Abstract
In various multicellular organisms, circadian clocks are present not only in the central nervous system, but also in peripheral organs and tissues. In mammals peripheral oscillators are not directly responsive to light, but are entrained by the central oscillator in the suprachiasmatic nucleus. These individual oscillators are diverse in their free-running periods and phases. In contrast, cultured peripheral tissues and cell lines from zebrafish are not only rhythmic, but can also be directly entrained by light. Because of the convenience of studying rhythms in cultured cells, however, little has been known about properties of individual oscillators in intact zebrafish. Here, we show the remarkable diversity and consistency of oscillator properties in various peripheral organs and tissues from the period3-luciferase (per3-luc) transgenic zebrafish. Tissue-dependent differences were found in free-running period, phase, response to light, and temperature compensation. Furthermore, cycling amplitudes were reduced at lower temperatures in some, but not all, of the organs tested. Finally, we found that per3-luc rhythms can free run in both constant dark and constant light with remarkably similar amplitudes, phases, and periods, despite the fact that the mRNA of per2 and per1 has been shown not to oscillate in constant light.
Keywords: circadian, period3, bioluminescence, temperature compensation
Circadian oscillators are essentially intracellular phenomena where transcriptional feedback loops regulate cycling expression of various genes (1–3). They are consistently present in unicellular organisms such as cyanobacteria. In higher metazoan species, it has been believed for a long time that circadian rhythms are present in specialized tissues, such as the suprachiasmatic nucleus (SCN) in mammals, the pineal gland in some lower vertebrates, and the optic lobe of some insects. This view changed radically after cloning of so-called clock genes and studies of their expression patterns. Various tissues all over the body of Drosophila have been shown to express the period (per) gene rhythmically (4, 5). Similarly, many tissues in mammals express per1 and per2 rhythmically (6, 7). However, among all of the tissues that possess oscillator properties in these organisms, there is a central oscillator that affects other oscillators, such as the SCN in mammals and some clock neurons in Drosophila (8, 9). The common properties of these central oscillators are: (i) they are neuronal cells in the central nervous system that are responsible for locomotor activity rhythms; and (ii) they can affect phases and/or amplitudes of other oscillators, possibly by chemical, physiological, or behavioral factors (10).
However, the extent to which the central oscillators affect peripheral oscillators varies among organisms. In Drosophila, most peripheral oscillators are directly responsive to light (5) and can be entrained independently of the central pacemakers in the brain (11, 12). The blue-light photoreceptor Cryptochrome (CRY) is supposed to be the molecule responsible for entrainment of peripheral oscillators (13, 14). Only some clock neurons in the brain and the prothoracic gland have been shown to depend on the small ventral lateral neurons for persistent rhythms in constant dark (DD) (15–17). Many of the peripheral oscillators seem to dampen quickly in DD (18), whereas some brain neurons other than the small ventral lateral neurons show persistent and independent molecular rhythms in DD (19).
In mammals, central and peripheral oscillators cannot be reset by light directly. Light-induced signals from the retina reset the clock in the SCN (20, 21), which in turn resets and/or sustains rhythmicity in many peripheral tissues (6, 7, 22, 23). Such tight regulation of peripheral oscillators by the central one in mammals may be the evolutionary consequence or, more likely, the prerequisite for the loss of direct photic response in peripheral oscillators. Yet, other cues such as food availability can entrain certain peripheral oscillators independently from the SCN (24), making the hierarchy of oscillators questionable. The persistence of rhythms in peripheral tissues can vary depending on the output studied; rat Per1-luciferase (luc)-mediated bioluminescence rhythms in peripheral tissues damp in several days, whereas those mediated by mouse Per2-luc persist for at least 20 cycles (6, 7). Recently, single-cell imaging techniques revealed that damping of bioluminescence rhythms in cultured cells is caused by desynchronization of individual cellular oscillators (25–27). However, the above-mentioned diversity in peripheral tissues suggests that damping in some peripheral oscillators could be caused by intrinsic properties of clocks in those tissues.
In zebrafish it has been shown that peripheral tissues and cell lines rhythmically express various clock-gene homologues such as clocks, bmals, crys, and pers (reviewed in ref. 28). It is also known that zebrafish possess more genes (including clock genes) than do mammals, because of a genomewise duplication during early teleost evolution (29). For example, there are six cry genes in zebrafish, whereas only two such genes are present in mammals (30). On the other hand, only three transcripts have been identified for zebrafish per genes, per1, per2, and per3 (per1 is also called per4) (31–34). Like in Drosophila, molecular rhythms in peripheral tissues from zebrafish are directly resettable by light even in culture (32, 35), although the photoreceptor molecule responsible for this resetting is not yet known. The location of the central pacemaker or even the presence of it in zebrafish is not yet known, but there may be such a pacemaker in the brain (28). Simultaneous ocular enucleation and pinealectomy in larval fish still leaves their locomotor activity rhythms intact, suggesting that neither the retina nor the pineal gland is necessary for this rhythm (28).
The fact that zebrafish rhythms can be studied in cultured cells facilitated studies of photic responses, promoter dissections of clock genes, single-cell monitoring of bioluminescence rhythms, and entrainment by temperature shift (25, 33, 36, 37). However, the use of cell lines also harbors the risk that these cells may not represent various cell types present in intact organisms, and the findings in cell lines may not be applicable to some tissues. Therefore, we used the per3-luc transgenic zebrafish to study the properties of various peripheral oscillators in adult zebrafish (38). Tissues and organs from this transgenic fish were dissected, cultured, and monitored for bioluminescence rhythms in various lighting and temperature conditions. Many tissues and organs showed bioluminescence rhythms in culture that were reset by light. These rhythms varied in free-running period, phase, response to light, and temperature compensation. Cycling amplitude was also affected by lower temperatures only in some organs tested. Furthermore, bioluminescence rhythms in DD and constant light (LL) were remarkably similar in period, amplitude, and phase. This is different from per1-luc-mediated bioluminescence rhythms in cultured cells, where the rhythm and overall level of luminescence were acutely suppressed by light (33).
Results and Discussion
Bioluminescence Rhythms in Isolated Tissues and Organs from per3-luc Zebrafish.
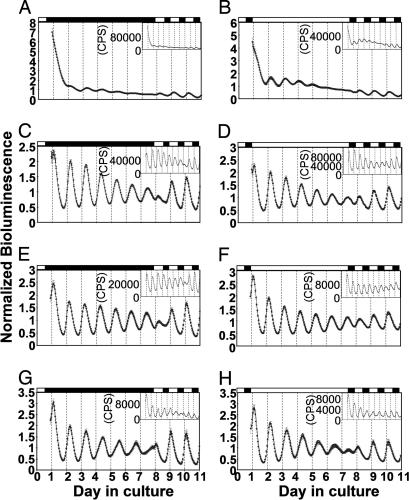
To test whether various peripheral organs from adult zebrafish produce per3-luc-mediated bioluminescence rhythms, they were dissected and monitored for bioluminescence as organ cultures in DD. Robust free-running rhythms that persisted for at least 6.5 days were found in the heart, spleen, and gall bladder (Fig. 1C, E, and G, respectively). The retina also showed fairly robust rhythms, although amplitudes and periods varied from experiment to experiment possibly because of the susceptibility of this organ to different batches of serum (Fig. 1A). The rhythms damped in DD and recovered during subsequent exposure to light-dark (LD) conditions (Fig. 1 A, C, E, and G). The phases and periods of the bioluminescence rhythms in DD varied depending on the organ. The gall bladder consistently had longer periods than did the heart and spleen (Table 1; P < 0.0001, α = 0.0051, Wilcoxon-Kruskal/Wallis test). The periods of retinal rhythms seemed more variable; they were >25 h in some experiments (see Fig. 5). The periods of the heart and spleen were not significantly different [P = 0.0094 by fast Fourier transform–nonlinear least squares (FFT-NLLS), and P = 0.2892 by maximum entropy spectral analysis (MESA), α = 0.0051, Wilcoxon-Kruskal/Wallis test]. Peaks in different organs occurred at various circadian times (CTs) in DD (Table 1). The retina peaked at later phases, and the gall bladder peaked at slightly earlier phases (P < 0.0001, α = 0.0051, Watson-Williams test). Peaks of these organs in LD conditions did not vary as much as in DD: retina, Zeitgeber time (ZT; ZT 12 corresponds to lights off, and ZT 22 to lights on) 4.4 ± 0.1, n = 23; heart, ZT 2.8 ± 0.2, n = 12; spleen, ZT 3.2 ± 0.1, n = 12; gall bladder, ZT 3.1 ± 0.2, n = 11.
Fig. 1.
Bioluminescence rhythms in cultured organs in DD (A, C, E, and G) and LL (B, D, F, and H). The retina (A and B), heart (C and D), spleen (E and F), and gall bladder (G and H) were tested for bioluminescence rhythms in DD and LL. After ≈6.5 days in constant conditions the cultured organs were transferred to 14:10 LD cycles and tested for 3 more days. For each type of organ, an averaged plot from an experiment is presented. Error bars represent SEM. The number of samples used for each averaged plot was: 19 (A), 15 (B), 11 (C–F and H), and 12 (G). (Insets) Representative single plots are shown for each type of organ. The first 12 h of each record was deleted from the plot. White and black bars represent the time when the lights were on and off, respectively. The 0 point on the x axis corresponds to ZT 0 (2 h after lights on) on the day the fish were dissected.
Table 1.
Rhythmicity, periods, and phases of bioluminescence rhythms in cultured peripheral organs in DD and LL
| Organ | Light condition | n, tested | n, rhythmic | Relative-amplitude error | Period by FFT-NLLS | Period by MESA | Phase (CT) |
|---|---|---|---|---|---|---|---|
| Retina | DD | 41 | 15 | 0.56 ± 0.05 | 24.0 ± 0.3 | 24.6 ± 0.1 (21) | 8.3 ± 0.7 |
| Heart | DD | 25 | 25 | 0.12 ± 0.00 | 25.0 ± 0.1 | 25.0 ± 0.1 | 4.1 ± 0.2 |
| Spleen | DD | 22 | 22 | 0.10 ± 0.00 | 24.7 ± 0.1 | 25.0 ± 0.1 | 3.8 ± 0.1 |
| Gall bladder | DD | 22 | 22 | 0.18 ± 0.01 | 26.4 ± 0.2 | 26.6 ± 0.2 | 2.9 ± 0.2 |
| Retina | LL | 32 | 22 | 0.45 ± 0.03 | 22.2 ± 0.2 | 23.5 ± 0.2 (14) | 8.8 ± 0.5 |
| Heart | LL | 20 | 20 | 0.18 ± 0.01 | 24.7 ± 0.1 | 24.9 ± 0.2 | 4.0 ± 0.1 |
| Spleen | LL | 20 | 20 | 0.12 ± 0.01 | 25.3 ± 0.1 | 25.5 ± 0.1 | 3.0 ± 0.1 |
| Gall bladder | LL | 19 | 19 | 0.18 ± 0.01 | 26.5 ± 0.2 | 26.7 ± 0.2 | 3.0 ± 0.2 |
For each group, the results of two experiments were pooled. The numbers of rhythmic samples judged by FFT-NLLS are given. Mean ± SEM of relative-amplitude error (53), periods, and peak phases were calculated for rhythmic samples only. Only the samples judged as rhythmic by autocorrelation (such numbers are given in parentheses in case they are different from those done by FFT-NLLS) were averaged for periods by MESA.
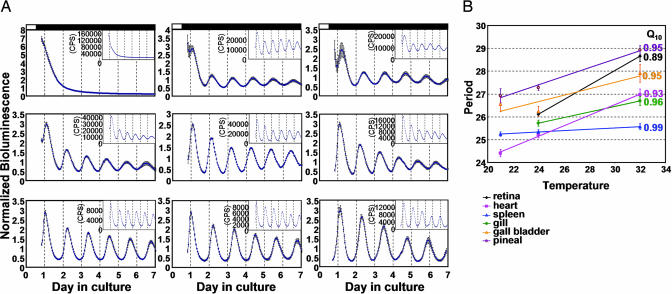
Fig. 5.
Effects of temperatures on periods of bioluminescence rhythms in various cultured tissues. (A) Averaged plots from an experiment for the retina (Top), spleen (Middle), and pineal gland (Bottom) at 21°C (Left), 24°C (Center), and 32°C (Right) are shown. The number of samples averaged for each plot was: retina, 9 at 21°C, 16 at 24°C, and 12 at 32°C; spleen, 6 at 21°C, 8 at 24°C, and 8 at 32°C; pineal gland, 6 at 21°C, 6 at 24°C, and 5 at 32°C. (B) Average period lengths for the six tissues tested are plotted. Period lengths were estimated by FFT-NLLS. Similar results were obtained by MESA (data not shown). For each temperature, results from two to three experiments are combined. The number of samples for each group was: retina, 24 at 24°C and 33 at 32°C; heart, 9 at 21°C, 25 at 24°C, and 23 at 32°C; spleen, 10 at 21°C, 24 at 24°C, and 22 at 32°C; gill, 21 at 24°C and 21 at 32°C; gall bladder, 6 at 21°C, 12 at 24°C, and 15 at 32°C; pineal gland, 7 at 21°C, 12 at 24°C and 9 at 32°C. The Q10 value for each tissue is given to the right. The retina and gill were mostly arrhythmic at 21°C. Error bars represent SEM. Q10 values that were obtained by MESA were: retina, 0.89; heart, 0.93; spleen, 0.99; gill, 0.94; gall bladder, 0.95; pineal, 0.95.
Besides the organs shown in Fig. 1, the brain, pineal gland, gill, kidney, liver, oocyte–follicle complex, caudal fin, and testis have been tested for bioluminescence rhythms. Only the oocyte–follicle complex and the pineal gland showed robust rhythmicity comparable to the heart, spleen, and gall bladder (see Fig. 5 and Fig. 6, which is published as supporting information on the PNAS web site). The gill, kidney, and caudal fin produced relatively low-amplitude rhythms in at least some specimens (see Figs. 5 and 6). The brain did not show rhythmicity when cultured as a whole brain, probably because of its large size. Different parts of the brain, however, showed bioluminescence rhythms in cultures (H. Borsetti and G.M.C., unpublished data). Bioluminescence from the testis did not last >3 days in culture, suggesting that this organ dies in the culture conditions used. The liver had very low bioluminescence that oscillated very little (Fig. 6). Importantly, similar bioluminescence rhythms in the retina, heart, spleen, gall bladder, and gill were observed in another line of per3-luc (line 86) with much lower overall luminescence counts (data not shown).
The low amplitude of the rhythms in some of these tissues and organs may be caused by their susceptibility to the culture condition used or to endogenous properties of their oscillators, such as weak coupling among cells in a given tissue and cells with variable periods, phases, and amplitudes within a tissue. Variability in cycling phase among different tissues has been documented in zebrafish for bmal1, bmal2, and clock (39). Tissue-dependent diversity in period and phase was also observed in the mPer2-luc knockin mouse (7). As has been suggested in their case, quantitative or qualitative difference in clock gene expression and/or cellular environment among different tissues may be responsible for the diversity. The tissue-dependent variability of clock-gene expression in zebrafish is not surprising considering that there are more clock genes present in zebrafish than in mammals (28).
Bioluminescence Rhythms in LL.
In LL, these organs showed bioluminescence rhythms remarkably similar to those in DD (Fig. 1). However, there were differences between DD and LL; the periods of the retinal rhythms were shorter in LL than in DD (Table 1), and the spleen showed slightly, but significantly, longer periods in LL than in DD (Table 1; P< 0.0006, α = 0.0051). Therefore, the effect of LL on period depends on the organ studied. The peaks occurred at similar CT times in LL and DD except that the spleen showed slightly, but significantly, earlier phases in LL than in DD (P = 0.0003; Table 1). Like in DD, rhythms damped gradually in LL, then recovered in subsequent LD cycles. The fact that per3-luc-mediated bioluminescence rhythms in some organs were not affected by LL was remarkable, because the other two zebrafish per genes are not rhythmic in LL at least in cell lines and some tissues; per2 mRNA is induced by light (32, 36), whereas per1 transcription is suppressed by light [per1 is also called per4 (33, 34)]. Although the exact elements responsible for per3 mRNA cycling are not known, three E-boxes have been found within 1-kb upstream sequences of this gene (38). If these E-boxes are responsible for per3 mRNA rhythms, constitutive levels of PER2 and PER1 could possibly dampen per3 mRNA rhythms. There are several possibilities to explain the results: (i) unlike in cell lines, per2 and per1 mRNA may cycle in LL in the organs studied here, although the light-dependent suppression of per1 was observed in the brain, and larval fish and the PAC-2 cell line (33), and induction of per2 has been observed in embryos (40); (ii) per2 and per1 mRNA may not cycle, but their protein products may do so by posttranscriptional mechanisms, although such mechanisms are unlikely to compensate for the very low level of per1 mRNA in LL; (iii) per2 and per1 may not be expressed in these organs, although per1 mRNA has been detected in the retina, heart and spleen (ref. 34 and M.K., unpublished data); and (iv) PER2 and PER1 are expressed, but do not affect the per3 promoter.
In any case, this study clearly shows that circadian clocks persist in LL in many zebrafish tissues. Adult zebrafish have persistent locomotor activity rhythms in LL (41). Because it is the only known zebrafish per gene that shows mRNA rhythms in LL, per3, which is a homologue of mammalian per3, may be the most important zebrafish per gene for the central pacemaker responsible for locomotor activity rhythms. If this is true, it is different from mammals where knocking out mPer3 has little effect on behavioral rhythms (42). Alternatively, there may be additional zebrafish per genes that are part of the central oscillator. In fact, there is a second per1-like sequence in the zebrafish genome although no cDNA corresponding to that sequence has been identified (G.M.C., unpublished data).
Resetting per3-luc Rhythms by Light.
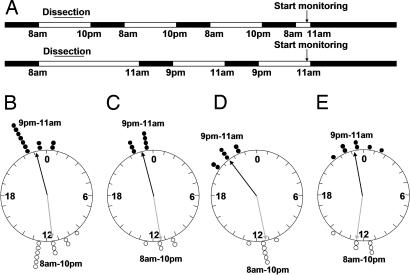
It has been shown that clocks in cultured tissues and cells from zebrafish are resettable by light (Fig. 1) (32, 35). We have also tested whether per3-luc-mediated bioluminescence rhythms can be re-entrained to an opposite LD cycle. The retina, heart, spleen, and gall bladder were entrained in two almost-opposite 14:10 LD cycles for 3 days in culture (lights on at 8 a.m. or 9 p.m.), and then tested for bioluminescence rhythms in DD (Fig. 2A). As a result, these two groups of organs had almost opposite phases, suggesting that these organs can be entrained by light in culture (Fig. 2 B–E). The phases of the spleen in the 9 p.m. to 11 a.m. LD group were significantly earlier than those of the other organs from the same group (P = 0.0008, α = 0.025), suggesting that spleen may not have been wholly shifted to a new phase by the 25-lux light used in this experiment (Fig. 2D). In another experiment that used 200-lux light all four organ types were shifted completely (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Entrainment of bioluminescence rhythms of cultured organs. The retina, heart, spleen, and gall bladder were cultured under 14:10 LD cycle of two almost opposite phases for ≈2.5 days. The intensity of lights used in this experiment was 25 lux. (A) The exact entrainment schedule is shown. The 8 a.m. to 10 p.m. LD cycle shown on top is the lighting condition in which the fish were kept. (B–E) The phase values of the peaks in real time (CST) were plotted on circular graphs for the retina (B), heart (C), spleen (D), and gall bladder (E) in empty circles (8 a.m. to 10 p.m.) and filled circles (9 p.m. and 11 a.m.). Gray and black arrows represent mean vectors for the 8 a.m. to 10 p.m. group and the 9 p.m. to 11 a.m. group, respectively. The lengths of mean vectors are: 8 a.m. to 10 p.m. group, 0.96; 9 p.m. to 11 a.m. group, 0.99 (B); 8 a.m. to 10 p.m. group, 0.99; 9 p.m. to 11 a.m. group, 0.96 (C); 8 a.m. to 10 p.m. group, 0.99; 9 p.m. to 11 a.m. group, 0.98 (D); and 8 a.m. to 10 p.m. group, 0.91; 9 p.m. to 11 a.m. group, 0.96 (E).
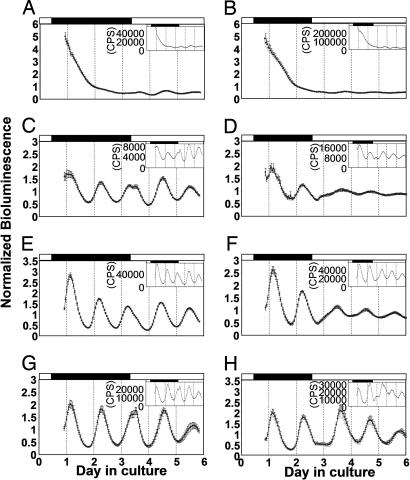
To test how bioluminescence rhythms respond to light stimuli, cultured organs were monitored in DD for 2–3 days and then transferred to LL. The results depended on the organ studied. The damped rhythms of the retina were somewhat recovered when they were shifted to LL, and the initial response was always a slight increase in bioluminescence (Fig. 3A and B). Bioluminescence in many of the hearts and gall bladders typically increased 1–2 h after the LL shift, and then decreased after a few hours (Fig. 3 C, D, G, and H). This response was observed whether LL shift occurred near the peak or trough time point in 10 of 19 hearts and 12 of 17 gall bladders. The rest of the hearts and gall bladders showed milder effects, slowing down of the decrease in bioluminescence in response to a shift that occurred near the peak time point, or delay of the increase in response to a shift near the trough time point. These milder effects were observed for all of the spleen samples (n = 10 for peak time point and n = 9 for trough; Fig. 3 E and F). Taken together, the initial responses of many of the organs to light seemed to be an increase in bioluminescence. However, this is not a long-lasting effect, but rather a transient effect that leads to a rhythm with a new phase. Initial induction of luc RNA by light was also suggested in per3-luc larval fish (38).
Fig. 3.
Responses of bioluminescence rhythms to LL shifts. The retina (A and B), heart (C and D), spleen (E and F), and gall bladder (G and H) were tested for bioluminescence rhythms in DD for 2–3 days, shifted to LL at approximate peak time points (A, C, E, and G) and trough time points (B, D, F, and H) and tested for 2–3 more days. The number of samples used for each averaged plot was: 20 (A), 15 (B), 10 (C, E, and G), 9 (D and F), and 7 (H).
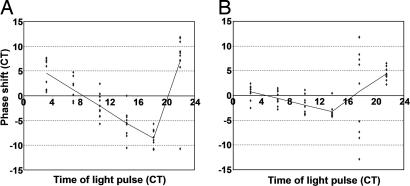
Finally, to test the effects of short light pulses on bioluminescence rhythms, phase response curves (PRCs) were constructed for the heart and spleen. The PRC for the heart showed large phase shifts and was close to type 0 (Fig. 4A). The melatonin production rhythm from cultured pineal glands, however, showed a typical type 1 PRC (34). The spleen showed much smaller phase shifts in most time points than did the hearts (Fig. 4). At one time point in the middle of the subjective night, the spleen showed all kinds of responses (Fig. 4B). The light pulse at this time point also made the rhythms in the spleen weaker (data not shown). These results are in contrast to the flat PRC of the cryb mutant flies (43), which are also rhythmic in LL. In the case of Drosophila, light degrades TIMELESS (TIM), possibly through the action of CRY, which leads to constantly low TIM, which in turn causes PER to be cytoplasmic and low (44). As a vertebrate, zebrafish may have molecular light responses very different from those in Drosophila. For instance, zebrafish crys may not function as circadian photoreceptors, and zebrafish tim may not be involved in the clock mechanism.
Fig. 4.
PRCs for the heart and spleen. Phase responses to 1-h light pulses were determined in CT for the heart (A) and spleen (B) and plotted against the time of light pulse in CT. Each dot represents a phase shift for each organ tested. The lines in each plot connect the average response for each time point. The results of two experiments are pooled.
As has been documented (32, 35), our data strongly support that cultured peripheral tissues from zebrafish are directly responsive to light. The response of per3-luc, however, is more transient in nature compared with those responses of the other zebrafish per genes. Our results on entrainment, LL shift, and PRC all point to lower light sensitivity of per3-luc rhythms in the spleen compared with photic responses of clocks in other organs such as the heart. This difference between the spleen and other organs may be caused by difference in photosensitivity, phototransduction pathways, or the pacemaking properties.
Effects of Temperature on Bioluminescence Rhythms.
Period lengths of circadian rhythms in various organisms are relatively invariant at varying temperatures. This is one of the basic characteristics of circadian clocks and is called temperature compensation. The per3-luc-mediated bioluminescence rhythm was also temperature-compensated in the six tissues tested (Fig. 5). As has been shown for the rhythms of melatonin production in cultured zebrafish pineal glands (34) and per1 promoter- and four E-box-mediated bioluminescence rhythms in cultured cells (37), bioluminescence rhythms in the retina, gill, heart, gall bladder, and pineal gland showed slight, but significant, overcompensation with Q10 values ranging from 0.89 in the retina to 0.96 in the gill (Fig. 5). These Q10 values are within the range of circadian rhythms in various organisms (45). The spleen showed almost perfect temperature compensation with a Q10 value of 0.99. The two-way ANOVA, performed on all of the six tissues and temperatures as variables, showed significant effects of tissue, temperature, and tissue × temperature interaction on period lengths (P < 0.0001 for each of the three effects).
Bioluminescence rhythms mediated by luciferase in Drosophila showed earlier phases at higher temperatures, and unstable luciferase activity in higher temperatures has been suggested as the mechanism responsible for this phenomenon (43). In zebrafish tissues, it is possible that lower enzyme stability at higher temperatures might have caused different period lengths. However, it is unlikely that this also caused tissue-dependent Q10 differences, because luciferase stability should be the same in all of the tissues at a given temperature.
We have documented tissue-dependent differences of Q10 values for circadian rhythms within species. The only comparable example may be a Q10 value of 0.99 for neuronal rhythms of the rat SCN (46) compared with a Q10 of 0.85–0.88 for mPer1::luc-mediated bioluminescence rhythms in Rat-1 fibroblasts (47). However, those studies dealt with rhythms of different biological processes in a homeothermic organism. Our studies showed that rhythms of the same output can vary in terms of temperature compensation in a poikilothermic organism. Because the mechanisms responsible for temperature compensation of circadian rhythms are not known, it is not easy to speculate what caused tissue-dependent variability of Q10 values. Because zebrafish has multiple homologues of many mammalian clock genes (28), it is possible that these different isoforms vary in terms of their sensitivity to temperature changes. In fact, changes in clock genes can affect temperature compensation, because some period-altering mutations in clock genes of Neurospora, Drosophila, and hamster also cause defects in temperature compensation (44, 48, 49). In Drosophila, even natural polymorphisms in a clock gene that are distributed as a highly significant latitudinal cline may account for differences in temperature compensation (50). A circadian period-altering mutation in zebrafish has been demonstrated to affect Q10 values of melatonin production rhythms from cultured pineal glands (34). Although the mutated gene has not been identified in this case, it has been speculated that the gene is involved in pace-making mechanisms of the zebrafish circadian rhythm, because the mutation affects both larval swimming and melatonin rhythms (34). Differences in relative expression levels of different clock genes in various tissues may account for the tissue-dependent variability in Q10 values.
Lahiri et al. (37) demonstrated that amplitudes of per1 promoter- and four E-box-mediated bioluminescence rhythms in cultured cells vary greatly depending on ambient temperature. In our study, we did not find such a dramatic amplitude differences between 24°C and 32°C except for the gill where significant amplitude difference between these two temperatures was observed (Table 2, which is published as supporting information on the PNAS web site). However, at 21°C, the gill and retina were mostly arrhythmic, and the heart, pineal gland, and spleen showed significant reduction in amplitudes (Fig. 5A and Table 2). The gall bladder did not show significant amplitude differences among the three temperatures. Therefore, cycling amplitudes seem to be stable over at least 8°C in most tissues and quickly deteriorate below certain temperatures in some tissues. Lahiri et al.’s studies on cycling amplitudes involved only two temperatures of rather large differences, 20°C and 30°C. Therefore, it is not clear whether the amplitude differences they found in their cell-based studies were caused by gradual changes over a range of temperature or sudden deterioration of rhythms below certain temperature. Sensitivity of cycling amplitude to temperature is a widespread phenomenon observed in diverse species such as Gonyaulax and chicken (51). Furthermore, our study shows that susceptibility of cycling amplitudes to ambient temperature varies depending on the tissue studied.
In summary, we have found that per3-luc-mediated bioluminescence rhythms differ among cultured peripheral tissues and organs in various ways such as period, phase, response to light, and temperature compensation. Furthermore, cycling amplitudes were reduced by lower temperatures in some, but not all, of the organs tested. This diversity suggests that those cell line-based studies successfully carried out in zebrafish should be treated as equivalent to studies based on one particular tissue (25, 33, 35, 37). This also raises the question of how these oscillators are orchestrated in intact animals. In zebrafish, individual oscillators can be entrained by light in LD conditions. Therefore, they may not need to be entrained by the central pacemaker in natural conditions. Do these oscillators desynchronize in constant conditions, or are they synchronized by the central oscillator in the brain as has been shown for mammals (6, 7)? This question is of particular interest in light of evolution of mammalian-like hierarchical circadian systems. The zebrafish, a vertebrate with Drosophila-like light-entrainable peripheral oscillators, could nonetheless have a more hierarchical system than do insects.
Materials and Methods
Animals.
Animals used in this study carried the per3-luc transgene in hemizygous condition and were originally derived from the University of Oregon *AB strain (38). Among five independent germ-line transformants, the strongest glowing line (line 23) was used unless otherwise stated (38). Animals were kept under 14-h light/10-h dark (14:10 LD; lights on 8 a.m.; lights off 10 p.m. CST) cycle, and group-housed in plastic tanks in a Z-MOD holding system (Marine Biotech, Beverly, MA) with recirculating filtered water at ≈28.5°C. They were fed commercial flake food in the morning, baby brine shrimp at midday, and adult brine shrimp in the evening. Experimental protocols were approved by the Institutional Animal Care and Use Committee.
Monitoring Bioluminescence Rhythms in Cultured Tissues and Organs.
Adult zebrafish between 9 and 14 months old were dissected for organs/tissues for cultures. Bioluminescence emitted from cultured organs and tissues were monitored as described (52). Details on culturing conditions and bioluminescence monitoring are in Supporting Text, which is published as supporting information on the PNAS web site.
Data Analysis.
Bioluminescence data from the Topcount were imported into Microsoft (Redmond, WA) Excel 2000 by the Import and Analysis macro (Steve Kay, The Scripps Institute, La Jolla, CA). The FFT-NLLS multicomponent cosine analysis was used to estimate rhythmicity, periods, and peak phases (53). Free-running periods were also estimated by MESA, and rhythmicity was estimated by autocorrelation (54). Details on these methods are in Supporting Text.
PRCs.
PRCs were constructed by a method similar to those described (33, 34). For details, see Supporting Text.
Statistics.
For period lengths and cycling amplitudes, either the Wilcoxon-Kruskal/Wallis test, one-way ANOVA, or two-way ANOVA was performed on JMP 3.1.5 or JMP 5.1 (SAS Institute, Cary, NC) (55). Phase values were treated by methods of circular statistics using Microsoft Exel 2000 (56–58). Details on statistical methods used are in Supporting Text.
Supplementary Material
Acknowledgments
We thank M. Hasegawa for advice on the light-entrainment experiments; M. Straume and J. D. Levine for help with rhythm analysis programs; H. Borsetti for discussion and general support on this project; and M. A. Rea and D. E. Wells for comments on the manuscript. M.K. was an O’Donnell Foundation Fellow of the Life Sciences Research Foundation. This work was supported by National Institutes of Health Grant MH60939 and Texas Advanced Research Program Grant 3652-761 (to G.M.C.).
Abbreviations
- SCN
suprachiasmatic nucleus
- per
period
- luc
luciferase
- DD
constant dark
- LL
constant light
- CRY
Cryptochrome
- LD
light-dark
- FFT-NLLS
fast Fourier transform–nonlinear least squares
- MESA
maximum entropy spectral analysis
- PRC
phase response curve
- ZT
Zeitgeber time
- CT
circadian time
Footnotes
The authors declare no conflict of interest.
References
- 1.Stanewsky R. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 2.Harmer SL, Panda S, Kay SA. Annu Rev Cell Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 3.Young MW, Kay SA. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 4.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 5.Plautz JD, Kaneko M, Hall JC, Kay SA. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 7.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helfrich-Forster C. J Comp Physiol A. 2004;190:601–613. doi: 10.1007/s00359-004-0527-2. [DOI] [PubMed] [Google Scholar]
- 9.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 10.Schibler U, Ripperger J, Brown SA. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 11.Hege DM, Stanewsky R, Hall JC, Giebultowicz JM. J Biol Rhythms. 1997;12:300–308. doi: 10.1177/074873049701200402. [DOI] [PubMed] [Google Scholar]
- 12.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 14.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 15.Myers EM, Yu J, Sehgal A. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoleru D, Peng Y, Nawathean P, Rosbash M. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 18.Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veleri S, Brandes C, Helfrich-Forster C, Hall JC, Stanewsky R. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 20.Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG, et al. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, Pletcher MT, Sato TK, Wiltshire T, Andahazy M, et al. Science. 2003;301:525–527. doi: 10.1126/science.1086179. [DOI] [PubMed] [Google Scholar]
- 22.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, et al. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 24.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 25.Carr AJ, Whitmore D. Nat Cell Biol. 2005;7:319–321. doi: 10.1038/ncb1232. [DOI] [PubMed] [Google Scholar]
- 26.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cahill GM. Cell Tissue Res. 2002;309:27–34. doi: 10.1007/s00441-002-0570-7. [DOI] [PubMed] [Google Scholar]
- 29.Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Toh H, Fukuda I, Tsujimura T, Terada N, Kamei Y, et al. Genes Cells. 2000;5:725–738. doi: 10.1046/j.1365-2443.2000.00364.x. [DOI] [PubMed] [Google Scholar]
- 31.Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B. Science. 2000;289:297–300. doi: 10.1126/science.289.5477.297. [DOI] [PubMed] [Google Scholar]
- 32.Pando MP, Pinchak AB, Cermakian N, Sassone-Corsi P. Proc Natl Acad Sci USA. 2001;98:10178–10183. doi: 10.1073/pnas.181228598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallone D, Gondi SB, Whitmore D, Foulkes NS. Proc Natl Acad Sci USA. 2004;101:4106–4111. doi: 10.1073/pnas.0305436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeBruyne J, Hurd MW, Gutierrez L, Kaneko M, Tan Y, Wells DE, Cahill GM. J Neurogenet. 2004;18:403–428. doi: 10.1080/01677060490894540. [DOI] [PubMed] [Google Scholar]
- 35.Whitmore D, Foulkes NS, Sassone-Corsi P. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 36.Cermakian N, Pando MP, Thompson CL, Pinchak AB, Selby CP, Gutierrez L, Wells DE, Cahill GM, Sancar A, Sassone-Corsi P. Curr Biol. 2002;12:844–848. doi: 10.1016/s0960-9822(02)00835-7. [DOI] [PubMed] [Google Scholar]
- 37.Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, Foulkes NS. PLoS Biol. 2005;3:e351. doi: 10.1371/journal.pbio.0030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko M, Cahill GM. PLoS Biol. 2005;3:e34. doi: 10.1371/journal.pbio.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cermakian N, Whitmore D, Foulkes NS, Sassone-Corsi P. Proc Natl Acad Sci USA. 2000;97:4339–4344. doi: 10.1073/pnas.97.8.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziv L, Levkovitz S, Toyama R, Falcon J, Gothilf Y. J Neuroendocrinol. 2005;17:314–320. doi: 10.1111/j.1365-2826.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- 41.Hurd MW, Debruyne J, Straume M, Cahill GM. Physiol Behav. 1998;65:465–472. doi: 10.1016/s0031-9384(98)00183-8. [DOI] [PubMed] [Google Scholar]
- 42.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 43.Stanewsky R, Lynch KS, Brandes C, Hall JC. J Biol Rhythms. 2002;17:293–306. doi: 10.1177/074873002129002609. [DOI] [PubMed] [Google Scholar]
- 44.Hall JC. Adv Genet. 2003;48:1–280. doi: 10.1016/s0065-2660(03)48000-0. [DOI] [PubMed] [Google Scholar]
- 45.Johnson C, Elliott JA, Foster RG, Honma KI, Kronauer R. In: Chronobiology: Biological Timekeeping. Dunlap J, Loros JJ, Decoursey PJ, editors. Sunderland, MA: Sinauer; 2003. pp. 66–105. [Google Scholar]
- 46.Ruby NF, Burns DE, Heller HC. J Neurosci. 1999;19:8630–8636. doi: 10.1523/JNEUROSCI.19-19-08630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izumo M, Johnson CH, Yamazaki S. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan LW, Feldman JF, Bell-Pedersen D. Philos Trans R Soc London B. 2001;356:1717–1724. doi: 10.1098/rstb.2001.0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tosini G, Menaker M. NeuroReport. 1998;9:1001–1005. doi: 10.1097/00001756-199804200-00009. [DOI] [PubMed] [Google Scholar]
- 50.Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 51.Rensing L, Ruoff P. Chronobiol Int. 2002;19:807–864. doi: 10.1081/cbi-120014569. [DOI] [PubMed] [Google Scholar]
- 52.Hirayama J, Kaneko M, Cardone L, Cahill G, Sassone-Corsi P. Methods Enzymol. 2005;393:186–204. doi: 10.1016/S0076-6879(05)93005-X. [DOI] [PubMed] [Google Scholar]
- 53.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 54.Levine JD, Funes P, Dowse HB, Hall JC. BMC Neurosci. 2002;3:1–25. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokal R, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Rhythms. New York: Freeman; 1995. [Google Scholar]
- 56.Batschelet E. Statistical Methods for the Analysis of Problems in Animal Orientation and Certain Biological Rhythms. Washington, DC: American Institute of Biological Sciences; 1967. [Google Scholar]
- 57.Batschelet E. Circular Statistics in Biology. London: Academic; 1981. [Google Scholar]
- 58.Fisher N. Statistical Analysis of Circular Data. Cambridge, UK: Cambridge Univ Press; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.