Abstract
Leptospirosis is one of the most common zoonotic diseases in the world, resulting in high morbidity and mortality in humans and affecting global livestock production. Most infections are caused by either Leptospira borgpetersenii or Leptospira interrogans, bacteria that vary in their distribution in nature and rely on different modes of transmission. We report the complete genomic sequences of two strains of L. borgpetersenii serovar Hardjo that have distinct phenotypes and virulence. These two strains have nearly identical genetic content, with subtle frameshift and point mutations being a common form of genetic variation. Starkly limited regions of synteny are shared between the large chromosomes of L. borgpetersenii and L. interrogans, probably the result of frequent recombination events between insertion sequences. The L. borgpetersenii genome is ≈700 kb smaller and has a lower coding density than L. interrogans, indicating it is decaying through a process of insertion sequence-mediated genome reduction. Loss of gene function is not random but is centered on impairment of environmental sensing and metabolite transport and utilization. These features distinguish L. borgpetersenii from L. interrogans, a species with minimal genetic decay and that survives extended passage in aquatic environments encountering a mammalian host. We conclude that L. borgpetersenii is evolving toward dependence on a strict host-to-host transmission cycle.
Keywords: genome sequence, leptospirosis, spirochete
Leptospirosis is one of the most widespread and significant zoonotic diseases in the world, with >500,000 human cases annually and a mortality rate up to 23% (1). Leptospirosis, caused by diverse species within the genus Leptospira (2), ranges in severity from a mild influenza-like disease to an acute potentially lethal infection (3). Human epidemics coincide with seasonal flooding in areas with insufficient sanitation, highlighting a need to improve disease control and prevention strategies (4). Maintenance host species rarely exhibit clinical signs of disease, whereas infection of nonmaintenance host species often results in fever, malaise, and hemorrhage, culminating in organ failure and death (5). The nearly ubiquitous distribution of pathogenic Leptospira in wild and domesticated animal species creates a persistent source of infection coupled with a direct impact on human health and livestock production (5).
Leptospira is one of several genera within the family Spirochaetales, an early branch in eubacterial evolution that, as a group, has unusual patterns of genetic organization and uses unique mechanisms of genetic regulation (6). Pathogenic Leptospira species share a common branch in evolution, distinct from saprophytic species (7, 8). Two of the largest phylogenetically distinct pathogenic species are Leptospira borgpetersenii and Leptospira interrogans, which, combined, cause most cases of leptospirosis and encompass 48% of the known 230 distinct antigenic types referred to as serovars (2). Although the clinical symptoms of infection due to these two species are similar, they are transmitted differently; L. interrogans is commonly acquired from contaminated surface water, whereas epidemiological data support a host-to-host transmission cycle for L. borgpetersenii.
To gain a better understanding of the genetic potential of Leptospira, we determined the complete genomic sequences of two strains of L. borgpetersenii serovar Hardjo, L550 and JB197. These virulent strains represent two distinct clonal subtypes (9) with different capacity to infect hamsters, which are commonly used for pathogenesis studies (5). Comparison of the L. borgpetersenii and L. interrogans genomes (10, 11) reveals substantial differences in genetic content and organization and suggests L. borgpetersenii is undergoing a process of insertion sequence (IS)-mediated genome reduction. We propose that the patterns of gene loss found in L. borgpetersenii provide a model useful for understanding early genetic events as bacteria evolve from generalists to host-dependent pathogens.
Results and Discussion
Genome Organization and General Features.
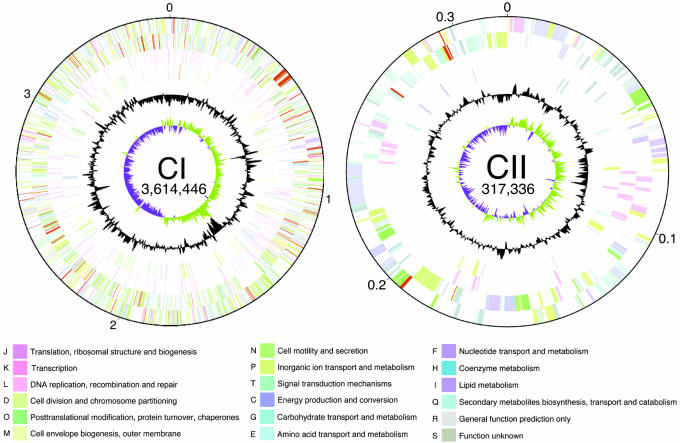
The genomes of two virulent strains of L. borgpetersenii serovar Hardjo, strains L550 and JB197, were sequenced and compared. Like other members of the genus Leptospira (10–12), L. borgpetersenii serovar Hardjo strains L550 and JB197 have two chromosomes; the large chromosomes (CI) contain 3,614,446 and 3,576,473 bp, and the small chromosomes (CII) contain 317,336 and 299,762 bp, respectively (Table 1). The putative replication origins of both CI and CII were localized by GC skews (Fig. 1). In CI, the GC skew coincides with genes common to bacterial chromosomal replication origins (e.g., dnaA and gidA; ref. 13), whereas in CII, this region is adjacent to parAB, a finding consistent with the ParAB partition proteins having a role in segregating new copies of this replicon to daughter cells (14). The CI replicons of L550 and JB197 have similar but distinct patterns of GC skew, likely because of the presence of several rearrangements.
Table 1.
Essential features of the L. borgpetersenii serovar Hardjo genome
| Feature | L550 |
JB197 |
||
|---|---|---|---|---|
| CI | CII | CI | CII | |
| Chromosome size, bp | 3,614,446 | 317,336 | 3,576,473 | 299,762 |
| G + C content, % | 40.23 | 40.16 | 40.23 | 40.43 |
| Protein coding, % | 74.0 | 73.9 | 73.8 | 74.8 |
| Functional CDS* | 2,607 | 235 | 2,540 | 230 |
| With assigned function | 1,647 | 134 | 1,594 | 131 |
| Conserved hypothetical | 363 | 31 | 363 | 30 |
| Unique hypothetical | 597 | 70 | 583 | 69 |
| Pseudogenes | 228 | 20 | 248 | 22 |
| Transposases (intact) | 114 | 7 | 121 | 5 |
| Total CDS | 2,949 | 262 | 2,909 | 257 |
| tRNAs | 37 | 0 | 37 | 0 |
| 23S rRNA | 2 | 0 | 2 | 0 |
| 16S rRNA | 2 | 0 | 2 | 0 |
| 5S rRNA | 1 | 0 | 1 | 0 |
*Not including transposases or pseudogenes.
Fig. 1.
Chromosome maps of L. borgpetersenii serovar Hardjo strain L550. Functional information for the large (CI) and small (CII) chromosomes is presented in concentric rings (maps are not drawn to scale; replicon sizes are shown in bp). Starting from the outside: genes encoded on the top strand (first ring), genes encoded on the bottom strand (second ring), transposases (third ring), and pseudogenes (fourth ring). Genes are colored according to the corresponding functional categories shown at the bottom. The fifth ring shows GC content deviations from the genomic average calculated by using a window of 10 kb in steps of 1 kb; values greater than the average fall on the outside of the ring, and values less than the average fall on the inside of the ring. The innermost ring shows GC skew calculated by using a window of 10 kb in steps of 1 kb; positive skew is shown in green, and negative skew is shown in purple.
The L. borgpetersenii CI replicon resembles a typical bacterial chromosome possessing most housekeeping genes, including those encoding all ribosomal and transfer RNA species, most ribosomal proteins, DNA synthesis, and chromosomal replication processes. As in other Leptospira spp., the rRNA genes are not organized in operons but are dispersed around the CI replicon (15).
Mobile Genetic Elements.
Mobile genetic elements, including IS elements and group II introns, collectively comprise >7% of the L. borgpetersenii genome. These elements contribute significantly to changes in genetic organization and gene function in Leptospira; IS elements often flank chromosomal rearrangements, and 17% of the pseudogenes are a result of IS-mediated disruption.
IS1533 comprises >3.5% of the L. borgpetersenii genome (16); there are 77 and 84 complete copies of IS1533 in strains L550 and JB197, respectively. Approximately 25 partial copies of IS1533 are distributed throughout each genome, providing evidence for frequent transposition and recombination events that alter genetic organization (9). For example, IS1533 insertion and subsequent recombination disrupted the single crc-like gene and created a 41-kb insertion relative to L. interrogans. In total, 15 genes in strain L550 and 25 genes in strain JB197 are disrupted by IS1533.
Fourteen new IS elements (Table 2, which is published as supporting information on the PNAS web site) were discovered in the L. borgpetersenii genome; these elements also promote pseudogene formation. At several loci, multiple copies of different IS elements are found in close proximity or are integrated with each other to generate complex loci up to 5 kb in length. Two of these loci are shared by both strains, indicating they are stable, and predate the divergence of the serovar Hardjo A and B subtypes.
A group II intron, belonging to a class of elements previously unidentified in spirochetes, is integrated within the CI replicon. Group II introns can be transferred between bacteria on conjugative elements and move from site to site within a bacterium by retrotransposition (17); their presence in L. borgpetersenii provides evidence for lateral transfer in Leptospira.
Comparative Genomics of L. borgpetersenii Strains.
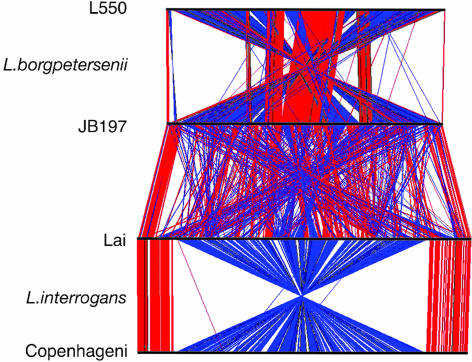
The CI replicons of L. borgpetersenii strains L550 and JB197 share extensive regions of synteny disrupted by IS-mediated rearrangements (Fig. 2). For example, recombination between copies of IS1533 ≈29 kb from the CI replication origin created a rearrangement that inverted the gene order over most of the length of the chromosome (Fig. 2). In addition, copies of IS1533 flank a unique 24-kb duplication of the S10-spc-α ribosomal protein operon in strain L550. By contrast, the CII replicons of strains L550 and JB197 are nearly collinear, except for an 18-kb insertion in strain L550 (Fig. 4, which is published as supporting information on the PNAS web site), flanked at both ends by IS elements.
Fig. 2.
Comparison of Leptospira CI replicons. The CI replicons of L. borgpetersenii serovar Hardjo strains L550 and JB197 and L. interrogans serovars Lai and Copenhageni were aligned by MegaBLAST and visualized with ACT (60). Colored lines drawn between two adjacent linearized chromosomes (horizontal black lines) show the location of homologous genes and indicate the same (red) or opposite (black) orientation relative to the chromosome immediately above. Note that the L550 CI replicon aligns with the CI replicons from either L. interrogans serovar with gene scattering similar to that shown for strain JB197.
Approximately 55% of the putative L. borgpetersenii proteins contained regions of similarity to recognized motifs that facilitated classification according to the Clusters of Orthologous Groups scheme (ref. 18; Fig. 5, which is published as supporting information on the PNAS web site). These data show both strains are similar, with the exception of translation proteins, the result of the S10-spc-α duplication in strain L550. The prominence of IS elements in L. borgpetersenii is evident in DNA recombination and repair functions associated with transposases. Approximately 12% of the L. borgpetersenii coding sequences (CDS) encode transposases (4%) or are pseudogenes or gene fragments (8%; Table 1 and Table 3, which is published as supporting information on the PNAS web site). Excluding pseudogenes or transposases, the L. borgpetersenii genome contains 2,842 (strain L550) and 2,770 (strain JB197) CDS.
L. borgpetersenii serovar Hardjo subtypes A and B establish chronic infections in cattle, their normal maintenance host, and can cause acute infections in humans but differ in their ability to infect hamsters. Strain JB197 (subtype B) causes acute lethal infections in hamsters characterized by extensive tissue hemorrhage (D.P.A. and R.L.Z., unpublished work), whereas subtype A strains capable of acute infections in hamsters have not been identified. Despite these differences, few genes unique to either strain were identified; strain L550 has 22 nonorthologous genes relative to strain JB197, whereas strain JB197 has eight nonorthologous genes relative to strain L550 (Table 4, which is published as supporting information on the PNAS web site). Most genetic diversity between strains L550 and JB197 is due to pseudogenes and genes harboring point mutations. Mutations leading to nonconservative amino acid changes occur in a wide range of L550 and JB197 genes, including those encoding thermotolerance, sensory transduction, chemotaxis, and transcriptional regulatory functions, and may account for the observed phenotypic differences between these strains.
Comparison of L. borgpetersenii to L. interrogans.
Substantial differences in the organization of the L. borgpetersenii and L. interrogans genomes (Fig. 2) are likely due to IS-mediated rearrangements. Surprisingly, most putative operons are not disrupted, but their location is altered. The L. borgpetersenii genome is 16% smaller than L. interrogans. As noted above, 12% of L. borgpetersenii CDS encode transposases or are pseudogenes or gene fragments, as compared with <4% in L. interrogans. We interpret these data as an indication that L. borgpetersenii is undergoing a process of IS-mediated genome reduction after the paradigm established in the genus Bordetella, where DNA loss through recombination events between IS elements may be the primary mechanism of speciation (19). In contrast, L. interrogans may be undergoing genome expansion by gene duplication and acquisition, as evidenced by the presence of novel genes relative to L. borgpetersenii, including potential phage genes (10, 11). We propose that simultaneous genome reduction in L. borgpetersenii and genome expansion in L. interrogans reflect differences in the environments these species traverse during transmission between hosts.
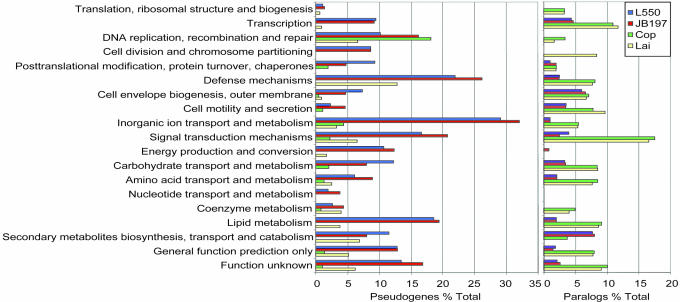
Evidence for this hypothesis came from comparison of the L. interrogans and L. borgpetersenii genomes, a process made possible only through reannotation of the two published L. interrogans genomes (see Materials and Methods), thereby providing consistent annotation criteria for both species. This comparison showed that functions lost by L. borgpetersenii are concentrated in genes encoding proteins that impair adaptation to, and survival in, diverse environments, e.g., sensory transduction and defense mechanisms. The presence of fewer metabolic and solute transport functions (Fig. 3) likely limits the diversity of nutrients that can be consumed by L. borgpetersenii. In contrast, L. interrogans has more signal transduction, transcriptional regulatory factors, and diverse metabolic and solute transport functions (Fig. 3), consistent with its capacity for extended survival in water (20). To test this model, we compared L. borgpetersenii and L. interrogans survival in water at 20°C and found that L. borgpetersenii lost >90% viability within 48 h, whereas L. interrogans retained 100% viability over the same time period. L. interrogans retained 30% viability over a 3-week incubation, by which time no viable L. borgpetersenii were detected (R.L.Z. unpublished data). These results support our hypothesis that L. borgpetersenii does not tolerate nutrient deprivation and does not survive passage through water. Two genes in L. interrogans that closely resemble the Myxococcus xanthus devR and devS genes and that are absent in the L. borgpetersenii genome may facilitate passage in water between mammalian hosts. The M. xanthus devRS genes are essential for fruiting-body development, a process induced by nutrient deprivation that prepares bacteria for long-term survival (21). We conclude that impairment of environmental sensing and metabolite transport and utilization functions in L. borgpetersenii limits its capacity to acquire nutrients and survive in environments external to a mammalian host. Conversely, L. interrogans appears well tailored for long-term survival in aquatic environments and can easily adapt to mammalian hosts. This interpretation is consistent with epidemiological studies that show L. interrogans infections usually result from exposure to contaminated surface water, whereas L. borgpetersenii infections occur by direct contact with bacteria-contaminated body fluids.
Fig. 3.
Quantitative analysis of functional groups. Genes were clustered from each Leptospira genome according to the functional groups shown. The percentage of pseudogenes (Left) or paralogs (Right) that comprise the total number of genes within each category is shown. For reference, a bar graph showing the total number of genes in each category is presented in Fig. 5.
Among the 137 genes in the L. borgpetersenii genome that lack orthologous counterparts in L. interrogans are several genes that offer insight into key differences between these two species. For example, L. borgpetersenii has a dam-methylase and several unique transcriptional regulators, which probably exert considerable influence on gene regulation. Surprisingly, L. borgpetersenii has two sets of cytochrome c-related genes, one set that shares extensive similarity with its L. interrogans counterpart, and a second that is similar to cytochrome-c genes from a broad group of eubacterial species. Unique potential virulence factors (see below) may also affect L. borgpetersenii pathogenesis.
Membrane Proteins and Protein Secretion.
Membrane proteins have an essential role in nutrient acquisition, environmental signaling, and cell homeostasis, yet there has been little functional analysis of most Leptospira membrane proteins. Approximately 31% of L. borgpetersenii genes encode proteins with a predicted membrane location, a value similar to L. interrogans. Identification of potential outer membrane proteins (OMPs) is essential for subunit vaccine development, and we focused on two classes of membrane proteins, β-barrel proteins and lipoproteins. Integral OMPs usually assume a β-barrel conformation (22), and 50 and 49 proteins were found in strains L550 and JB197, respectively, with potential to assume this conformation, including OmpLI, a proven transmembrane porin (23), and TonB- and TolC-related proteins (Table 5, which is published as supporting information on the PNAS web site). Lipoproteins activate host cells to induce a proinflammatory response (24) and may elicit protective immunity (25). We identified 128 and 121 putative lipoproteins (not including pseudogenes) in strains L550 and JB197, respectively, including all known Leptospira lipoproteins and an OmpA homolog (Table 6, which is published as supporting information on the PNAS web site). In Escherichia coli, lipoproteins are processed by a Type II signal peptidase (SPase) and sorted to the inner or outer membrane by the Lol system (26). L. borgpetersenii has one Type II SPase, but only the inner membrane components of the Lol transport system (LolCDE) were identified, suggesting L. borgpetersenii may use an alternative system to transport lipoproteins to the outer membrane.
L. borgpetersenii utilizes Type I and Type II secretory systems to export proteins from the cytoplasm. The Type I secretion system, exemplified by ABC transporter proteins is Sec-independent (27); there are 18 distinct permeases associated with ABC transporters identified in L. borgpetersenii. The Type II secretion genes are organized in a single cluster. This translocation system interacts with and relies on Sec translocase to deliver proteins, including toxins and hydrolytic enzymes, to a variety of extracytoplasmic destinations (27). The signal peptides of Sec-translocase exported proteins are removed by Type I SPases. L. borgpetersenii resembles Gram-positive bacteria by the presence of three Type I SPases. Gram-negative bacteria typically have one Type I SPase, (28), raising the intriguing possibility that L. borgpetersenii may export virulence proteins by a specific Type I SPase as recently reported for Listeria monocytogenes (29).
Environmental Sensing and Gene Regulation.
Leptospira differentially regulates transcription of several genes, but the regulatory mechanisms have not been characterized. For example, the virulence proteins LigA and LigB are induced by increased osmolarity (30), and LipL36 is down-regulated during infection (31). Because differential gene regulation can directly affect bacterial functions during infection, we focused our analysis on gene regulation as it relates to environmental sensing and discovered intriguing differences between L. borgpetersenii and L. interrogans.
L. borgpetersenii genes encoding the transcription initiation factors σ70 and σ54 were identified, suggesting that these factors are used for generalized (32) and differential transcription (33), respectively, as in other bacterial genera. Three classes of signal transduction factors used by bacteria to adapt to changes in the extracellular environment were identified in L. borgpetersenii: extracytoplasmic function (ECF) σ factors, two-component response regulators (34), and GGDEF response regulators (35). We predict that the 11 L. borgpetersenii ECF genes either have unusual patterns of regulation or may be nonfunctional, because the genes encoding cognate regulatory factors including the anti-σ factor antagonist and σ factor regulator (36), which are commonly cotranscribed with the ECF, are instead dispersed around the genome. Likewise, only 18 of the two-component response regulators are paired with the cognate histidine kinases in L. borgpetersenii, as compared with 24 complete response regulator-histidine kinase pairs in L. interrogans. Additionally, five GGDEF response regulators were found in L. borgpetersenii, which is less than half the number found in L. interrogans. Consistent with few functional response regulators, L. borgpetersenii has 23% less putative transcriptional regulatory proteins than L. interrogans. However, two transcriptional regulators important for survival in a mammalian host, Fur, the iron response regulator, and HcrA, which regulates response to thermal stress (37), are present in both species. Collectively, these findings are consistent with our hypothesis that L. borgpetersenii is deficient in the ability to sense and therefore adapt to diverse external environments that may be encountered between mammalian hosts.
Potential Virulence Traits.
L. borgpetersenii and L. interrogans are capable of causing serious human infections and yet establish chronic infections in maintenance hosts. Many potential virulence traits are shared between both species, but several important differences were identified. Shared traits include the presence of several well conserved proteases that may facilitate tissue invasion, including 10 trypsin-like serine proteases, four zinc-dependent proteases, and a collagenase. An unusual vitamin K-dependent γ carboxylase, vgc, may affect hemostasis during infection (38). Attachment of factor H to the surface of Leptospira spp. by the conserved protein LfhA (39) may inactivate C3b and interfere with the antibacterial action of complement.
L. borgpetersenii has two unique N-acetylneuraminic acid synthetases that may prevent detection by the host immune system by coating the cell surface with sialic acid; homologous genes were not detected in the L. interrogans genome. There may also be species-specific differences in how Leptospira disrupt cellular integrity through the action of sphingomyelinases; L. borgpetersenii has three sphingomyelinase genes vs. five found in L. interrogans.
Differences in potential adhesins may alter how L. borgpetersenii and L. interrogans interact with their hosts. L. borgpetersenii has a unique HA-like protein that may facilitate attachment to host tissue. Other potential adhesins include integrin α proteins with FG-GAP repeat motifs (10) and leucine repeat proteins (40). Two of the three integrin α-like proteins identified in L. interrogans serovar Copenhageni (LIC12259 and LIC13101; ref. 10) are pseudogenes in L. borgpetersenii. We identified 12 unique extracytoplasmic leucine-rich repeat proteins in L. interrogans and none in L. borgpetersenii. The L. interrogans bacterial Ig-like proteins LigA, LigB, and LigC may facilitate host cell attachment (41), but the L. borgpetersenii genome has only the ligB gene. Collectively, these results suggest L. borgpetersenii and L. interrogans may differ in the mechanisms used to attach to host tissue.
Conclusions
Several lines of evidence suggest that L. borgpetersenii is undergoing IS-mediated genome reduction that affects transmission between hosts. IS elements are a prominent feature of the L. borgpetersenii genome, contributing to numerous chromosomal rearrangements, pseudogene formation, and consequently a lower coding density than L. interrogans. However, changes in genetic organization and gene loss do not impair virulence; both L. borgpetersenii and L. interrogans can cause lethal infections in nonmaintenance hosts. Instead, gene loss appears to impair L. borgpetersenii tolerance of nutrient deprivation, increasing host dependence relative to L. interrogans, and consistent with evidence that L. borgpetersenii relies on direct contact for disease transmission. We propose that L. borgpetersenii represents an intermediate stage in evolution from generalist to host dependence. Characterization of changes leading to this stage should help identify early events leading to host dependence among diverse bacterial genera.
Bacteria that rely on a eukaryotic host for survival include pathogens, e.g., Treponema pallidum (42), and symbionts, e.g., Buchnera aphidicola (43), and often have reduced genomes with limited metabolic potential and limited capacity to respond to changes in the extracellular environment (44). The initial steps leading to this level of host dependence are unclear. Our findings comparing the L. borgpetersenii to L. interrogans genomes suggest that loss of sensory transduction functions impairing host-free survival is a critical step early in the evolution of host dependence. Reliance on direct contact for transmission between hosts removes selective pressure on bacteria to retain genes required for survival outside the host but also limits access to gene pools available in diverse environments. This model is consistent with our observation that L. borgpetersenii does not survive nutrient deprivation and is limited to a direct host-to-host transmission cycle, whereas L. interrogans, by virtue of its superior coding capacity, can withstand prolonged nutrient deprivation and maintain a transmission cycle that often involves passage through surface water between mammalian hosts.
Materials and Methods
Bacteria.
L. borgpetersenii serovar Hardjo strain L550 was isolated in Australia from a human patient with leptospirosis contracted from exposure to infected cattle. Strain JB197 was isolated in the U.S. from a beef steer at slaughter (45). Standard methods were used to confirm the A and B subtypes of both strains (9). Genomic DNA was isolated from a colony-purified stock of strain L550 after seven passages following initial isolation. A colony-purified seed stock of strain JB197 was passed through a hamster and isolated from liver, and genomic DNA was isolated directly from the expanded culture.
Sequencing.
Plasmid libraries were prepared in pSmart (LucigenMiddleton, WI; strain L550) and pZERO-1 (Invitrogen, Carlsbad, CA; strain JB197) by using sheared DNA with insert sizes of 3–5 and 5–7 kb. Whole-genome shotgun sequencing of randomly picked clones, followed by primer walking and PCR amplification was used to complete each sequence. In total, 40,686 and 60,391 sequencing reads were used in the final assembly providing coverage of 6× and 9× (46), with overall error rates of 0.9 and 0.02 errors per 10,000 bp for strains L550 and JB197, respectively. Phred, Phrap, and Consed programs were used for genome assembly (47–50), supported by scripts written in Perl for various tasks.
Data Analysis.
Annotation was done by using Wasabi (T.S., unpublished data; available on request), an interactive platform that links several data analysis programs to identify putative coding regions, provides biochemical information for each protein, identifies homologs, and stores these data in a retrievable format by using a MySQL database structure. Putative CDS were identified with GeneMarkS (51), Glimmer, and Artemis (52), and then clustered according to the position of stop codons. Database searches for each CDS were done by using National Center for Biotechnology Information BLASTP analysis against the GenBank nonredundant protein database. Features for each predicted protein were determined by using RPS-BLAST against the Conserved Domain Database, PSORT (53), PSORTb, CELLO (54), LipoP, SpLip (55), SignalP, TMHMM, and TMpred (56). β-Barrel protein structures were predicted by using BOMP (57).
Orthologous and paralogous genes were identified within spirochete genomes by using BLASTP analysis of databases of L. borgpetersenii serovar Hardjo strain L550 and strain JB197 genomes and spirochete-specific databases for B. burgdorferi (58), L. interrogans serovars Copenhageni (10) and Lai (11), T. denticola (42), and T. pallidum (59) by using a cutoff e value of 1 × 10−6.
To facilitate direct comparison of the L. borgpetersenii and L. interrogans genomes, we revised the annotation features of both serovars Copenhageni and Lai to apply consistent annotation terms across all four genomes. This process removed 1,206 putative CDS and added 52 previously unidentified CDS to the previous Lai annotation (11), resulting in 3,613 recognized CDS. The revised L. interrogans serovar Copenhageni annotation contains 3,530 recognized CDS after removal of 287 CDS and addition of 91 CDS (10). These revised annotations are accessible at http://vbc.med.monash.edu.au/genomes.
Supplementary Material
Acknowledgments
We thank Ami Frank, Karen Halloum, Richard Hornsby, Amanda Toot, and John Foley for excellent technical assistance and David Miller (National Veterinary Services Laboratory, Ames, IA) for L. borgpetersenii strains. This work was supported by the Medical Genomics Program of the Australian National Health and Medical Research Council and by the U.S. Department of Agriculture.
Abbreviations
- CDS
coding sequences
- IS
insertion sequences
- SPase
signal peptidase.
Footnotes
References
- 1.World Health Organization. Wkly Epidemiol Rec. 1999;74:237–242. [Google Scholar]
- 2.Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, Weyant RS. Int J Sys Bacteriol. 1999;49:839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 3.Levett PN. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride AJ, Athanazio DA, Reis MG, Ko AI. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- 5.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 6.Saint Girons I, Norris SJ, Gobel U, Meyer J, Walker EM, Zuerner R. Res Microbiol. 1992;143:615–621. doi: 10.1016/0923-2508(92)90119-9. [DOI] [PubMed] [Google Scholar]
- 7.Haake DA, Suchard MA, Kelley MM, Dundoo M, Alt DP, Zuerner RL. J Bacteriol. 2004;186:2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, Stanton TB, Zablen L, Mandelco L, Woese CR. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuerner RL, Ellis WA, Bolin CA, Montgomery JM. J Clin Microbiol. 1993;31:578–583. doi: 10.1128/jcm.31.3.578-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nascimento ALTO, Ko AI, Martins EAL, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, et al. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, et al. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 12.Zuerner RL, Herrmann JL, Saint Girons I. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogasawara N, Yoshikawa H. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 14.Dubarry N, Pasta F, Lane D. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukunaga M, Mifuchi I. J Bacteriol. 1989;171:5763–5767. doi: 10.1128/jb.171.11.5763-5767.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuerner RL. Plasmid. 1994;31:1–11. doi: 10.1006/plas.1994.1001. [DOI] [PubMed] [Google Scholar]
- 17.Conlan LH, Stanger MJ, Ichiyanagi K, Belfort M. Nucleic Acids Res. 2005;33:5262–5270. doi: 10.1093/nar/gki819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston A, Parkhill J, Maskell DJ. Nat Rev Microbiol. 2004;2:379–390. doi: 10.1038/nrmicro886. [DOI] [PubMed] [Google Scholar]
- 20.Trueba G, Zapata S, Madrid K, Cullen P, Haake D. Int Microbiol. 2004;7:35–40. [PubMed] [Google Scholar]
- 21.Thony-Meyer L, Kaiser D. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berven FS, Flikka K, Jensen HB, Eidhammer I. Nucleic Acids Res. 2004;32:W394–W399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang ES, Exner MM, Summers TA, Martinich C, Champion CI, Hancock RE, Haake DA. Infect Immun. 1995;63:3174–3181. doi: 10.1128/iai.63.8.3174-3181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sellati TJ, Bouis DA, Kitchens RL, Darveau RP, Pugin J, Ulevitch RJ, Gangloff SC, Goyert SM, Norgard MV, Radolf JD. J Immunol. 1998;160:5455–5464. [PubMed] [Google Scholar]
- 25.Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, Wagar EA. Infect Immun. 1999;67:6572–6582. doi: 10.1128/iai.67.12.6572-6582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz N, Kahne D, Silhavy TJ. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 27.Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. J Bacteriol. 2005;187:4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anne J. Biochim Biophys Acta. 2004;1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Bonnemain C, Raynaud C, Reglier-Poupet H, Dubail I, Frehel C, Lety MA, Berche P, Charbit A. Mol Microbiol. 2004;51:1251–1266. doi: 10.1111/j.1365-2958.2004.03916.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsunaga J, Sanchez Y, Xu X, Haake DA. Infect Immun. 2005;73:70–78. doi: 10.1128/IAI.73.1.70-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haake DA, Martinich C, Summers TA, Shang ES, Pruetz JD, McCoy AM, Mazel MK, Bolin CA. Infect Immun. 1998;66:1579–1587. doi: 10.1128/iai.66.4.1579-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paget MS, Helmann JD. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazmierczak MJ, Wiedmann M, Boor KJ. Microbiol Mol Biol Rev. 2005;69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galperin MY, Nikolskaya AN, Koonin EV. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 35.Romling U, Gomelsky M, Galperin MY. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 36.Helmann JD. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 37.Ballard SA, Go M, Segers RP, Adler B. Gene. 1998;216:21–29. doi: 10.1016/s0378-1119(98)00329-1. [DOI] [PubMed] [Google Scholar]
- 38.Rishavy MA, Hallgren KW, Yakubenko AV, Zuerner RL, Runge KW, Berkner KL. J Biol Chem. 2005;280:34870–34877. doi: 10.1074/jbc.M504345200. [DOI] [PubMed] [Google Scholar]
- 39.Verma A, Hellwage J, Artiushin S, Zipfel PF, Kraiczy P, Timoney JF, Stevenson B. Infect Immun. 2006;74:2659–2666. doi: 10.1128/IAI.74.5.2659-2666.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikegami A, Honma K, Sharma A, Kuramitsu HK. Infect Immun. 2004;72:4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, et al. Mol Microbiol. 2003;49:929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, et al. Proc Natl Acad Sci USA. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 44.Moran NA, Dunbar HE, Wilcox JL. J Bacteriol. 2005;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller DA, Wilson MA, Beran GW. Am J Vet Res. 1991;52:1761–1765. [PubMed] [Google Scholar]
- 46.Lander ES, Waterman MS. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 47.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 48.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 49.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 50.Gordon D, Desmarais C, Green P. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Besemer J, Lomsadze A, Borodovsky M. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 53.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu CS, Lin CJ, Hwang JK. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Setubal JC, Reis M, Matsunaga J, Haake DA. Microbiology. 2006;152:113–121. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofmann K, Bucher P, Falquet L, Bairoch A. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berven FS, Flikka K, Jensen HB, Eidhammer I. Nucleic Acids Res. 2004;32:W394–399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 59.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, et al. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 60.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.