Abstract
The phytopathogenic bacterium Ralstonia solanacearum encodes a family of seven type III secretion system (T3SS) effectors that contain both a leucine-rich repeat and an F-box domain. This structure is reminiscent of a class of typical eukaryotic proteins called F-box proteins. The latter, together with Skp1 and Cullin1 subunits, constitute the SCF-type E3 ubiquitin ligase complex and control specific protein ubiquitinylation. In the eukaryotic cell, depending on the nature of the polyubiquitin chain, the ubiquitin-tagged proteins either see their properties modified or are doomed for degradation by the 26S proteasome. This pathway is essential to many developmental processes in plants, ranging from hormone signaling and flower development to stress responses. Here, we show that these previously undescribed T3SS effectors are putative bacterial F-box proteins capable of interacting with a subset of the 19 different Arabidopsis Skp1-like proteins like bona fide Arabidopsis F-box proteins. A R. solanacearum strain in which all of the seven GALA effector genes have been deleted or mutated was no longer pathogenic on Arabidopsis and less virulent on tomato. Furthermore, we found that GALA7 is a host-specificity factor, required for disease on Medicago truncatula plants. Our results indicate that the GALA T3SS effectors are essential to R. solanacearum to control disease. Because the F-box domain is essential to the virulence function of GALA7, we hypothesize that these effectors act by hijacking their host SCF-type E3 ubiquitin ligases to interfere with their host ubiquitin/proteasome pathway to promote disease.
Keywords: plant pathogen, SCF complex, type III secretion system, virulence
The soil-borne pathogen Ralstonia solanacearum uses a type III secretion system (T3SS) to inject effector proteins into plant cells (1). The T3SS is a major determinant controlling the virulence of bacterial pathogens on both mammal and plant hosts. The function of T3SS effectors in controlling the host invasion and the onset of the disease has remained elusive for years. Discovering the function of these effectors will prove invaluable for understanding bacterial invasion strategies and seeking new ways to prevent disease. It is only recently that molecular functions of several T3SS effectors have been uncovered. An example of well characterized T3SS effectors are the cysteine proteases encoded by both human and plant pathogenic bacteria (2–5), allowing them to cleave or degrade specific host substrates (6–9). Genome sequencing of several bacteria using the T3SS strategy has revealed that there are several sets of numerous and diverse putative T3SS effectors within and between species (10–13). The extent of this diversity is likely to provide a complex arsenal for bacteria to promote disease on their different hosts.
We have shown that the plant pathogen R. solanacearum encodes a large repertoire (up to 80 candidates) of putative T3SS effectors (13–15). Several effectors are part of multigene families. Among these, we identified a group of seven genes that have homologies with plant-specific leucine-rich repeats (LRR) (14). These T3SS effectors were named “GALA” proteins after a conserved GAxALA sequence in their LRR (14). GALA6 has been shown to be translocated into plant cells (14), and the first 50 aa of the six other GALA proteins show the characteristic features of T3SS substrates (14, 16).
Recent draft sequences of two other R. solanacearum strains adapted to specific host plants revealed the presence of orthologous genes for six of the seven GALA genes in both strains (Molk2 banana-specific strain (N.P., C. Boucher, and S.G., unpublished data) and UW551, a potato-specific strain) (17). The conservation of these effectors among evolutionarily distant strains (18) suggests that these T3SS effectors are selected for and are, therefore, probably important for R. solanacearum pathogenicity.
Here, we present data that contribute to the understanding of the role of this important family of T3SS effectors, uncovering a strategy developed by plant pathogens to colonize their hosts.
Results
R. solanacearum Encodes for Type III Effectors Containing F-Box Domains.
To investigate the function of the GALA LRR family of T3SS effectors (14), a yeast two-hybrid (Y2H) screen was carried out with GALA6 on an Arabidopsis cDNA library (Arabidopsis Col-0 whole plants, 6.4 × 106 yeast colonies screened; Clontech, Mountain View, CA). This initial screen enabled us to identify the Arabidopsis SKP1-like ASK1 as a potential GALA6 interactor. After a careful sequence analysis of GALA6 and the other GALA members, we discovered an F-box domain in each of these bacterial proteins [Pfam analysis (PF00646)]. The F-box is a short domain (48 aa) characteristic of the eukaryotic F-box proteins, allowing them to interact directly with the SKP1-like proteins (19). SKP1-like proteins interact with Cullin1, and, all together, they form the SCF-type E3 ubiquitin ligase (20). E3 ubiquitin ligases and, particularly, F-box proteins, are a very large protein family in Arabidopsis (21). These proteins recognize specific target proteins via protein–protein-interaction domains and recruit these target proteins into the SCF complex, thanks to their interaction with the core SKP1-like proteins. Subsequent ubiquitinylation of the target proteins can modulate their activity or lead to their proteasome-dependent degradation, depending on the nature of the ubiquitin chain (22). For all GALA proteins except GALA2, this F-box motif is located in the N terminus of the protein, whereas most of the rest of the protein is constituted by the LRR (Fig. 1A). Also, from the F-box sequences of the GALA proteins (Fig. 1B), we can infer that GALA4 F-box might not be functional because of the phenylalanine (F) amino acid at position 65. Indeed all F-box domains (plant and other eukaryotes) have a conserved leucine at that position, which has been shown to be important for the interaction with the SKP1-like protein (19). Interestingly this F65 is reverted to the conserved leucine residue in the GALA4 orthologous gene in strains Molk2 (N.P., C. Boucher, and S.G., unpublished data) and UW551 (17).
Fig. 1.
Structure of the GALA F-box proteins. (A) Structure of the seven GALA proteins. In parentheses is the length of the protein in amino acids, followed by the number of LRR. The black box represents the 48-aa-long F-box domain as defined by Pfam. Gray boxes represent 24-aa-long LRR (14). (B) Sequence alignment of GALA F-box domains, with the GALA consensus (GALA cons.). The size of the F-box domains aligned is as defined (25). SKIP cons. stands for the consensus defined on the alignment of all F-box domains from plant proteins known to interact with ASK1 and/or ASK2 (25). hSKP2 is the human SKP2 F-box protein (Q13309). Asterisks indicate hSKP2 residues that contact hSKP1 (19). Amino acid substitution groups were numbered as follows: 1, ED; 2, NQ; 3, ST; 4, KR; 5, FYW; and 6, LIVM. Residues that are conserved >80% are dark shaded, residues that are 60–80% conserved are light shaded. Shading of hSKP2 residues according to the most conserved residue in either GALA or SKIP consensus.
GALA6 Binds to Arabidopsis ASK1 and ASK2.
When expressed as a fusion protein with the yeast GAL4-binding domain (BD), GALA6 was able to interact in yeast with ASK1 fused to the activation domain (AD). As a control, this interaction was not observed when the F-box domain was deleted from GALA6 (Fig. 2A). The lack of Y2H interaction observed was not because of the absence of BD-GALA6ΔF-box protein, because the protein could be detected by Western blot (results not shown). To further confirm that GALA6 has a functional F-box domain, we tested the ability of GALA6 to physically interact with plant-expressed ASK proteins. For that purpose, a GST-tagged version of GALA6 was used in a pull-down assay to evaluate the ability of this protein to bind a 6myc-tagged version of ASK2 produced by a transgenic Arabidopsis line (23). An antiserum, directed against ASK1 and cross-reacting with ASK2, was used to analyze the proteins that were bound to GST-GALA6. This experiment revealed a 37-kDa band corresponding to the 6myc-tagged form of ASK2 and a 18-kDa band corresponding to the native forms of ASK1 and/or ASK2 present in the transgenic lines (Fig. 2B). The reprobing of the stripped Western-blot with an anti-myc antibody identified the same 37-kDa band (result not shown). When GST-GALA6ΔF-box was used, the interaction with ASK2 was no longer observed.
Fig. 2.
Interaction between R. solanacearum GALAs and Arabidopsis ASKs. (A) Y2H among BD-GALA6, BD-GALA6ΔF-box, and AD-ASK1. Serial 10× dilutions were spotted from left to right on nonselective medium (−LT) and selective medium lacking His (−LTH). (B) Western blot probed with anti-ASK1 antibody after GST pull-down of Arabidopsis total protein extract. Equal amounts of GST, GST-GALA6, and GST-GALA6ΔF-box (as estimated by PAGE/Coomassie, data not shown) were incubated with total protein extract from Arabidopsis seedlings expressing 6myc-tagged ASK2 (23). (C) Y2H interaction pattern between ASKs and GALAs. Positive interactions were observed only when the BD was fused to GALA and the AD was fused to ASK.
GALA Proteins Specifically Interact with Subsets of Arabidopsis SKP1-Like Proteins.
Unlike yeast, Drosophila, or human, Arabidopsis has an expanded SKP1-like (ASK) gene family with 19 functional genes (24, 25). We systematically tested the interaction between the six cloned GALAs and the different ASKs by Y2H. Results summarized in Fig. 2C show the existence of interaction specificities. With the exception of GALA3 and GALA4, for which no interaction was detected with any of the ASK proteins, each of the four remaining effectors tested has its own specific ASK-interaction pattern. These four GALA proteins interact with ASK1 and ASK2, the two most expressed ASKs. Only GALA1 interacts with the third most expressed, ASK4. Interestingly, GALA1, 5, 6, and 7 interact with ASK11 and/or ASK13 and do not interact with ASK12, even though ASK11 and ASK12 are 98% identical. As for the GALA6–ASK1 interaction (Fig. 2A), interactions between GALA6 and ASK2 or ASK13 were abolished when the F-box domain was removed from GALA6. The lack of Y2H interaction observed with BD-GALA4 was not because of the absence of BD-fusion protein, because its expression was confirmed by Western blot (results not shown).
GALA Genes Are Essential for Disease on Arabidopsis and Tomato.
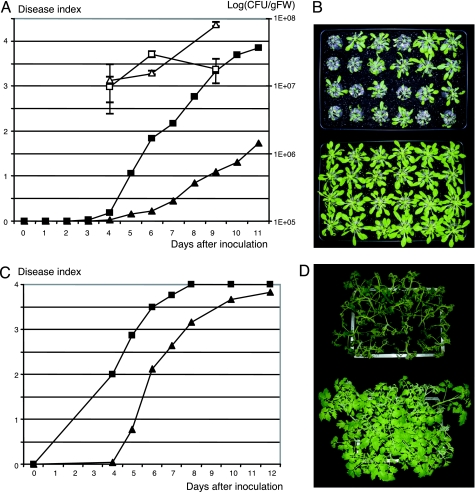
Previous experiments have shown that mutations generated in each of the seven GALA genes did not affect R. solanacearum’s pathogenicity toward Arabidopsis and tomato (14). The lack of phenotype of single mutants could be explained by a functional overlap of these proteins. To test this hypothesis, we constructed a R. solanacearum strain (GRS447) in which all of the seven GALA genes were deleted or mutated. This mutant was then tested for virulence on Arabidopsis and tomato. Fig. 3A shows the progression of the wilting symptoms on Arabidopsis over 11 days. Plants inoculated with the GRS447 mutant show a very reduced wilting (Fig. 3B). Even after 11 days, the wilting symptoms recorded were very low, and plants could be kept alive for a long period (up to a month after inoculation). We tested the presence of the inoculated bacteria by measuring the internal bacterial load at three time points (Fig. 3A, open symbols). Even though the mutant-inoculated plants do not display wilting symptoms, they contain similar amounts of R. solanacearum as the control wilting plants.
Fig. 3.
Virulence test of GRS447 on Arabidopsis and tomato. (A) Progression of the average wilting symptoms on Arabidopsis Col-0 plants over 11 days; filled squares, wild-type strain GMI1000; filled triangles, GRS447. IGCs are represented as a mean and standard error calculated on three data points, at days 4, 6, and 9; open squares, GMI1000; open triangles, GRS447. (B) Plant-wilting phenotype of Arabidopsis 6 days after inoculation; GMI1000-inoculated (Upper) and GRS447-inoculated (Lower) plants. (C) Progression of the average wilting symptoms on tomato (cultivar Super Marmande) over a 12-day period, symbols as in A. (D) Tomato wilting at day 5 after inoculation; GMI1000-inoculated (Upper) and GRS447-inoculated (Lower) plants.
When tested on tomato, the absence of all seven GALA genes resulted in a significant reduction of virulence (2-days delay), but, unlike Arabidopsis, all of the tomato plants inoculated with GRS447 eventually wilted (Fig. 3 C and D).
GALA7 Is an Essential Host-Specificity Factor on Medicago truncatula.
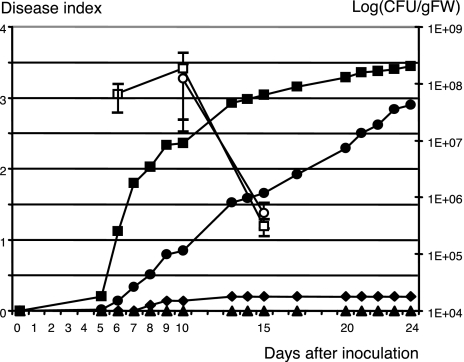
To circumvent the lack of phenotype for the single mutants on Arabidopsis and tomato, we also tested the seven single-mutant strains on M. truncatula, a recently characterized host plant for R. solanacearum (26). Among these strains, the GALA7 mutant strain GRS138 (14) shows a dramatic reduction in virulence on this host (Fig. 4). Fourteen-day-old plants inoculated with the wild-type strain GMI1000 start wilting after 6 days, whereas the mutant strain GSR138 barely displays any symptoms on M. truncatula (the symptoms in Fig. 4 correspond to 2 wilting plants of 39 tested; in similar experiments, GRS138 didn’t display any symptoms at all). A complementation test was performed by using pNP221 carrying the GALA7 gene. Although the appearance of the symptoms was delayed compared with the wild-type strain, the presence of the complementing plasmid significantly restored the pathogenicity of the mutant strain GRS138. Bacterial internal growth at different time points was checked (Fig. 4, open symbols), and, at day 6, only one in three repetitions shows the presence of infecting R. solanacearum when the complemented strain GRS138(pNP221) was used. At later stages of appearance of wilting symptoms, the internal growth curves (IGCs) were similar with the wild-type strains GMI1000 and GRS138(pNP221). In plants inoculated with the other strains, no bacteria could be detected.
Fig. 4.
Virulence of GRS138 on M. truncatula line A17. Plants were inoculated at 108 cfu/ml. The progression of wilting symptoms was assessed by average disease-index score. The wild-type strain GMI1000 (filled squares) induces wilting 6 days after inoculation. Strain GRS138 (filled diamonds), which carries a disruption of the GALA7-encoding gene (14), is severely impaired in its virulence on M. truncatula. GRS138 complemented with the GALA7 gene (pNP221, filled circles) has a restored virulence, whereas the inoculation with GRS138 expressing a GALA7 gene with its F-box deleted (pNP222, filled triangles) did not produce any wilting on M. truncatula. Open symbols represent the IGC (mean and standard error bars) analyzed for GMI1000 (squares) and GRS138(pNP221) (circles) at day 6 [no R. solanacearum was detected for GRS138(pNP221)] and days 10 and 15. No R. solanacearum could be detected in GRS138 or GRS138(pNP222) at these same time points.
BLAST and alignment analysis enabled us to identify MSKa and MSKb as the probable homologues of ASK1 and ASK2 in M. truncatula EST libraries. We showed, in a Y2H assay, that BD-GALA1, 3, 5, 6, and 7 were capable of interacting with both AD-MSKa and AD-MSKb. As expected, BD-GALA6ΔF-box and BD-GALA7ΔF-box did not interact with the AD-MSKs (results not shown). We then showed that the complementation of GRS138 did not occur with a plasmid expressing the GALA7 gene with its F-box deleted (pNP222, Fig. 4). Using MS analysis on bacterial culture supernatant, we were able to identify GALA7 by using GMI1000 and GRS138pNP221, but GALA7 was not identified in the supernatant of GMI1694 (hrp mutant strain) (14), GRS138, or GMI1694pNP221. In GRS138pNP222 (but not GMI1694pNP222), we were able to identify GALA7ΔF-box (results not shown). This result shows that GALA7ΔF-box, like GALA7, can be secreted by the T3SS. We also showed, using the adenylate cyclase reporter system (27), that GALA7ΔF-box as well as GALA7 could be injected by R. solanacearum in M. truncatula mesophyll cells. Indeed, the efficiency of GALA7-CyaA′ and GALA7ΔF-box-CyaA′ calmodulin-dependent cAMP production in plant cells was not significantly different (Student test, P = 0.05), whereas disruption of the T3SS (mutant hrcV, GMI1694) significantly reduced cAMP production of either GALA7 versions (Student test, P = 0.05) (see Table 1, which is published as supporting information on the PNAS web site). All together, these data demonstrate that the F-box domain of GALA7 is required for the virulence function of GALA7 on M. truncatula.
Discussion
Different bacteria, interacting with different hosts, have independently evolved T3SS effectors capable of interfering with similar functions in their host cells. These bacteria manage to do so by mimicking components of their eukaryotic hosts (28, 29), and some have been shown to target key cell mechanisms: There are examples of T3SS effectors that interfere with the transcription machinery (30, 31), whereas others are functional cysteine proteases and can cleave plant proteins (2, 3, 5, 32). Bacterial T3SS effectors that directly interfere with their host ubiquitin/proteasome pathway have recently been identified and studied in Shigella flexneri (OspG, inhibiting E2 ubiquitin-conjugating enzyme) (33) and Pseudomonas syringae (AvrPtoB, U-box type E3-ubiquitin ligase) (34, 35). The work on AvrPtoB highlights that bacterial T3SS effectors can mimic a eukaryotic E3 ubiquitin ligase. Furthermore, it has been shown that viruses (36, 37) and type 4 secretion system bacteria (38–40), can mimic F-box proteins, part of the SCF type of E3 ubiquitin ligase. We provide evidence that R. solanacearum uses T3SS effectors that contain a plant-like F-box domain and contribute to the virulence of this organism on various host plants. In Arabidopsis, F-box proteins are the essential adaptor proteins linking the protein to be ubiquitinated to the SCF E3-ubiquitin ligase via its interaction with the ASK core unit. F-box proteins interact mostly with ASK1 and ASK2, the two majors ASKs (41), but also with ASK4, 11, and 13 (21, 25). ASK4 is the third most abundant ASK and, together with ASK1 and ASK2, is expressed in all plant tissues (42). Although many Arabidopsis F-box proteins bind ASK11, they often do not bind ASK12, even though these two proteins are 98% identical. We found that the GALA F-box proteins have a similar interaction pattern, suggesting that these virulence proteins were once acquired by R. solanacearum, probably through lateral transfer of plant DNA, and that the plant characteristic features of the F-box domain were kept by selective pressure.
The fact that single GALA mutants were not affected in their virulence on Arabidopsis and tomato and that the septuple GALA mutant (GRS447) is affected could be explained by either (i) one GALA protein is a virulence determinant, but its absence in the single mutants is complemented by one or more of the other GALAs or (ii) at least two nonfunctionally overlapping GALAs are required. Future complementation studies of GRS447 should enable us to identify the key GALA players on Arabidopsis and tomato. Interestingly, even though GRS447 does not induce a wilting phenotype on Arabidopsis, this mutant can colonize the plant similarly to the wild-type strain, indicating that symptoms and bacterial internal growth could be dissociated. This observation has already been reported to be the case for two P. syringae T3SS effectors (43, 44). For R. solanaceraum, development of wilt symptoms is directly associated with the obstruction of the xylem vessels by the exopolysaccharides (EPS) produced by the bacteria. Because GRS447 is not defective in EPS production, our observation suggests that GALA T3SS effectors may also be involved in disease symptom development in a way that remains unknown.
R. solanacearum GMI1000 has a large repertoire of T3SS effectors (13–15). We can imagine that this characteristic allows the bacterium to colonize a wide variety of plants. In this work, we have pinned down GALA7 as a T3SS virulence effector directly responsible for host-specific colonization. The only other known examples of T3SS effectors that positively extend the host specificity are HsvG and HsvB of Pantoea agglomerans (45).
GALA7’s function in the interaction between R. solanacearum and M. truncatula requires the presence of the F-box, suggesting that GALA7’s function in virulence could be associated with the specific ubiquitinylation and, possibly, degradation of a plant protein. One could imagine that the bacterium succeeds in colonization by degrading a positive regulator of the plant’s basal defense. But, because we have not yet identified any targets of the SCFGALA, we cannot formally rule out other possibilities for GALA effector function, such as the specific interference with plant SCF complexes or depletion of ASK subunits affecting the general ubiquitin/proteasome pathway. However, because we showed that GALA7 is not the only GALA effector that can interact with M. truncatula SKP1-like proteins, it seems more likely that the GALA7 virulence role relies on the capacity of its LRR to recruit specific plant components to a SCFGALA7 E3 ubiquitin ligase for subsequent ubiquitination and possible degradation.
Materials and Methods
GALA and SKP1-Like cDNA Cloning.
All GALA genes except GALA2 were cloned by PCR on R. solanacearum genomic DNA with Pfx (Invitrogen) into the pENTR/SD/D-TOPO or pDONR207 (Invitrogen). Oligonucleotides were designed to obtain the ORFs from the start to the last codon. Repeated attempts in cloning GALA2 have been unsuccessful. Arabidopsis ASK ORFs were cloned by using the same strategy. The M. truncatula SKP1-like cDNAs MSKa and MSKb were PCR amplified from root cDNA preparations and cloned like ASK cDNAs. For more details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
For the complementation test, we PCR amplified the GALA7 gene and the F-box-deleted version from the whole promoter region (410 bp) to the last codon and cloned the PCR fragment in pSC154 (14). pNP219, 220, 221, and 222 are pSC154-derived plasmids cloned into the KpnI site of pLAFR6 (46). pNP220 contains the GALA7 ORF in fusion with CyaA′ (27), and pNP219 contains the GALA7ΔF-box ORF in fusion with CyaA′. pNP221 and pNP222 are, respectively, the equivalents of pNP220 and pNP219, with a stop codon as the last codon of GALA7 or GALA7ΔF-box. All of the oligonucleotides used in this work are listed in Table 2, which is published as supporting information on the PNAS web site.
Secretion and Injection of GALA7.
For MS analysis, the bacterial supernatants were prepared (47) and analyzed by LC-MS-MS as described (48). The T3SS-dependent injection assay of GALA7 and GALA7ΔF-box was performed with pNP219 and pNP220. Both plasmids were introduced into GRS138 and the hrp secretion mutant GMI1694 (14). Each strain was infiltrated in M. truncatula (A17) leaves at 108 cfu/ml. After 6 h, four infiltrated leaves were harvested and frozen in liquid nitrogen. cAMP quantification was assessed as described (14). Each experiment was repeated three times.
Y2H Analysis.
GALA and ASK ORFs were fused to the BD and AD of yeast GAL4 protein by Gateway LR recombination into pACT and pAS2 (Matchmaker1; BD Biosciences, Palo Alto, CA) previously made Gateway-compatible. Saccharomyces cerevisiae strain AH109 (BD Biosciences) was used for the two-hybrid tests. Yeast cells were cotransformed with a GALA-BD fusion and an ASK-AD fusion containing vectors. Double transformants were then tested for interaction by plating six successive 10× dilutions on synthetic minimal medium lacking Leu, Trp, and His. Each interaction test was performed on three different colonies.
GST Pull-Down.
Recombinant GST-GALA6 and GST-GALA6ΔF-box proteins were incubated together with total protein extract prepared from Arabidopsis seedlings expressing 6myc-tagged ASK2 (23). The bound proteins were probed on a Western blot with anti-ASK1. For more details, see Supporting Materials and Methods.
Multiple GALA Mutant Strain.
A septuple-mutant strain for each GMI1000 GALA gene was generated by an iterative disruption process. First, a disruption strain for the GALA3 gene was generated by insertion of the Ω interposon (49) in the unique HindIII site of a DNA fragment amplified with primers 2003UP and 2003DW and introduced in the GMI1000 genome by natural transformation as described (14). The resulting strain, GRS417, was used as a recipient for introduction of the deletion mutation of GALA1 already described (14). The remaining GALA genes (GALA2, 4, 5, 6, and 7) were deleted in this double mutant by using a cre-lox system for antibiotic marker recycling (50). For more details, see Supporting Materials and Methods and Table 2. At every disruption step, the genomic structure of the transformants was verified by PCR analysis and Southern hybridization. The septuple mutant was also checked for displaying the typical hypersensitive-response phenotype upon infiltration into tobacco leaves.
Virulence Tests and Bacterial IGC.
R. solanacearum GMI1000, GRS447, GRS138, and the complemented GRS138 strains were cultured overnight in the B medium (51). Four-week-old Arabidopsis Col-0 plants were inoculated (with 107 cfu/ml) after root clipping (52). The test involved 49 plants for each bacterial inoculation. For each IGC time point, 3 plants were collected in the order they were placed in the tray and pooled; this procedure was repeated three times. The IGC was performed as described (52), except that 5 μl of the diluted solutions were spotted on SMSA plates (53). The typical R. solanacearum phenotype of the bacteria counted on the different dilutions was checked on B rich medium. Twenty-four, 4-week-old tomato plants (cult. Super Marmande) were inoculated by soaking the soil with R. solanacearum suspension at 107 cfu/ml. Thirty-nine 14-day-old M. truncatula (A17) plants, grown on Jiffy pellets, were inoculated with the bacterial suspension (108 cfu/ml) just after root clipping, as described (26). IGC on M. truncatula were performed by collecting the first true leaf of three successive plants. This procedure was repeated three times, and the subsequent steps were carried out as for Arabidopsis. For all these plant inoculations, the symptoms were scored according to a scale ranging from 0 for no symptom to 4 for completely wilted plants.
Supplementary Material
Acknowledgments
We thank J. Uhrig (Max-Planck-Institut für Züchtungsforschung, Cologne, Germany) for the Gateway-compatible Y2H vectors. We also thank M. Rossignol and the proteomic facility staff of the IFR40 in Toulouse for the MS analysis. This work was funded by Genoplante Grant GENER006 and by a French Government PhD grant (to A.A.).
Abbreviations
- AD
activation domain
- BD
binding domain
- IGC
internal growth curve
- LRR
leucine-rich repeat
- T3SS
type III secretion system
- Y2H
yeast two-hybrid
Note Added in Proof.
While this article was under review, Nomura et al. (54) reported on the T3SS virulence effector HopM1 of P. syringae.This effector, without having classical E3 ubiquitin ligase features, is capable of directing the proteasome-dependent degradation of AtMin7 and preventing plant defense-associated callose deposition.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences in this paper have been deposited in the GenBank database [accession nos. RSp0914 (GALA1), RSp0672 (GALA2), RSp0028 (GALA3), RSc1800 (GALA4), RSc1801 (GALA5), RSc1356 (GALA6), and RSc1357 (GALA7) (R. solanacearum) and DQ641945 (MSKa) and DQ641946 (MSKb) (M. truncatula)].
References
- 1.Genin S, Boucher C. Annu Rev Phytopathol. 2004;42:107–134. doi: 10.1146/annurev.phyto.42.011204.104301. [DOI] [PubMed] [Google Scholar]
- 2.Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- 3.Alfano JR, Collmer A. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 4.Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 5.Viboud GI, Bliska JB. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 6.Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 7.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 8.Orth K. Curr Opin Microbiol. 2002;5:38–43. doi: 10.1016/s1369-5274(02)00283-7. [DOI] [PubMed] [Google Scholar]
- 9.Shao F, Vacratsis PO, Bao Z, Bowers KE, Fierke CA, Dixon JE. Proc Natl Acad Sci USA. 2003;100:904–909. doi: 10.1073/pnas.252770599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, et al. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 11.Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, Brinkac LM, Daugherty SC, Deboy R, Durkin AS, Giglio MG, et al. J Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, et al. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 14.Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. Mol Microbiol. 2004;53:115–128. doi: 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- 15.Occhialini A, Cunnac S, Reymond N, Genin S, Boucher C. Mol Plant–Microbe Interact. 2005;18:938–949. doi: 10.1094/MPMI-18-0938. [DOI] [PubMed] [Google Scholar]
- 16.Chang JH, Urbach JM, Law TF, Arnold LW, Hu A, Gombar S, Grant SR, Ausubel FM, Dangl JL. Proc Natl Acad Sci USA. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, et al. Mol Plant–Microbe Interact. 2006;19:69–79. doi: 10.1094/MPMI-19-0069. [DOI] [PubMed] [Google Scholar]
- 18.Poussier S, Prior P, Luisetti J, Hayward C, Fegan M. Syst Appl Microbiol. 2000;23:479–486. doi: 10.1016/S0723-2020(00)80021-1. [DOI] [PubMed] [Google Scholar]
- 19.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 20.Vierstra RD. Trends Plant Sci. 2003;8:135–142. doi: 10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 21.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirkpatrick DS, Denison C, Gygi SP. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kipreos E, Pagano M. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. reviews3002.1–reviews3002.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. Plant J. 2003;34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- 26.Vailleau F, Sartorel E, Jardinaud MF, Chardon F, Genin S, Huguet T, Gentzbittel L, Petitprez M. Mol Plant–Microbe Interact. 2006 doi: 10.1094/MPMI-20-2-0159. in press. [DOI] [PubMed] [Google Scholar]
- 27.Sory MP, Cornelis GR. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 28.Desveaux D, Singer AU, Dangl JL. Curr Opin Plant Biol. 2006;9:376–382. doi: 10.1016/j.pbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Annu Rev Microbiol. 2006 doi: 10.1146/annurev.micro.60.080805.142251. in press. [DOI] [PubMed] [Google Scholar]
- 30.Szurek B, Marois E, Bonas U, Van den Ackerveken G. Plant J. 2001;26:523–534. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- 31.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotson A, Mudgett MB. Curr Opin Plant Biol. 2004;7:384–390. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Proc Natl Acad Sci USA. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 36.Banks L, Pim D, Thomas M. Trends Biochem Sci. 2003;28:452–459. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 37.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. Proc Natl Acad Sci USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 39.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 40.Tzfira T, Vaidya M, Citovsky V. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D. Plant Cell. 2004;16:5–20. doi: 10.1105/tpc.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H. Plant Physiol. 2003;133:203–217. doi: 10.1104/pp.103.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, He SY. Mol Microbiol. 2003;49:1239–1251. doi: 10.1046/j.1365-2958.2003.03647.x. [DOI] [PubMed] [Google Scholar]
- 44.Lopez-Solanilla E, Bronstein PA, Schneider AR, Collmer A. Mol Microbiol. 2004;54:353–365. doi: 10.1111/j.1365-2958.2004.04285.x. [DOI] [PubMed] [Google Scholar]
- 45.Nissan G, Manulis-Sasson S, Weinthal D, Mor H, Sessa G, Barash I. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05301.x. in press. [DOI] [PubMed] [Google Scholar]
- 46.Huynh TV, Dahlbeck D, Staskawicz BJ. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 47.Gueneron M, Timmers AC, Boucher C, Arlat M. Mol Microbiol. 2000;36:261–277. doi: 10.1046/j.1365-2958.2000.01870.x. [DOI] [PubMed] [Google Scholar]
- 48.Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerre-Tugaye MT, Pont-Lezica R. Proteomics. 2005;5:212–221. doi: 10.1002/pmic.200400882. [DOI] [PubMed] [Google Scholar]
- 49.Prentki P, Krisch HM. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 50.Marx CJ, Lidstrom ME. BioTechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 51.Boucher CA, Barberis PA, Trigalet AP, Demery DA. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- 52.Deslandes L, Pileur F, Liaubet L, Camut S, Can C, Williams K, Holub E, Beynon J, Arlat M, Marco Y. Mol Plant–Microbe Interact. 1998;11:659–667. doi: 10.1094/MPMI.1998.11.7.659. [DOI] [PubMed] [Google Scholar]
- 53.Elphinstone J, Hennessy J K. W. J., Stead DE. EPPO Bull. 1996;26:663–678. [Google Scholar]
- 54.Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. Science. 2005;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.