROCK-1 cleavage. Specificity of the clipped subspecies was confirmed by a second antibody that is specific only for the C-terminal end of ROCK-1 and that revealed only the 160-kDa full-length species of ROCK-1 and not the 130-kDa cleaved species (Fig. 2F). Because increased cleaved ROCK-1 correlated well with a high level of caspase-3 activity observed in bigenic Gq-HGK mice, we then asked whether ROCKΔ1, a ROCK-1 cleavage mutant, would activate caspase-3.
ROCKΔ1 Supported Caspase-3 Activation.
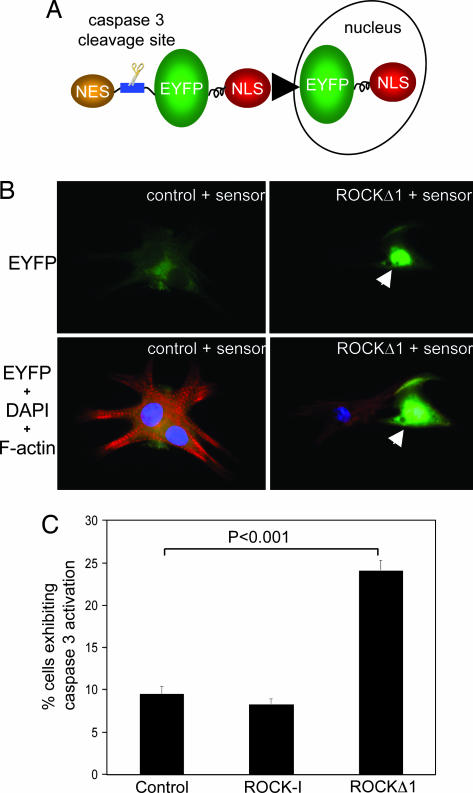
Cardiomyocytes were transfected with a ROCKΔ1 expression vector, a C-terminally truncated mutant (amino acids 1–1080), which closely corresponded to the caspase-3 cleaved activated species (amino acids 1–1113) (10, 11). A second plasmid, pCaspase-3-sensor, encoding an EYFP fusion protein (Fig. 3A), was cotransfected, which provided a visual readout of cleaved ROCK-1 after caspase-3 activation. Caspase-3-dependent cleavage was quantified by counting the number of transfected myocytes in which the fluorescent fusion proteins were translocated to nuclei. Expressed ROCKΔ1 caused a 3-fold increase of caspase-3 activity and inflicted myofilament damage, which were not observed with full-length ROCK-1 or the control expression vector (Fig. 3 B and C). Thus, the truncated, constitutively active ROCK-1 was sufficient to induce caspase-3 activation, indicative of a feed-forward regulatory loop, whereby ROCK-1 cleavage caused further caspase-3-dependent cleavage and further activation of ROCK-1.
Fig. 3.
Activated mutant, ROCKΔ1, induced caspase-3 activation and apoptosis in neonatal rat cardiomyocytes. (A) Schematic diagram of a caspase-3 sensor, which incorporated a caspase-3-specific cleavage site located between EYFP and NES. When caspase-3 is inactive, the dominant NES targets EYFP to the cytosol. After the induction of apoptosis, the export signal is removed by active caspase-3, which triggers the redistribution of EYFP from cytosol to the nucleus via NLS (nuclear localization signal). (B) Representative images showed cardiac myocytes subjected to ROCKΔ1 accumulated EYFP in myocyte nuclei indicated by arrows and developed apoptosis, as shown by disorganized myofilaments. Green, EYFP-fusion protein; red, rhodamine-conjugated phalloidin staining for F-actin; blue, DAPI nucleus staining. (C) The prevalence of caspase-3 activation was evaluated by the percentage of transfected cardiomyocytes exhibiting caspase-3 activation (nuclear localization of EYFP-fusion protein). Results are the average ± standard error of four separate experiments.
ROCK-1 Knockout Prevented Apoptosis Induced by Ceramide in Cultured Cardiomyocytes or in Mice Subjected to Systolic Overload.
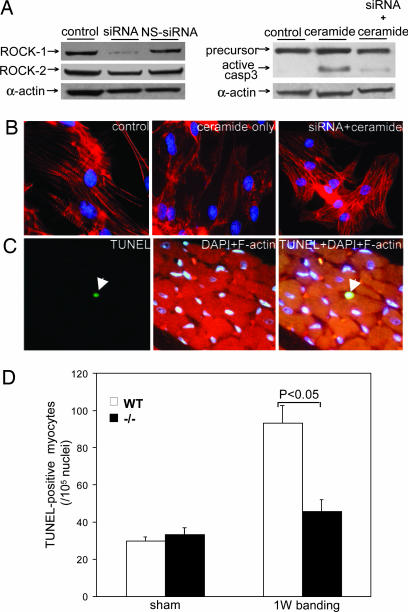
Would blocked expression of endogenous ROCK-1 protect cardiomyocytes against apoptosis induced by apoptotic stimuli? We applied a specific siRNA to significantly knock down ROCK-1 expression in cardiomyocytes without blocking expression of the other Rho kinase isoform, ROCK-2 (Fig. 4A Top Left). ROCK-1 siRNA inhibited caspase-3 activation induced by ceramide (Fig. 4A Upper Right). Fluorescent staining showed that pretreatment with siRNA-protected cardiomyocytes against ceramide-induced apoptosis and protected myofilament organization in comparison to the disorganized and damaged myofilaments in non-siRNA-treated cells (Fig. 4B). Without siRNA treatment, ceramide strongly induced caspase-3 activation along with apoptosis.
Fig. 4.
Blocked ROCK-1 expression reduced cardiomyocyte apoptosis. (A) A specific siRNA knocked down ROCK-1 expression (Top Left). ROCK-1 siRNA inhibited caspase-3 activation induced by ceramide (Upper Right). (B) Fluorescent staining showed that pretreatment with the siRNA protected cardiomyocytes from ceramide-induced apoptosis. (C) A representative ROCK-1−/− mouse myocardium image showed a TUNEL-positive cardiac myocyte revealed by TUNEL green staining after 1 week of aortic banding. (D) Comparison of TUNEL-positive cardiac myocytes (per 105 nuclei) in left ventricle myocardium from WT and ROCK-1−/− mice after 1 week of aortic banding or surgical sham. n = 5 mice for each group. NS, nonspecific; casp3, caspase-3. Red, phalloidin staining for F-actin; blue, DAPI nucleus staining.
To substantiate the role of ROCK-1 in this proteolytic cascade process in vivo, ROCK-1-null mice, which are viable (15), were subjected to severe pressure overload as a pathophysiological signal for apoptosis (15, 20). Consistent with the siRNA results, we found that ROCK-1−/− mice exhibited significantly reduced TUNEL-positive cardiac myocytes (46 myocytes per 105 nuclei) compared with WT mice (93 myocytes per 105 nuclei, P < 0.05) when subjected to pressure overload through aortic banding for 1 week (Fig. 4 C and D). Therefore, both in vitro and in vivo data suggest that ROCK-1 played an essential role in cardiac apoptosis, and disrupted ROCK-1 expression contributed to myocardial protection.
Caspase-3 Activation by ROCKΔ1 Induced PTEN Activation and Akt Inhibition.
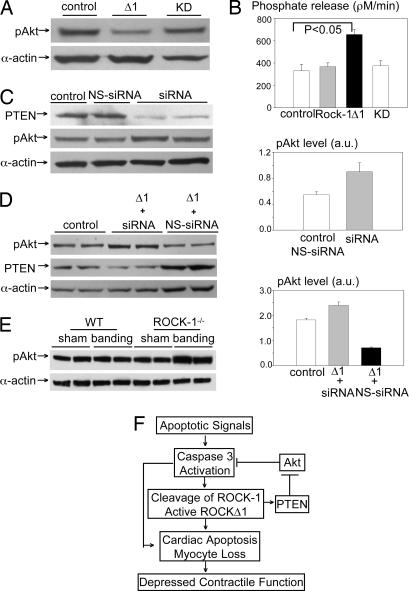
The PI3K/Akt is a central component of several receptor-mediated survival pathways (21). PTEN is a PI3K upstream negative regulator (22, 23) and is phosphorylated by ROCK-1 in vitro (24). In addition, bioactive lipid sphingosine 1-phosphate activates PTEN via a Rho GTPase-dependent pathway (25). To elucidate a mechanism involved in ROCK-1-dependent caspase activation, we tested for potential changes in the PI3K/Akt cell survival pathway (21) and for PTEN activity after transfection of ROCKΔ1 expression vectors into human HEK cells. ROCKΔ1 decreased the phospho-Akt (pAkt) level by 30% (Fig. 5A) and increased PTEN activity by 2-fold in comparison with the control (P < 0.05). No change in activity was found with either full-length ROCK-1 or a kinase-deficient mutant, ROCK-1KD expression vectors (Fig. 5B). Therefore, the repression of Akt activity by ROCKΔ1 may contribute to its proapoptotic effect through increased PTEN activity. This hypothesis was substantiated by a 65% increase in the basal phosphorylation level of Akt after blocked PTEN expression by siRNA treatment (Fig. 5C). In addition, decreased pAkt activity caused by ROCKΔ1 was reversed by blocked PTEN expression (Fig. 5D). Furthermore, the levels of pAkt showed a 55% increase in ROCK-1-null mouse hearts when subjected to 1 week of aortic banding compared with the banded WT hearts (Fig. 5E). Therefore, the increase in PTEN activity and the subsequent decrease in Akt activity (dephosphorylation) may contribute to the proapoptotic effect of ROCKΔ1.
Fig. 5.
Regulation of PTEN and Akt by ROCK-1. (A) HEK cells were transfected with ROCKΔ1 and its kinase mutant KD (kinase-deficient mutant ROCK-1). Cellular lysates were assessed for phospho-Akt (pAkt) by Western blot analysis. (B) PTEN activities were assessed by Malachite green assay after cells were transfected with full-length ROCK-1 and its mutants. (C) PTEN and pAkt were detected by Western blots after cells were treated by PTEN-specific siRNA. Densitometry analysis for pAkt is shown at Right after normalization by actin expression. (D) In cotransfection with PTEN-specific siRNA and ROCKΔ1, PTEN and pAkt levels were analyzed. The expression levels were normalized to actin and are shown at Right. All experiments were conducted in human HEK cells. Δ1, active Rho kinase ROCK-1; a.u., artificial unit; NS-siRNA, nonspecific siRNA. (E) Levels of pAkt were assessed by Western blot analysis in hearts from WT and ROCK-1−/− mice subject to either sham surgery or 1 week aortic banding pressure overload. (F) A proposed model outlining the role of caspase-3 cleavage activation of ROCK-1 on cardiac dysfunction. ROCK-1 may be the leading target for activated caspase-3 in failing myocardium. Cleavage-activated ROCK-1 kinase generated a positive feed-forward loop for caspase cascade activation. This proapoptotic effect is associated with the activation of PTEN and the dephosphorylation of Akt.
Discussion
ROCK-1 is one of two isoforms, ROCK-1 and ROCK-2, of a serine/threonine kinase identified as effectors of Rho GTPase proteins, which mainly include RhoA, Rac1, and Cdc42. They regulate a wide variety of cellular processes including apoptosis (26, 27). The potential proapoptotic effect of Rho kinase has been suggested by others (28), and pharmacologic inhibition of Rho kinase significantly reduced the level of myocyte apoptosis induced by ischemia/reperfusion in an isolated perfused rat heart preparation (29). However, a limitation of these studies is the low specificity of pharmacological Rho kinase inhibitors that fail to distinguish ROCK-1 from ROCK-2 as well as other kinases (30–32). By contrast, a genetic approach shown here readily revealed specific roles for ROCK-1 versus ROCK-2. Highly selective blockades of ROCK-1 expression in cultured cardiomyocytes by application of siRNA, as well as loss-of-function studies of the intact mouse heart revealed cardiac protection against apoptosis. ROCK-1 deficiency supported an essential role of ROCK-1 cleavage and activation in facilitating myocyte apoptosis associated with cardiac overload and/or failure.
Rho kinase's proapoptotic role may be mediated via regulation of actin cytoskeletal rearrangement, which in turn induced the activation of the caspase cascade via the assembly of the death-inducing signaling (28, 29, 33, 34). In addition, we demonstrated that constitutively active ROCK-1 generated by cleavage was sufficient to activate caspase-3 and lead to myocyte apoptosis, as outlined in a schematic model (Fig. 5F). Thus, we provided evidence that cleaved activated ROCK-1 directly activated the caspase cascade machinery.
Consistent with our observation, other ROCK-1 mutant mice were also viable, with no detectable cardiac phenotype under physiological conditions (35, 36). Furthermore, null ROCK-1 mouse mutants did not impair the compensatory hypertrophic response induced by pressure overload but developed less perivascular and interstitial fibrosis (15), a finding supported by a recent study in which the ROCK-1 haploid-insufficient mice showed less perivascular fibrosis after hypertrophic stimuli (36).
The precise relationship among the multiple factors controlling apoptosis in cardiac overload, hypertrophy, and failure may depend on the fundamental initiating factors. It is obvious that increased apoptosis precedes the onset of cardiac dysfunction in a discrete systolic overload model. Other models beginning with genetic induction of myocardial dysfunction are followed by systolic overload resulting from cardiac dilation. What is evident is that augmented apoptosis contributes to the cleaved activated ROCK-1 that plays a synergistic role in cardiac myocyte apoptosis. Cleaved activated ROCK-1 in failing hearts may be one of the mechanisms that facilitates myocyte apoptosis and initiates a further feed-forward activation of caspase-3. Inhibition or deletion of ROCK-1 appears to abort this proteolytic cycle. The therapeutic potential of these observations is attractive because the inhibition of ROCK-1 may be relatively safe.
Materials and Methods
Source of Human Myocardium.
Failing myocardium came from 13 patients with end-stage heart failure at the time of transplant. An additional group of 10 patients had been maintained on LVAD until transplant. Samples obtained from 7 patients who died of noncardiac causes were used as a control group. All of the myocardium was obtained from the left ventricle apex. The Institutional Review Board of Baylor College of Medicine approved the protocol.
Cell Culture, Plasmid Constructs, and Recombinant Adenoviruses.
Neonatal rat cardiomyocytes were isolated and cultured in DMEM/F12 medium with 10% horse serum (DF10). For immunostaining, cells were cultured on precoated (with 0.2% gelatin) coverslips. Transfections were performed by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Human embryonic kidney (HEK) A293T cells were cultured in DMEM with 10% FBS. The cDNA for full-length human ROCK-1 cloned into the pCAG-myc vector (pCAG-ROCK-1) was kindly provided by Shuh Narumiya (Kyoto University, Kyoto, Japan). The Asp718–MscI cDNA fragment encoding ROCKΔ1 mutant (residues 1–1080) was cloned into the pCAG-myc vector (pCAG-ROCKΔ1) (37). A single lysine at residue 105 was replaced by alanine to generate a kinase-deficient mutant, pCAG-ROCK-1KD. The adenoviral vector, Ad-G/iCasp 3, expressing inducible caspase-3, was generously provided by David Spencer and Kevin Slawin (Baylor College of Medicine). Addition of CID, AP20187 (50 nM, 24 h; ARIAD, Cambridge, MA), provoked the aggregation and activation of caspase-3 along with rapid apoptosis (18).
Apoptosis Assay and Immunofluorescence Analysis.
Apoptosis was evaluated in caspase-3 activity and poly(ADP-ribose) polymerase (PARP) cleavage by Western blot analysis. With regards to apoptotic cultured neonatal cardiomyocytes, a caspase-3 sensor (BD Biosciences Clontech, Mountain View, CA) was introduced to detect the onset of caspase-3 activity (Fig. 3A). The vector encodes EYFP (enhanced yellow fluorescent protein) fused with NES (nuclear export signal) and NLS (nuclear localization signal). A caspase-3-specific cleavage site is located between EYFP and NES. When caspase-3 is inactive, the dominant NES directs EYFP to the cytosol. When apoptosis is induced, the export signal is removed by active caspase-3, which triggers the redistribution of EYFP from the cytosol to the nucleus via NLS, and is quantified by counting the number of transfected cells by the nuclear localization of fluorescent fusion protein over the total transfected cells. Alexa Fluor 594 phalloidin staining (Invitrogen Molecular Probes, Carlsbad, CA) and DAPI staining were applied for F-actin and cellular nuclei visualization, respectively.
Animal Models with Different Apoptotic Levels.
Three transgenic mouse lines with different apoptotic levels were used. HGK was overexpressed in mouse myocardium by using the αMHC promoter (38). Gerald Dorn (University of Cincinnati Medical Center, Cincinnati, OH) kindly provided αMHC-Gq mice (19). Bigenic mice overexpressing Gq-HGK mice were generated by breeding HGK with Gq mice. All experiments were performed in 10-week-old mice with an isogenic FVB/N background. No early lethality resulted from cardiac overexpression of exogenous HGK and Gq alone.
Application of siRNAs for ROCK-1 and PTEN Knockdown and Ceramide-Induced Apoptosis.
To knock down ROCK-1 expression in neonatal rat cardiomyocytes, 100 nM siRNA (Ambion, Austin, TX) was used with cotransfection of caspase-3 sensor vector as described above. The same amount of siRNA specific for PTEN (Ambion) was applied to human HEK cells. Cells were treated with ceramide for 40 h after transfection at 50 μg/ml for 2 h to induce apoptosis. For each specific siRNA knockdown, three different siRNAs were tested. Two of the effective siRNAs were chosen to conduct each experiment.
ROCK-1 Knockout Mice.
Generation and characterization of ROCK-1−/− mice have been described by Zhang et al. (15).
Transverse Aortic Banding.
Transverse aortic banding was conducted in 12-week-old adult WT and ROCK-1−/− mice, N ≥ 6 mice for each group (39). Briefly, both ROCK-1−/− and WT mice received a comparable load, based on the right-to-left carotid artery flow velocity ratio (from 5:1 to 10:1) after constricting the transverse aorta. As a control, a sham operation without occlusion was performed on respective age-matched littermate mice.
Western Blot and Malachite Green Assay.
The antibodies purchased were anti-phospho-Aktser473 (Cell Signaling Technology, Danvers, MA), anti-PTEN (Upstate, Lake Placid, NY), anti-ROCK-1 (N-terminal, epitope maps between residues 755 and 840), caspase-3, PARP, and α-actin (Santa Cruz Biotechnology, Santa Cruz, CA). To verify the cleaved ROCK-1, an anti-ROCK-1 C-terminal antibody (epitope maps between residues 1300 and 1354) was applied (Bethyl Laboratories, Montgomery, TX). Protein samples for Western blot analysis were prepared and separated as described in ref. 40. Even loadings were confirmed by an antibody probed for α-actin. PTEN activity was evaluated by Malachite green assay, used to measure the rate of phosphate release (Upstate). Five hundred micrograms of cell lysate protein was used for immunoprecipitation.
Statistical Analyses.
We analyzed all data by using one-way ANOVA (SigmaStat; SYSTAT, Richmond, CA). A P < 0.05 was considered significant. Data are presented as mean ± SEM.
Acknowledgments
We thank Drs. David Spencer and Kevin Slawin for supplying adenoviral vector Ad-G/iCasp 3; Shuh Narumiya for supplying ROCK-1 vector; Dr. Gerald W. Dorn II for supplying Gq mice; and Ms. Susan Tang and Mr. Weitao Song for providing technical assistance. This work was supported by American Heart Association Scientist Development Grant 0335155N (to J.C.); American Heart Association Scientist Development Grant 0130015N (to L.W.); and National Institutes of Health Grants R01-HL72897 (to L.W.), P01-HL49953 (to R.J.S.), R01-HL64356 (to R.J.S.), and P01-HL42550 (to M.L.E.).
Abbreviations
- ROCK-1
Rho-associated coiled-coil protein kinase 1
- HGK
hematopoietic progenitor kinase-/germinal center kinase-like kinase
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
- Akt
protein kinase B
- pAkt
phospho-Akt
- LVAD
left ventricle assist device
- PARP
poly(ADP-ribose) polymerase
- CID
chemical inducer of dimerization
- EYFP
enhanced yellow fluorescent protein
- NES
nuclear export signal.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Narula J, Arbustini E, Chandrashekhar Y, Schwaiger M. Cardiol Clin. 2001;19:113–126. doi: 10.1016/s0733-8651(05)70198-3. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN, Schneider MD. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 3.Foo RS, Mani K, Kitsis RN. J Clin Invest. 2005;115:565–571. doi: 10.1172/JCI24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narula J, Haider N, Virmani R, DiSalvo TG, Kolodgie FD, Hajjar RJ, Schmidt U, Semigran MJ, Dec GW, Khaw BA. N Engl J Med. 1996;335:1182–1189. doi: 10.1056/NEJM199610173351603. [DOI] [PubMed] [Google Scholar]
- 5.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara JA, Quaini E, Di Loreto C, Beltrami CA, Krajewski S, et al. N Engl J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 6.Reed JC, Paternostro G. Proc Natl Acad Sci USA. 1999;96:7614–7616. doi: 10.1073/pnas.96.14.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, Shirani J, Armstrong RC, Kitsis RN. J Clin Invest. 2003;111:1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Circ Res. 2003;92:139–150. doi: 10.1161/01.res.0000053618.86362.df. [DOI] [PubMed] [Google Scholar]
- 9.Chang J, Wei L, Otani T, Youker KA, Entman ML, Schwartz RJ. Circulation. 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- 10.Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 11.Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 12.Ueda H, Morishita R, Itoh H, Narumiya S, Mikoshiba K, Kato K, Asano T. J Biol Chem. 2001;276:42527–42533. doi: 10.1074/jbc.M102529200. [DOI] [PubMed] [Google Scholar]
- 13.Amano M, Fukata Y, Kaibuchi K. Exp Cell Res. 2000;261:44–51. doi: 10.1006/excr.2000.5046. [DOI] [PubMed] [Google Scholar]
- 14.Riento K, Ridley AJ. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YM, Bo J, Taffet GE, Chang J, Shi J, Reddy AK, Michael LH, Schneider MD, Entman ML, Schwartz RJ, Wei L. FASEB J. 2006;20:916–925. doi: 10.1096/fj.05-5129com. [DOI] [PubMed] [Google Scholar]
- 16.Kishi T, Hirooka Y, Masumoto A, Ito K, Kimura Y, Inokuchi K, Tagawa T, Shimokawa H, Takeshita A, Sunagawa K. Circulation. 2005;111:2741–2747. doi: 10.1161/CIRCULATIONAHA.104.510248. [DOI] [PubMed] [Google Scholar]
- 17.Narula J, Pandey P, Arbustini E, Haider N, Narula N, Kolodgie FD, Dal Bello B, Semigran MJ, Bielsa-Masdeu A, Dec GW, et al. Proc Natl Acad Sci USA. 1999;96:8144–8149. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shariat SF, Desai S, Song W, Khan T, Zhao J, Nguyen C, Foster BA, Greenberg N, Spencer DM, Slawin KM. Cancer Res. 2001;61:2562–2571. [PubMed] [Google Scholar]
- 19.Adams JW, Sakata Y, Davis MG, Sah VP, Wang Y, Liggett SB, Chien KR, Brown JH, Dorn GW., II Proc Natl Acad Sci USA. 1998;95:10140–10145. doi: 10.1073/pnas.95.17.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh H, Wang SC, Prahash A, Sano M, Moravec CS, Taffet GE, Michael LH, Youker KA, Entman ML, Schneider MD. Proc Natl Acad Sci USA. 2003;100:5378–5383. doi: 10.1073/pnas.0836098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantley LC. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 22.Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Backx PH. J Mol Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Schwartzbauer G, Robbins J. J Biol Chem. 2001;276:35786–35793. doi: 10.1074/jbc.M102479200. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, Wu D. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez T, Thangada S, Wu MT, Kontos CD, Wu D, Wu H, Hla T. Proc Natl Acad Sci USA. 2005;102:4312–4317. doi: 10.1073/pnas.0409784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall A. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 27.Van Aelst L, D'Souza-Schorey C. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 28.Lai JM, Hsieh CL, Chang ZF. J Cell Sci. 2003;116:3491–3501. doi: 10.1242/jcs.00660. [DOI] [PubMed] [Google Scholar]
- 29.Bao W, Hu E, Tao L, Boyce R, Mirabile R, Thudium DT, Ma XL, Willette RN, Yue TL. Cardiovasc Res. 2004;61:548–558. doi: 10.1016/j.cardiores.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, Narumiya S. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 31.Ishizaki T, Uehata M, Tamechika I, Keel J, Nonomura K, Maekawa M, Narumiya S. Mol Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- 32.Breitenlechner C, Gassel M, Hidaka H, Kinzel V, Huber R, Engh RA, Bossemeyer D. Structure (London) 2003;11:1595–1607. doi: 10.1016/j.str.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Lai JM, Wu S, Huang DY, Chang ZF. Mol Cell Biol. 2002;22:7581–7592. doi: 10.1128/MCB.22.21.7581-7592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrache I, Crow MT, Neuss M, Garcia JG. Biochem Biophys Res Commun. 2003;306:244–249. doi: 10.1016/s0006-291x(03)00945-8. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu Y, Thumkeo D, Keel J, Ishizaki T, Oshima H, Oshima M, Noda Y, Matsumura F, Taketo MM, Narumiya S. J Cell Biol. 2005;168:941–953. doi: 10.1083/jcb.200411179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rikitake Y, Oyama N, Wang CY, Noma K, Satoh M, Kim HH, Liao JK. Circulation. 2005;112:2959–2965. doi: 10.1161/CIRCULATIONAHA.105.584623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 38.Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 39.Hartley CJ, Taffet GE, Reddy AK, Entman ML, Michael LH. ILAR J. 2002;43:147–158. doi: 10.1093/ilar.43.3.147. [DOI] [PubMed] [Google Scholar]
- 40.Chang J, Knowlton AA, Wasser JS. Am J Physiol. 2000;278:R209–R214. doi: 10.1152/ajpregu.2000.278.1.R209. [DOI] [PubMed] [Google Scholar]