Abstract
To successfully propagate and cause disease, pathogenic bacteria must modulate their transcriptional activities in response to pressures exerted by the host immune system, including secreted immunoglobulins such as secretory IgA (S-IgA), which can bind and agglutinate bacteria. Here, we present a previously undescribed flow cytometry-based screening method to identify bacterial genes expressed in vitro and repressed during infections of Vibrio cholerae, an aquatic Gram-negative bacterium responsible for the severe diarrheal disease cholera. We identified a type IV mannose-sensitive hemagglutinin (MSHA) pilus that is repressed specifically in vivo. We showed that bacteria that failed to turn off MSHA biosynthesis were unable to colonize the intestines of infant mice in the presence of S-IgA. We also found that V. cholerae bound S-IgA in an MSHA-dependent and mannose-sensitive fashion and that binding of S-IgA prevented bacteria from penetrating mucus barriers and attaching to the surface of epithelial cells. The ability of V. cholerae to evade the non-antigen-specific binding of S-IgA by down-regulating a surface adhesin represents a previously undescribed mechanism of immune evasion in pathogenic bacteria. In addition, we found that repression of MSHA was mediated by the key virulence transcription factor ToxT, indicating that V. cholerae is able to coordinate both virulence gene activation and repression to evade host defenses and successfully colonize intestines.
Keywords: mannose-sensitive hemagglutinin, repression, secretory IgA, toxT
The Gram-negative bacterium Vibrio cholerae is normally found in association with plankton in surface water. It is also the causative agent of cholera, a devastating diarrheal disease that affects millions of people in the world each year (1). Environmental persistence and infection of human hosts pose very different challenges for V. cholerae. In its estuarine and riverine reservoirs, Vibrios must express proteins capable of mediating attachment to and use of nutritive substrates and promoting persistence in the face of nutrient limitation and temperature and osmolaric stress (2). In the host, V. cholerae, and indeed all other mucosal pathogens, must contend with many host factors excluding them from the epithelium, especially secreted immunoglobulins and a thick glycocalyx of mucins (3). The ability of V. cholerae to adapt to these different challenges shapes its potential as a human pathogen.
Extensive studies have demonstrated that the ability of V. cholerae to colonize and cause disease depends on the expression of a number of virulence factors during infection. This process is highly coordinated and is directly controlled by ToxT, a member of the AraC family of transcriptional regulators. toxT transcription is induced upon entry of V. cholerae into the host upper intestines (4) and regulated by the ToxRS and TcpPH protein complexes in response to environmental signals (5). Key virulence genes up-regulated by ToxT include those for the toxin-coregulated pilus (tcpA), necessary for colonization of the small bowel, and those encoding the cholera toxin (ctxAB), responsible for the diarrhea associated with acute cholera. However, the vast bulk of transcriptional changes taking place in V. cholerae as it enters the lumen of the human host is undefined.
We hypothesized that V. cholerae, in addition to activating a set of virulence genes to facilitate colonization of the host, also may specifically repress genes to evade the effects of the innate immune system, which is initially the greatest barrier to colonization of the intestine by enteric pathogens. We developed a FACS-based screening method to identify V. cholerae genes that are expressed in vitro and repressed during infection of the infant mouse, a model mammalian host. We found that repression of genes responsible for the synthesis of a type IV mannose-sensitive hemagglutinin (MSHA) pilus was critical for V. cholerae infection. We show that repression of this structure allows V. cholerae to evade nonspecific interactions with glycan moieties on one of the primary host mucosal immune factors, secretory IgA (S-IgA), thus facilitating colonization of the intestinal epithelium. We further demonstrate that the virulence regulator ToxT mediated the repression of msh expression under in vitro virulence-inducing conditions (6), suggesting that V. cholerae is able to employ both positive and negative transcriptional regulation to shift from a marine microorganism to a successful human pathogen.
Results
Screening for V. cholerae Genes Expressed in Vitro and Repressed in Vivo.
We hypothesize that V. cholerae requires the repression of certain genes to transition from environmental growth to host infection. To identify V. cholerae-repressed genes in vivo, we constructed a V. cholerae strain containing a PLtetO-1-controlled gfp and subsequently introduced a mariner transposon bearing a promoterless tetR gene. TetR is able to tightly repress the LtetO-1 promoter (7). Thus, gfp expression is repressed if the transposon inserts in an active promoter and derepressed when this promoter is inactive. The resulting transposon library was screened for GFP− cells (tetR is expressed in such mutants) after in vitro growth by using FACS sorting. These isolated bacteria then were inoculated intragastrically into infant mice. Bacteria were isolated from the mouse intestines 12 h after infection and screened for GFP+ bacteria. We obtained ≈2,000 colonies that were GFP+ (tetR is not expressed in such mutants) in the intestine and used arbitrary PCR to identify the transposon insertion sites (8). We identified 89 genes that are highly expressed during in vitro growth and repressed during colonization (Table 1, which is published as supporting information on the PNAS web site). Among them were many genes involved in various metabolic and membrane transport pathways, which was not surprising, given the drastic differences in nutrient content in vitro and in vivo. A large set of hypothetical genes also were down-regulated in vivo. Interestingly, many genes related to bacterial motility, such as flagellar biosynthesis and chemotaxis genes, also were identified as being down-regulated in vivo in our screen. These results agree with previous reports that demonstrate that chemotaxis actually inhibits the ability of V. cholerae to colonize the small intestine of infant mice (9). Of note, our screen for genes that were expressed in LB medium and repressed in infant mice was not saturated because the library pools that were inoculated into mice were too complex, and even strains with unimpaired colonization ability would be lost randomly from the pools.
MSHA Pilus Biosynthesis Genes Were Repressed in Vivo.
From the screen described above, we identified a set of in vivo-repressed genes belonging to the msh operon (VC0398 to VC0414). This operon is required for the biosynthesis of a type IV MSHA pilus (10, 11). MSHA is required for the formation of V. cholerae monolayers before biofilm formation, playing a role in the attachment of bacteria to surfaces such as cellulose (12) and is shared by evolutionarily diverged phages as a common receptor (13, 14). This pilus appears to be constitutively expressed during in vitro growth but was thought to play no role in virulence because msh mutants, as well as WT bacteria, were shown to colonize (15, 16). To confirm that expression of the msh operon differed in vivo from in vitro, we constructed a plasmid containing Pmsh-tetR and introduced it into the V. cholerae strain harboring a chromosomal PLtetO-1-gfp. The resulting strain was grown in rich LB medium and inoculated into mice. FACS analysis indicated that all bacterial cells were GFP− in the culture, but some of the bacteria were GFP+ when grown in vivo (Fig. 1A). Note that many pieces of bacteria-sized tissue debris were observed as nonfluorescent events. However, FACS analysis of intestines from uninfected mice did not reveal a GFP+ population (data not shown). In addition, quantitative real-time PCR detected an ≈10-fold decrease in msh transcripts in vivo as compared with growth in broth (Fig. 1B). Taken together, these data indicated that the msh gene expression was down-regulated during infection.
Fig. 1.
V. cholerae msh expression is repressed during infection. (A) FACS analysis of strains contain PLtetO-1-gfp and either Pmsh-tetR or Plac-tetR grown in vitro and in vivo. Bacteria were grown in LB at 37°C or incubated in 5-day-old infant mice for 12 h and were analyzed by flow cytometry. (B) Real-time PCR analysis of msh transcription in vitro and in vivo. RNA isolated from in vitro growth and in vivo growth was normalized against 16S RNA, and the ratio of in vivo to in vitro expression was presented. The results are representative of three experiments ± SD, and representative fields are shown.
Constitutive Expression of msh Genes in V. cholerae Affects Colonization.
To investigate whether the repression of msh genes is important for V. cholerae pathogenesis, we interrupted the normal in vivo repression of MSHA and examined the effects on V. cholerae colonization. We generated a V. cholerae strain that constitutively expresses MSHA by replacing the WT msh promoter with a constitutive Plac promoter. This strain (annotated mshC) demonstrated increased mannose-sensitive hemagglutination when grown in LB (see Fig. 5A) and actively transcribed msh genes during infection, as assayed by using quantitative real-time PCR (Fig. 1). We then performed a competitive colonization assay by using infant mice, where mshC mutants were >10-fold defective in colonization (Fig. 2, column 1). Strikingly, when an mshC and WT bacterial mixture was inoculated into mice that were allowed to suckle, no mshC bacteria were recovered (Fig. 2, column 2). These data suggest that repression of msh gene expression is critical for V. cholerae colonization, and that breast milk may contain factors responsible for inhibition of colonization.
Fig. 5.
ToxT represses msh expression. (A) In vitro virulence-inducing conditions lead to loss of mannose-sensitive hemagglutination. V. cholerae strains were grown in LB or AKI medium and assayed for MSHA production by hemagglutination and TcpA production by a Western blot. Two-fold dilutions of mid-log cultures of bacteria (left to right on each image) were assayed for their ability to agglutinate washed sheep erythrocytes. Assay was repeated three times, and representative results are shown. (B) ToxT mediates repression of msh transcription. V. cholerae containing msh-lacZ transcriptional fusions were grown in LB and AKI and assayed for β-gal activity. The results are representative of three experiments ± SD. (C) ToxT represses the msh promoter in E. coli. E. coli strains harboring plasmids containing msh-lacZ and either PBAD-toxT (17) or the vector control (18) were incubated in the absence or in the presence of 0.1% arabinose until stationary phase and assayed for β-gal activity. The results are representative of three experiments ± SD.
Fig. 2.
Constitutive expression of MSHA (mshC) impairs colonization of infant mice. Infant WT and S-IgA knockout mice were infected intragastrically with mixed suspensions of WT and mshC bacteria and were either isolated from their mother or allowed to suckle. The symbols indicate mutant/WT output ratios for individual mice (n = 3–6). Output ratios have been normalized to input ratios.
To explain the colonization defect of mshC V. cholerae in the suckling mouse model, we hypothesized that interactions occur between the MSHA pilus and host protective factors found in mucosal secretions, provided to the immunologically naïve suckling mouse via breast milk. In mucosal secretions, including the colostrum, secretory immunoglobulins are the primary factors responsible for defense against pathogens (3). S-IgA is the major isotype of secreted antibody (19). S-IgA is heavily modified by N- and O-linked glycosylation, which include mannose residues (20). These glycan moieties allow S-IgA to participate in innate and adaptive immunity by expanding its binding range to include bacterial lectin-like structures, independent of an antigen-specific immune response (21). Binding of S-IgA antibodies has been shown to decrease the ability of bacteria to adhere to epithelial cells (22), and it is thought that antibody binding leads to aggregation of bacteria, promoting their clearance in mucosal secretions (3).
To determine whether S-IgA was responsible for the reduced colonization of mshC bacteria, we repeated the competitive colonization assay by using IgA−/− mice (ref. 23; Fig. 2). In these mice, which do not produce secreted IgA antibodies at the epithelium or in milk, mshC bacteria colonized almost as well as wild-type strains. The presence of milk from IgA−/− mothers did not alter this pattern of colonization. These data strongly suggest that V. cholerae represses the MSHA pilus to prevent interactions with host secretory IgA.
MSHA-Dependent Interaction of S-IgA with V. cholerae.
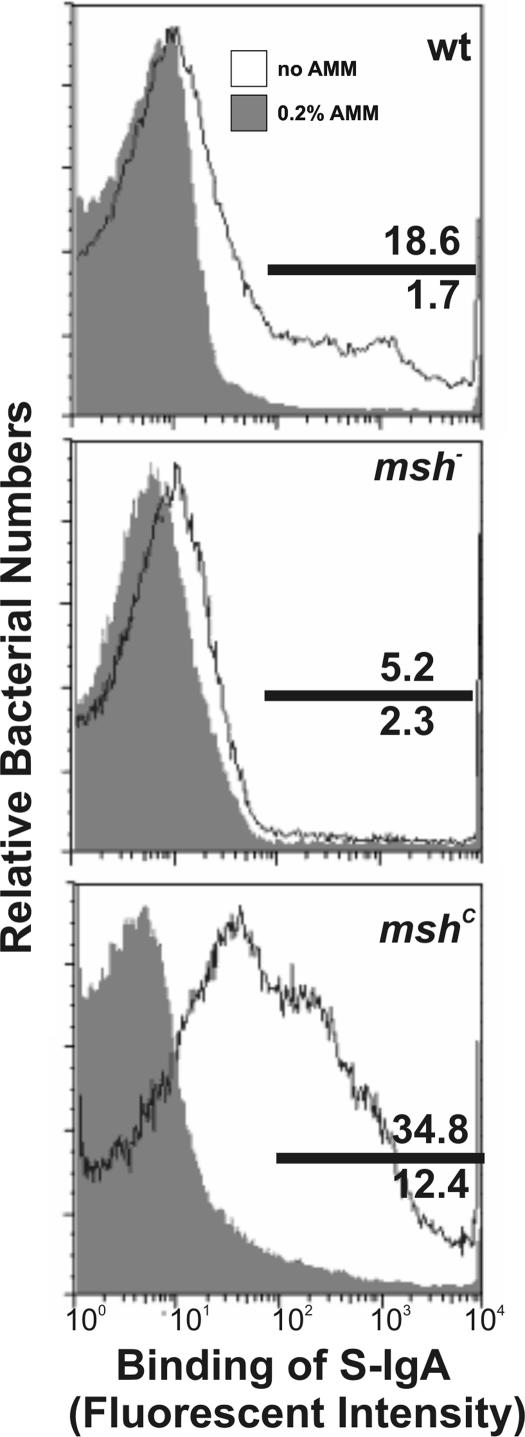
The mshC colonization assays above indicated that V. cholerae producing MSHA may interact with S-IgA. The MSHA pilus agglutinates erythrocytes in a process that can be blocked by the addition of exogenous mannose or mannose analogues, which led us to speculate that MSHA may mediate V. cholerae binding of S-IgA through interactions with the antibody glycan moieties. Precedent for such an interaction is provided by Escherichia coli type 1 pili, which bind S-IgA in a mannose-sensitive and non-antigen-specific manner (24). To assay for interactions between S-IgA and V. cholerae and to test whether this interaction is MSHA-dependent, we incubated in vitro cultures of WT bacteria, bacteria that constitutively express msh (mshC), and msh deletion mutants with purified human colostral S-IgA. Cells then were stained with goat anti-human-S-IgA IgG fraction conjugated to fluorescein isothiocyanate and analyzed via flow cytometry to reveal any binding of V. cholerae to S-IgA. Mannose-specific interactions were assayed by the addition of a nonmetabolizable mannose analogue, α-methyl-mannoside (AMM), known to disrupt the hemagglutinating activity of MSHA (25). Bacteria constitutively expressing MSHA were more highly bound by S-IgA than WT bacteria, which also express MSHA in vitro, whereas msh mutant bacteria did not bind (Fig. 3), indicating that binding of S-IgA was MSHA-dependent. The presence of 0.2% AMM abolished binding of V. cholerae to human S-IgA, indicating that binding was mannose-sensitive. Together, these data suggest that V. cholerae MSHA pili can bind mannose moieties on S-IgA.
Fig. 3.
S-IgA binding to V. cholerae is MSHA-dependent and mannose-sensitive. Mid-log V. cholerae strains were incubated with or without S-IgA for 1 h. Samples then were stained with FITC-conjugated IgG fraction anti-S-IgA in the absence (open histogram) or presence (filled histogram) of α-methyl-mannoside (AMM) and analyzed by flow cytometry. Binding of S-IgA is represented as the fluorescent intensity. Representative FACS plots are shown.
Physiological Roles of S-IgA Binding of V. cholerae.
We then investigated the physiological significance of MSHA-mediated S-IgA binding for V. cholerae pathogenesis. The importance of antigen-specific IgA in neutralizing cholera toxins has been hinted at in previous studies (26), but nonspecific binding via carbohydrate residues may play an important role in the process of immune exclusion mediated by secreted Ig.
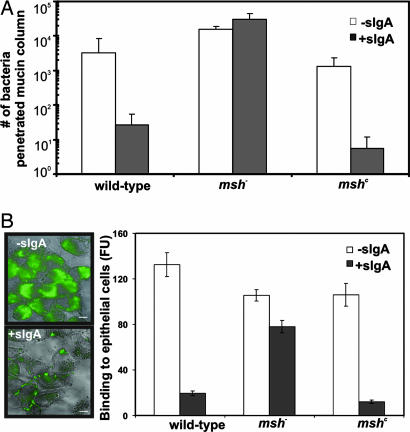
After entering the host intestine, V. cholerae need to penetrate mucus layers to colonize. Mucins may synergize with secretory antibodies to aid in the agglutination or exclusion bacteria (27). To examine the ability of S-IgA to modulate V. cholerae movement through mucous, we used a mucin penetration assay as described in ref. 28. Briefly, V. cholerae strains containing WT, mshC, or an msh deletion were incubated with and without S-IgA and layered on top of a column of bovine submaxillary mucin. Bottom fractions were collected after 20 min, and the number of bacteria penetrating the mucous layer were quantified by serial dilution. Bacteria producing MSHA (both WT and mshC) treated with S-IgA migrated more slowly through the column, whereas S-IgA binding did not affect the migration of msh− cells (Fig. 4A).
Fig. 4.
The effects of S-IgA binding on V. cholerae. (A) S-IgA inhibits passage of V. cholerae through mucous barriers. V. cholerae strains with or without S-IgA treatment were loaded on top of 500-μl mucin columns for 20 min. Fractions were collected from the bottom of the column after 20 min, and bacteria were quantified by serial dilution. (B) S-IgA inhibits binding of V. cholerae to HEp-2 epithelial cells. V. cholerae strains expressing GFP were treated with or without S-IgA and added to confluent HEp-2 cell culture grown on coverslips at multiplicity of infection = 100. Cells were then washed of unbound bacteria and examined by fluorescence microscopy. Cells also were lysed, and GFP fluorescence was measured by using a plate reader. FU, arbitrary fluorescence unit. The results are representative of three experiments ± SD; representative fields are shown.
Bacteria that penetrate the mucus layer then must adhere to the epithelium to colonize. To determine the effect of antibody binding on adherence, we incubated V. cholerae strains expressing GFP (29) with and without S-IgA on a confluent monolayer of HEp-2 epithelial cells. S-IgA-treated WT and mshC bacteria were not able to adhere to epithelial cells as efficiently as those bacteria that were not incubated with S-IgA (Fig. 4B). This process is MSHA-mediated, because the binding capacity of msh− bacteria was not affected by S-IgA. These results suggest that S-IgA can interact with bacteria that fail to repress MSHA, reducing their capacity for mucus layer penetration and attachment to the epithelial surface.
Virulence Regulator ToxT Represses msh Expression.
Our results suggest that during infection, V. cholerae is able to down-regulate msh expression, thus avoiding the effects of S-IgA binding. To examine msh regulation during infection, we examined MSHA production by using hemagglutination assays under the in vitro virulence-inducing conditions (AKI medium), which leads to the up-regulation of virulence genes in V. cholerae, including toxT, tcpA, and ctxAB (6). Under these conditions, we observed that TcpA was produced in both WT and mshC bacteria (Fig. 5A). WT bacteria could not hemagglutinate, as compared with LB-grown bacteria, whereas mshC strains did hemagglutinate (Fig. 5A). Thus, we hypothesized that the virulence regulatory machinery of the bacterium caused a loss of MSHA function. Indeed, in toxR, tcpP, or toxT mutant bacteria, mannose-sensitive hemagglutination was preserved after growth in AKI conditions (Fig. 5A and data not shown), indicating that all three virulence regulators play roles in regulation of MSHA. Because ToxR and TcpP function upstream of ToxT, these data suggested that ToxT may be responsible for repression of msh. To test whether ToxT represses the msh operon transcriptionally, we constructed a chromosomal msh-lacZ transcriptional reporter fusion in both WT and toxT mutants. The expression of msh was ≈5-fold higher in wild type grown in LB versus AKI medium, whereas in the toxT mutant, the msh expression level was higher than that of wild type in both LB and AKI (Fig. 5B), indicating that toxT represses the transcription of the msh operon. The inhibition of ToxT on msh expression is likely to be direct, because overexpression of ToxT (pBAD-toxT; ref. 17) repressed msh expression in E. coli MC4100 (Fig. 5C). Taken together, these data imply that V. cholerae can coordinately activate a set of virulence factors to facilitate colonization and repress a set of genes to avoid attack by the host mucosal innate immune system.
Discussion
V. cholerae has developed the ability to survive and colonize despite wide variations in environmental conditions. During the transition from the aquatic environment to the human body, bacterial cells retool their transcriptional profiles to adjust to the conditions of the host. Most studies in the bacterial pathogenesis field are focused on bacterial “virulence genes” whose activation during infection is critical for pathogenesis. In this study, we developed a FACS-based genetic screen to identify V. cholerae genes that are repressed during infection. Our findings, which indicate that repression of MSHA pilus production is critical for V. cholerae colonization, suggest that in vivo gene repression also plays an important role in bacterial pathogenesis.
The ability of ToxT to repress transcription of msh genes suggests a model of MSHA expression in the host in which V. cholerae coregulates the expression of virulence and colonization determinants with the repression of the MSHA pilus early in infection through transcriptional control mediated by ToxT. Repression of MSHA enables the bacterium to evade binding of S-IgA at the mucosa, allowing for colonization mediated by the toxin-coregulated pilus and other virulence factors also regulated by ToxT. Upon exit from the host and return to its environmental reservoir, V. cholerae loses virulence gene expression and reactivates the expression of genes important for persistence outside the host. Thus, the infectious cycle of this very successful human pathogen can be thought of in terms of a cycle of transcriptional regulation, mediating a set of diverse responses that are flexibly applied to respond to an ever-changing environmental milieu. Interestingly, the respiratory bacterium Bordetella bronchiseptica employs the BvgAS two-component system to enhance virulence by both activating and repressing genes during colonization (30).
MSHA pili are produced by a wide variety of V. cholerae strains (31). MSHA biogenesis and structural genes are organized as an operon on the large chromosome of V. cholerae. Although the first gene in the operon, mshH (VC0398), is not required for MSHA production in growth in LB (11), a constitutive Plac promoter inserted upstream of mshH produced more MSHA than that of WT in vitro (Fig. 5A). Previous studies have stressed the role of MSHA as that of an environmental adhesin, because it participates in a variety of functions likely to be important in the natural aquatic habitat of V. cholerae. Within the host, MSHA had appeared to play little role in pathogenesis, because V. cholerae deficient in MSHA production showed no loss of virulence compared with wild type (15, 32). However, our data demonstrate that V. cholerae unable to repress MSHA in vivo are dramatically impaired in colonization of infant mice provided with milk and that this phenotype is caused through interaction with S-IgA (Fig. 2).
Binding interactions through the MSHA pilus are sensitive to carbohydrates, which led us to hypothesize that MSHA may mediate V. cholerae binding of S-IgA through interactions with antibody glycan moieties. Indeed, V. cholerae binding to S-IgA depends on the formation of MSHA pili, and this interaction can be inhibited by the addition of exogenous mannose analogues (Fig. 3). Our data indicates that the binding of S-IgA to V. cholerae inhibits the movement of these bacteria through the protective glycocalyx of mucins at the intestinal epithelium and the adherence of bacteria to epithelial cells (Fig. 4).
Our data suggest that the key virulence regulator ToxT plays a role in MSHA repression (Fig. 5). ToxT is a member of the AraC family of transcriptional regulators, several of which can serve as both activators and repressors (33). Previous microarray analysis of WT and toxT transcriptional profiles did not detect msh repression (34). However, it is possible that microarray analysis is not sensitive enough to detect the levels of msh gene transcription and modulation by ToxT or that this is due to strain and growth condition differences. The exact mechanisms of ToxT repression awaits further investigation. Interestingly, expression of ToxT in mshC bacteria still leads to reduced levels of hemagglutination (Fig. 5A and data not shown), suggesting that in addition to directly repressing the msh expression, ToxT may initiate posttranscriptional mechanisms to inhibit MSHA production. In addition, exactly when MSHA repression occurs is still unknown, except that it occurs within the first 12 h of infection, as sampled in our screen. This result is consistent with previous reports, which indicated that virulence gene induction in the infant mouse was maximal 5 h after infection (4).
Although MSHA is transcriptionally repressed in the infant mouse model, anti-MSHA antibodies have been detected in cholera patient blood samples, suggesting that MSHA may be produced at some point during V. cholerae infection (32, 35). The expression pattern of MSHA in humans, or in later stages of infections in suckling mice, is unknown. It is possible that MSHA production resumes after colonization of the small bowel. Indeed, one study comparing the transcriptional profiles of V. cholerae in early and late stages of infection of humans suggests that virulence genes activated by ToxT are turned off late in infection (36). However, MSHA repression for evasion of the host innate immune response appears to be critical during early stages of infection, because V. cholerae penetrates the mucous layer and begins to express the virulence factors required for colonization.
Methods
Strains, Plasmids, and Culture Conditions.
V. cholerae El Tor C6706 (16) was used as the WT strain in this study. E. and V. cholerae strains were propagated in LB containing appropriate antibiotics at 37°C, unless otherwise noted. AKI media and growth conditions were used to induce virulence gene expression (6). PLtetO-1-gfp was cloned into the plasmid pCVD442 (37) and integrated into the lacZ locus of the C6706 chromosome, resulting in the strain JZV338. A mariner transposon containing the promoterless tetR gene (pJZ320) was constructed by cloning the tetR-coding sequence into pSC123 (38). Constitutive expression of the msh operon was achieved by cloning a fragment including VC0397 and VC0398 into pWM91 (39) and, subsequently, inserting a Plac promoter between VC0397 and VC0398. The resulting plasmid then was transferred into C6706 and cells selected for double crossover events. The plasmid containing Pmsh-tetR was constructed by cloning the msh promoter region (chI 422929 to 423580) fused with the tetR coding sequence into pBBR1-MCS2 (40). The msh deletion mutant was constructed by using the plasmid pHT1 as published in ref. 16. Pmsh-lacZ was constructed by cloning msh promoter region into pJZ244 (41) and then integrated into the C6706lacZ− chromosome by homologous recombination. V. cholerae strains labeled with GFP were constructed by inserting a Plac-gfp in the lacZ locus on the chromosome (28). Deletion mutants in toxR, tcpP, and toxT were constructed as described in ref. 34.
Screen V. cholerae Genes Expressed in Vitro and Repressed in Vivo.
A transposon library (JZV338::pJZ320) was inoculated into LB and grown at 37°C for 12 h. Bacteria then were washed with PBS and submitted for FACS. GFP− bacteria were collected by FACS, and the recovered bacteria then were pooled and ≈106 cells were used to infect 5-day-old infant CD-1 mice. After 12 h, mice were killed, and the intestines were homogenized. Samples were then submitted for FACS. GFP+ bacteria were collected by FACS and plated on LB agar with appropriate antibiotics. Arbitrary PCR (8) then was performed to identify transposon insertion sites. See Supporting Materials, which is published as supporting information on the PNAS web site, for detailed protocol.
RNA Extraction and Real-Time PCR.
CD-1 suckling mice were infected with V. cholerae as described above. After 12 h, mice were killed, and bacteria were recovered by homogenizing intestines. RNA from homogenates was purified by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA from bacteria grown in vitro was isolated by using the standard method described in ref. 17. RNA reverse transcription was performed by using the SuperScript II kit (Invitrogen) with 4 μg of RNA. Reactions without reverse transcriptase were performed to confirm the absence of contaminating DNA. Quantitative real-time PCR by using primers hybridized to VC0399 was performed on an ABI7700 Prism system (Applied Biosystems, Foster City, CA). 16S ribosomal RNA was used for an internal control in all reactions. Data were collected by using an Apple (Cupertino, CA) Macintosh G4 desktop computer linked to the ABI7700 system with the SDS 1.5 software.
Infant Mouse Competition Assay.
Approximately 105 C6706 (lacZ−) and mshC (lacZ+) (1:1) were inoculated intragastrically into 5-day-old CD-1, C57/B6 (Charles River Laboratories, Wilmington, MA) or C57/B6 IgA−/− mice (22). Infected pups were placed alone (unfed) or with the mother mouse (fed). Mice were killed after 20 h, and intestinal colonized bacteria were quantified as described in ref. 42. Competitive index was reported as mshC/WT and normalized for input ratio.
FACS Quantitation of S-IgA Binding.
Approximately 105 mid-logarithmic phase bacteria were collected by centrifugation and resuspended in 100 μl of Krebbs-Ringer-Tris (KRT) buffer (42). Bacteria were allowed to bind purified human colostral S-IgA (MP Biochemicals, Solon, OH) for 1 h at 37°C, added at a final concentration of 0.5 mg/ml. When indicated, α-methyl-mannoside was added for a final concentration of 0.2% wt/vol. Bacteria then were collected by centrifugation and resuspended in 100 μl of 1% BSA in PBS, containing 1:250 dilution of fluorescein-conjugated goat anti-human S-IgA IgG (MP Biochemicals). Samples were allowed to stain for 20 min at 22°C and then were analyzed by flow cytometry on a BD Biosciences (San Jose, CA) FACScan instrument.
Mucin Penetration Assay.
Mucin columns were prepared in 1-ml syringes with 500 μl of 3% bovine submaxillary mucin (wt/vol) in KRT buffer. Approximately 1 × 106 logarithmic phase bacteria were collected by centrifugation and resuspended in KRT. Purified human colostral S-IgA then was added to bacteria at a final concentration of 0.5 mg/ml and allowed to bind at 37°C for 1 h. After binding, bacteria were layered on top of mucin columns and allowed to settle for 20 min at 37°C. After settling, 100-μl fractions were collected from the bottom of the mucin columns, and bacteria were quantified by serial dilution.
Cell Culture and Quantification of Epithelial Cell Binding.
HEp-2 human epithelial cells were propagated in DMEM supplemented with 10% FBS (Sigma, St. Louis, MO) under 5% CO2. V. cholerae strains with a Plac-gfp inserted in the chromosomal lacZ locus (28) were grown to mid-log phase and resuspended in 100 μl of KRT with 0.5 mg/ml S-IgA. After incubation with antibody for 1 h at 37°C, bacterial suspensions were added to HEp-2 cells at a multiplicity of infection of 100 in KRT. Epithelial cells and bacteria then were incubated for 30 min at 37°C with 5% CO2. The media then was removed, and epithelial cells were washed three times with KRT to remove unbound bacteria. Coverslips were examined with a Leica (Deerfield, IL) fluorescent microscope and quantitation of the binding of bacteria to HEp-2 cells was measured by fluorescence per well after lysis with Triton X-100 with a Synergy HT spectrophotometer (Bio-Tek, Burlington, VT).
Hemagglutination Assays.
Mannose-sensitive hemagglutination by V. cholerae was measured as described in ref. 42. Briefly, sheep erythrocytes were washed in PBS and resuspended in KRT buffer for a final concentration of 10% vol/vol. Bacteria were grown to mid-logarithmic phase in AKI media and serially diluted in KRT buffer. Erythrocyte suspensions then were added, and samples were checked for hemagglutination after 2 h. Blood alone and MSHA-negative cultures settled to form a dark pellet; hemagglutination by V. cholerae (MSHA-positive) caused blood in the well to remain diffuse.
β-Gal Activity Assays.
V. cholerae strains containing transcriptional LacZ reporter fusions were grown at 37°C in LB or AKI media. Samples were withdrawn at indicated time points, and β-gal activity was determined as described in ref. 43.
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Flow Cytometry Core Facility (Philadelphia, PA) for aid with flow-cytometry screening and Dr. Yimin Yu (University of Pennsylvania) for providing IgA−/− knockout mice. This study was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases K22 Award AI060715 and a Penn Genomics Institute seed grant. A.H. is supported by the Bacteriology T32 training grant.
Abbreviations
- KRT
Krebbs-Ringer-Tris
- MSHA
mannose-sensitive hemagglutinin
- S-IgA
secretory IgA.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reidl J, Klose KE. FEMS Microbiol Rev. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 3.Woof JM, Kerr MA. J Pathol. 2006;208:270–282. doi: 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, Hava DL, Waldor MK, Camilli A. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 5.Krukonis ES, DiRita VJ. Curr Opin Microbiol. 2003;6:186–190. doi: 10.1016/s1369-5274(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 6.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 7.Lutz R, Bujard H. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Judson N, Mekalanos JJ. Nat Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, Butler SM, Camilli A. Proc Natl Acad Sci USA. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonson G, Lebens M, Holmgren J. Mol Microbiol. 1994;13:109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 11.Marsh JW, Taylor RK. J Bacteriol. 1999;181:1110–1117. doi: 10.1128/jb.181.4.1110-1117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watnick PI, Fullner KJ, Kolter R. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque SM, Bin Naser I, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M, Qadri F, Yamasaki S, Ghosh AN, Mekalanos JJ. J Bacteriol. 2005;187:4095–4103. doi: 10.1128/JB.187.12.4095-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jouravleva EA, McDonald GA, Marsh JW, Taylor RK, Boesman-Finkelstein M, Finkelstein RA. Infect Immun. 1998;66:2535–2539. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attridge SR, Manning PA, Holmgren J, Jonson G. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thelin KH, Taylor RK. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman LM, Belin D, Carson MJ, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macpherson AJ, Hunziker L, McCoy K, Lamarre A. Microbes Infect. 2001;3:1021–1035. doi: 10.1016/s1286-4579(01)01460-5. [DOI] [PubMed] [Google Scholar]
- 20.Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, El-Redwan RM, Wilson IA, Daha MR, Dwek RA, Rudd PM. J Biol Chem. 2003;278:20140–20153. doi: 10.1074/jbc.M301436200. [DOI] [PubMed] [Google Scholar]
- 21.Moshier A, Reddy MS, Scannapieco FA. Curr Microbiol. 1996;33:200–208. doi: 10.1007/s002849900100. [DOI] [PubMed] [Google Scholar]
- 22.Williams RC, Gibbons RJ. Science. 1972;177:697–699. doi: 10.1126/science.177.4050.697. [DOI] [PubMed] [Google Scholar]
- 23.Harriman GR, Bogue M, Rogers P, Finegold M, Pacheco S, Bradley A, Zhang Y, Mbawuike IN. J Immunol. 1999;162:2521–2529. [PubMed] [Google Scholar]
- 24.Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, Edén CS. Infect Immun. 1990;58:3073–3077. doi: 10.1128/iai.58.9.3073-3077.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorthy S, Watnick PI. Mol Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass RI, Svennerholm AM, Stoll BJ, Khan MR, Hossain KM, Huq MI, Holmgren J. N Engl J Med. 1983;308:1389–1392. doi: 10.1056/NEJM198306093082304. [DOI] [PubMed] [Google Scholar]
- Mestecky J, Russell MW. Microbial Pathogenesis and the Intestinal Epithelial Cell. Washington, DC: Am Soc Microbiol; 2003. pp. 95–112. [Google Scholar]
- 28.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguenec C, Gounon P, Phillips A, Nataro JP. J Clin Invest. 2002;110:1329–1337. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bomchil N, Watnick P, Kolter R. J Bacteriol. 2003;185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akerley BJ, Cotter PA, Miller JF. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 31.Albert MJ, Bhuiyan NA, Talukder KA, Faruque AS, Nahar S, Faruque SM, Ansaruzzaman M, Rahman M. J Clin Microbiol. 1997;35:2588–2592. doi: 10.1128/jcm.35.10.2588-2592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tacket CO, Taylor RK, Losonsky G, Lim Y, Nataro JP, Kaper JB, Levine MM. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, Mekalanos J. Proc Natl Acad Sci USA. 2003;100:2801–2806. doi: 10.1073/pnas.2628026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hang L, John M, Asaduzzaman M, Bridges EA, Vanderspurt C, Kirn TJ, Taylor RK, Hillman JD, Progulske-Fox A, Handfield M, et al. Proc Natl Acad Sci USA. 2003;100:8508–8513. doi: 10.1073/pnas.1431769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larocque RC, Harris JB, Dziejman M, Li X, Khan AI, Faruque AS, Faruque SM, Nair GB, Ryan ET, et al. Infect Immun. 2005;73:4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donnenberg MS, Kaper JB. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang SL, Mekalanos JJ. Infect Immun. 2000;68:6391–6397. doi: 10.1128/iai.68.11.6391-6397.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 40.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Hsiao A, Joelsson A, Zhu J. J Bacteriol. 2006;188:2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardel CL, Mekalanos JJ. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JH. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.