Abstract
We used quantitative phase tomography with synchrotron radiation to elucidate the 3D structure of Arabidopsis seeds in their native state. The cells are clearly distinguished, and their internal structure is revealed through local variations in electron density. We visualized a 3D network of intercellular air space that might allow immediate gas exchange for energy supply during germination and/or serve for rapid water uptake and distribution during imbibition.
Keywords: mature seed structure, synchrotron radiation, x-ray imaging, embryo organs, cell components
Our understanding of seed structure and germination depends critically on insights about the structural arrangement of organs and tissues within the seed. Germination starts with water uptake by the dry seed and is complete when the elongating radicle traverses the seed coat. One of the first changes during imbibition is reestablishment of respiration in mitochondria. This activity is connected to a remarkable initial rise in oxygen consumption that declines afterward until germination is complete; i.e., the radicle traverses the seed coat, and atmospheric oxygen can enter the seed and reach the young growing plantlet (1–3). In seeds, limited seed coat permeability is a strong obstacle to gas exchange (4, 5). Consequently, the oxygen needed for germination to start should be rapidly available within the seed. Oxygen might originate from inside the seed and/or from water entering the seed at the beginning of imbibition. At present, no data are available concerning the internal oxygen content in mature seeds because the rigidity of the seed coat at that stage precludes measuring this content. However, stored internal oxygen should exist and be important for germination because oxygen production by endogenous photosynthesis is known to influence seed viability and germination. Unlike animals, plants lack specialized circulation systems for oxygen transport, and the question arises whether air (and oxygen) is stored in seeds and by which means it circulates.
To answer these questions it is important to know the fine structure of the mature seeds. Histology of seeds is usually based on optical or electron microscopy of slices, the results from adjacent sections being then combined. The cutting and fixation steps alter the tissues, and only one-directional (i.e., 2D) processing is possible. Thus, methods based on slicing and fixation procedures cannot give reliable visualization of small cellular interspaces or indicate the existence of networks of spaces. We therefore looked for noninvasive tomographic techniques that could improve the visualization of cell-to-cell organization in the seed tissues. The tomographic approach consists of acquiring projections of an object along different directions and combining them computationally to obtain a 3D reconstruction of the object. This approach has been successfully used in soft x-ray microscopy, with Fresnel zone plates as objective lenses, providing unique visualization of a whole cell at 60-nm resolution (6, 7). The emerging field of “lensless imaging” or “coherent diffraction imaging,” based on phasing a coherent Fraunhofer diffraction pattern (8, 9), is expected, in the 3D imaging of tiny isolated biological objects, to overcome the resolution limit set by the x-ray optical devices. However, to get a 3D image of all of the cells in a mature seed, with a minimum average size of 0.1 mm, other techniques are needed. Hard x-rays are easily transmitted through objects of this size because the absorption they incur is low. Nevertheless, the local variations in attenuation are also very small. Far better sensitivity to compositional changes, hence to the object structure, is obtained from the variations in the phase of the beam. Our experiment schematically shown in Fig. 1 involves quantitative phase contrast tomography (holotomography) using synchrotron radiation with good spatial coherence. In this approach, spatial variations in the phase of the beam, related to the real part of the material’s refractive index, are turned into contrast through Fresnel diffraction, as discussed in Materials and Methods. Because the refractive index of a material for hard x-rays is directly related to its electron density, the result is a 3D representation of the local electron density. This method permits us to analyze at room temperature an autonomous living object, viz whole seeds as they are, without any sample-modifying preparation procedure.
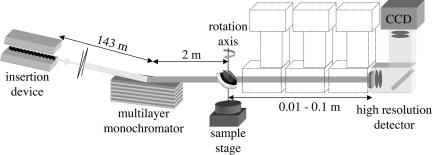
Fig. 1.
Sketch of the experimental setup for quantitative phase tomography: a multilayer monochromator selects the photons with an energy close to 21 keV from the synchrotron radiation emitted by an insertion device. A high-resolution detector system records Fresnel diffraction patterns at varying distances with respect to the sample. During the tomographic scan that is acquired in air and at room temperature, the specimen is rotated around the vertical axis.
Results
3D Images of the Whole Seed.
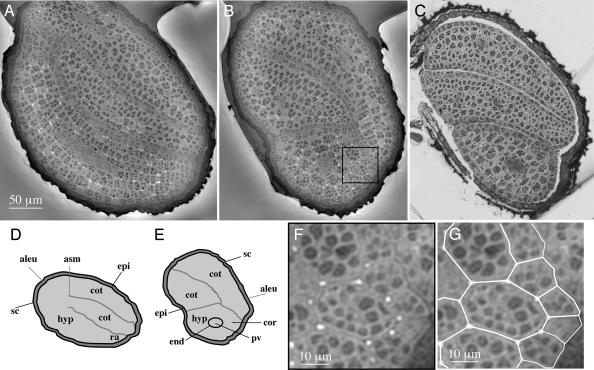
The embryo, enclosed together with food reserves within the seed coat, is composed of cotyledons, hypocotyl and radicle. These parts are readily visible on the images of mutually perpendicular tomographic slices shown in Fig. 2A and B and are schematically represented in Fig. 2 D and E. The two cotyledons and the hypocotyl are densely packed within the seed coat, leaving no intermediary space (Fig. 5, which is published as supporting information on the PNAS web site). On average, the measured difference in the refractive index of the dense structures relative to the matrix is 1.2 × 10−7, similar to the value of 1.57 × 10−7 calculated, at an x-ray energy of 21 keV, for the pair BSA–lipid, BSA being taken as representative of a protein. The corresponding difference in mass densities is about Δρm = 0.28 × 103 kg·m−3. Because the water content of dry seeds is very low and water is not present as free molecules, the matrix corresponds to tightly packed oil bodies, whereas the higher-density structures correspond to regions with a high protein content. For comparison, a light microscopic observation is reported in Fig. 2C representing a 1.2-μm section of a seed in similar orientation as the virtual tomographic slice shown in Fig. 2B. For fixation and cutting it was necessary to introduce an opening into the coat allowing the resin to penetrate. This opening is visible and introduces intermediary space between the embryo organs (Fig. 2C).
Fig. 2.
Architecture of an Arabidopsis seed. Shown are a longitudinal virtual slice (A) and a transverse virtual slice (B) through the same seed, 0.3 μm thick, reconstructed by quantitative phase tomography, along with their schematic representations (D and E). Part of the slice in B, viewed at higher magnification, is shown in F with a duplication (G) with white lines to indicate the cell boundaries. For comparison, a classical light microscopic observation of a transverse section of a resin-embedded open-coat seed is shown (C), brought to a similar orientation and scale as the virtual slice shown in B. With the exception of the coat openings, the schematic representation (E) is applicable to the classical microscopic observation (C). The key components are marked as follows: aleu, aleuron; cot, cotyledon; hyp, hypocotyl; ra, radicle; asm, apical shoot meristem; sc, seed coat; cor, cortex; end, endodermis; epi, epidermis; pv, provasculature.
3D Images of Single Cells.
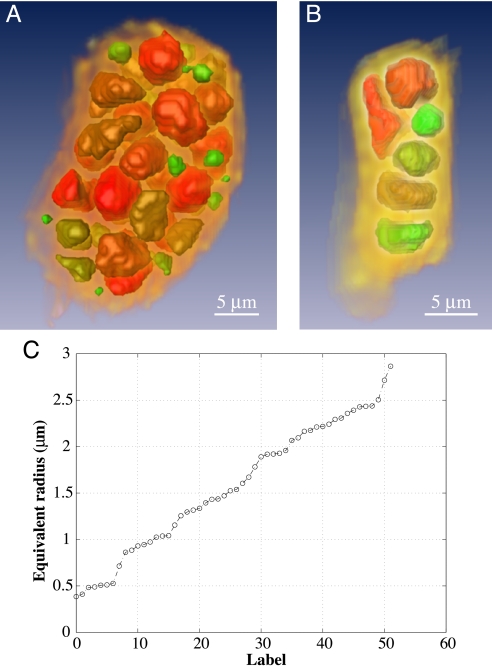
Inspection at high magnification of two single cells (one mesophyll and one palisade cell) reveals well separated dense structures, heterogeneous in shape and size, embedded in a matrix of lower density (Fig. 3A and B). The sizes of these dense structures are evenly distributed over the range 0.4–3.0 μm (Fig. 3C). This cellular organization might correspond to optimal space occupation, e.g., to an optimal distribution between hydrophilic and hydrophobic structures within the seed cells. It is not possible to identify these structures as protein bodies, plastids, mitochondria, or the nucleus through their size distribution. When observed in slices at high magnification, as in Fig. 2F, these denser structures are seen to be inhomogeneous in contrast, and hence in density. It is therefore likely that each single dense structure consists of several organelles or complexes grouped together.
Fig. 3.
3D rendering of single cells. (A) A mesophyll cell. (B) A palisade cell. Higher-density structures in the cells are classified and colored according to their volume such that green and red correspond, respectively, to the smallest and largest cell structure. (C) Equivalent radii of the structures in the mesophyll cell, arranged in ascending order. An animated 3D image of the mesophyll cell, corresponding to varying angles of view, can be seen in Movie 1, which is published as supporting information on the PNAS web site.
Void Network.
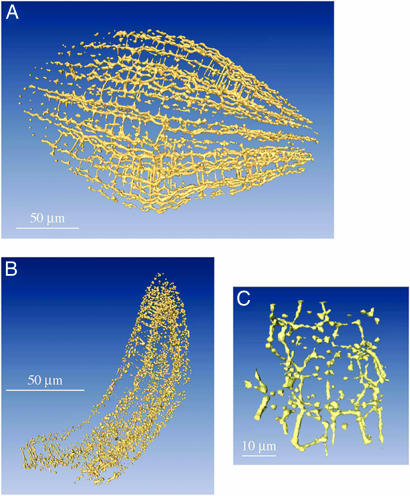
At lower magnification (see Fig. 2 A and B), many cell corners appear as white spots with a density near 0, indicating air-filled cavities. This intercellular air space, as visualized here by white spots at the cell corners and lines between cells, extends throughout all of the tissues, except for the epidermal cell layers and the provascular axial tissue. Surprisingly, air space is ≈2.5 times more abundant in the hypocotyl compared with cotyledons, the corresponding volume fractions, to be considered as upper bounds because of the finite spatial resolution, being 1.5 × 10−2 and 0.58 × 10−2, respectively. In the hypocotyl, many of these air spots are connected, forming white lines localized between three adjacent cells. To better characterize the air space structure, we also show it in a 3D presentation (Fig. 4). Air channels that traverse the hypocotyl in the longitudinal direction are observed. In many cases, neighboring channels are laterally connected, thus leading to the formation of an interconnected network of air channels criss-crossing the hypocotyl cortex (Fig. 4A). In cotyledon tissues, air spots seem to be only occasionally connected by lateral air channels (Fig. 4B). However, higher magnification (Fig. 4C) shows connectivity between corner-positioned air spots there too. Animated 3D images of this void network can be seen in Movies 2 and 3.
Fig. 4.
3D rendering of intercellular air space in the hypocotyl (A) and in a cotyledon (B) and air space in cotyledon at higher magnification (C). Animated 3D images of the void network can be seen in Movies 2 and 3, which are published as supporting information on the PNAS web site.
Discussion
The structure and function of interstices between plant cells still contains riddles, and oxygen supply to internal tissues is not yet well understood. The structure and composition of cell corners are tissue-dependent and vary with plant development. Close cell contacts are observed within meristematic and embryonic tissues. The highest degree of cell separation at corners is observed in the spongy parenchyma of leaf cells (10). Although the large spaces at cell corners seem to be important for vertical and lateral diffusion of CO2 inside leaves, from substomatal cavities to the assimilating tissues (11), other functions for intercellular spaces have been discussed, e.g., connected to plant secretion (10). Cell corners can be filled with glycine-rich proteins in the protoxylem (12), with prolin-rich proteins in the hypocotyl cortex (13, 14), and with lignins between wood cells (15). Actually, little is known on the functional implications of these variations in corner structure and composition. It is believed that pectins glue cells together, alongside their cell walls and at the corners (16). Turgor pressure forces plant cells toward a spherical form (17); i.e., turgor pressure might be considered as the driving force leading to the separation of cells at the corners. However, our results show that corner cavities exist in dry seeds (Fig. 2 A and B), where the notion of turgor pressure is not relevant, thus raising the question of the formation and the function of these cavities in seeds.
We suggest that the air space network that is imaged in Fig. 4 serves to provide the necessary oxygen at the onset of germination. Gas diffusion inside plant tissues has so far been studied in leaves (11, 18), where it is influenced by atmospheric gas composition and exchange via opening/closure of stomata. In seeds, stomata are not yet present in cotyledons (19), and the low seed coat permeability strongly restricts gas exchange (4, 5). Thus, as outlined in the introduction, the oxygen involved in the first stage of germination should be provided from within the seed. Internal oxygen concentration is lowest during early stages of seed development. It increases considerably in maturing cotyledons because of embryonic photosynthesis of seed plastids (5, 20). We can assume that at least part of the oxygen that is produced by embryonic photosynthesis is stored in the intercellular space network until germination.
The results presented here show structural details in the mature seed that were not observed before. We suggest that the air space network in Arabidopsis seeds, visualized through the nondestructive approach of quantitative phase tomography, provides a transport system for easy gas exchange in embryos and serves as storage space for the oxygen that will be needed at the onset of germination. In addition, during imbibition it could serve for rapid water transport from the micropylar seed end to the various seed tissues and organs (21).
Materials and Methods
Holotomography.
The approach used in the present work, holotomography, is related to, albeit more elaborate than, the usual form of x-ray microtomography (7, 22) based on absorption, i.e., attenuation contrast. Small, light objects such as the Arabidopsis seeds investigated in the present work have very small absorption for hard x-rays. Thus, they are very close to pure phase objects. The effect of the sample on an x-ray beam going through it is described by the refractive index n(x,y,z) = 1 – δ(x,y,z) + i·β(x,y,z). Its real part, in the hard x-ray range, differs from unity by the small (typically 10−6) decrement δ = roρλ2/(2π), with ro = 2.8 fm the classical electron radius, (roρ) the mean scattering length density of the material, and λ the x-ray wavelength. The fact that δ is much larger than the imaginary part β is the reason why the phase provides more sensitivity than the absorption. Except for photon energies close to absorption edges of elements in the sample, ρ is actually the mean electron density. Because the latter is in turn nearly proportional to the mass density ρm, the relevant quantity, leading to the images, can be considered to be either density.
Local variations in the phase of the beam do not produce changes in intensity, i.e., contrast, if the detector is placed right after the sample. They do produce contrast on a radiograph, through Fresnel diffraction, if the detector is placed at a distance, and if an x-ray beam with appreciable spatial coherence is used (23–26). The imaging process leading to the 2D phase radiographs can be described as simple propagation, defocusing, or in-line holography. Ignoring, in this simplified description, several correction operations, two further steps lead from simple radiographs to holotomography, the 3D reconstruction of the real part of the refractive index n(x,y,z) (27, 28). First the 2D distribution of the phase shift suffered by the beam is retrieved. This is not a straightforward step because the relationship between the intensity and the phase is not one-to-one as it is for the absorption case. Therefore, various approximation and iteration schemes must be used: in the present work we use the adaptation to the hard x-ray case of a method developed for electron microscopy (29). Actually, several sample-detector distances are required to obtain a reliable phase map. The second step, tomographic reconstruction, computes the 3D distribution from many 2D phase maps by using the filtered back-projection algorithm. From there, either 3D renditions of the refractive index, or virtual slices along any direction, one pixel (in the present case 0.3 μm) thick, can be obtained. Thus, tomographic slices show the different densities of the analyzed seed. From differences in densities we can conclude on the composition of particles and/or structures. In the reproduction scheme we used, dark zones corresponded to more dense structures than lighter zones.
Positioning of the Seed.
Mature seeds of Arabidopsis thaliana, ecotype Columbia, were used. The seeds were mounted on the surface of an aluminum cylinder with cyanoacrylate glue, and the cylinder was placed on a set of rotation stages (Fig. 1). The stage with vertical axis was used to change the orientation of the sample, in 0.225° intervals, with respect to the incident beam. Between angular scans, a translation stage moved the detector system with respect to the sample, varying the sample-to-detector or defocusing distance, an essential parameter in the visualization of phase objects.
Data Acquisition and Processing.
The present work used the experimental station ID19 of the European Synchrotron Radiation Facility (Grenoble, France), a long (150 m) imaging beamline where the spatial coherence of the beam is especially large. It used 21-keV photons selected from the radiation from a wiggler through a multilayer set to reflect in the vertical plane, resulting in an energy band ΔE/E of 2 × 10−2, considerably broader than from a perfect-crystal monochromator. In the detection device used, visible light produced by an x-ray-sensitive converter is imaged onto a cooled charge-coupled device in a FReLoN camera (30). The effective pixel size at the converter screen position was 0.3 μm, resulting in a field of view of 0.6 × 0.6 mm2. The data set for a single sample comprised four angular scans of 800 images each, recorded at different sample-detector distances: 13 mm, 33 mm, 63 mm, and 103 mm, chosen to avoid consistently low values of the optical transfer function for some spatial frequencies. The exposure time was 2 s per picture. The volume renderings were obtained by using isosurface representations and volume texture renderings with Amira from Mercury Computer Systems (Chelmsford, MA). Geometrical and quantitative measurements on the sample are performed with ImageJ (available from http://rsb.info.nih.gov/ij).
Light Microscopy.
Seeds were imbibed on a moistened filter for 2 h, and incisions were made to open the coat. Seeds were then fixed at room temperature with 3% glutaraldehyde in 0.1 M Na-cacodylate buffer (pH 7.0) for 90 min, dehydrated through a graded ethanol series (50%, 70%, 90%, and three changes of 100%; 15 min each), infiltrated overnight with ethanol/Spurr’s resin (vol/vol; Agar Scientific, Stansted, U.K.), and embedded with Spurr’s resin for 24 h with several changes. Polymerization was performed at 70°C over the weekend, and 1.2-μm-thick sections were made by using a Leica UC6 ultramicrotome (Leica Microsystems, Wetzlar, Germany). Sections were colored with toluidine blue and mounted on a glass slide before observations using a Nikon E 600 microscope (Nikon, Kawasaki, Japan).
Supplementary Material
Acknowledgments
We thank E. Gentilhomme and B. Vachez-Collomb for the preparation of thin microtome slices for optical observations and Prof. Michel Robert-Nicoud for the critical reading of the manuscript. This work was supported by European Synchrotron Radiation Facility Project LS-1737 through allocation of beam time.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Botha FC, Potgieter GP, Botha A-M. J Plant Growth Regul. 1992;11:211–216. [Google Scholar]
- 2.Bewley JD, Black M. Seeds: Physiology of Development and Germination. 2nd Ed. New York: Plenum; 1994. [Google Scholar]
- 3.Bewley JD. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolletschek H, Borisjuk L, Koschorreck M, Wobus U, Weber H. J Exp Bot. 2002;53:1099–1107. doi: 10.1093/jexbot/53.371.1099. [DOI] [PubMed] [Google Scholar]
- 5.Rolletschek H, Weber H, Borisjuk L. Plant Physiol. 2003;132:1196–1206. doi: 10.1104/pp.102.017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider G, Anderson E, Vogt S, Knochel C, Weiss D, Legros M, Larabell CA. Surf Rev Lett. 2002;9:177–183. [Google Scholar]
- 7.Larabell CA, Le Gros MA. Mol Biol Cell. 2004;15:957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao J, Hodgson KO, Ishikawa T, Larabell CA, LeGros MA, Nishino Y. Proc Natl Acad Sci USA. 2003;100:110–112. doi: 10.1073/pnas.232691299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro D, Thibault P, Beetz T, Elser V, Howells MR, Jacobsen C, Kirz J, Lima E, Miao H, Nieman AM, Sayre D. Proc Natl Acad Sci USA. 2005;102:15343–15346. doi: 10.1073/pnas.0503305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knox JP. Plant J. 1992;2:137–141. [Google Scholar]
- 11.Pieruschka R, Schurr U, Jahnke S. J Exp Bot. 2005;56:857–864. doi: 10.1093/jxb/eri072. [DOI] [PubMed] [Google Scholar]
- 12.Ryser U. Planta. 2003;216:857–864. doi: 10.1007/s00425-002-0938-7. [DOI] [PubMed] [Google Scholar]
- 13.Marcus A, Greenberg J, Averyhart-Fullard V. Physiol Plant. 1991;81:273–279. [Google Scholar]
- 14.Ye Z-H, Song Y-R, Marcus A, Varner JE. Plant J. 1991;1:175–183. doi: 10.1111/j.1365-313x.1991.00175.x. [DOI] [PubMed] [Google Scholar]
- 15.Fromm J, Rockel B, Lautner S, Windeisen E, Wanner G. J Struct Biol. 2003;143:77–84. doi: 10.1016/s1047-8477(03)00119-9. [DOI] [PubMed] [Google Scholar]
- 16.Willats WGT, McCatney L, Mackie W, Knox JP. Plant Mol Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- 17.Jarvis MC. Plant Cell Environ. 1998;21:1307–1310. [Google Scholar]
- 18.Evans JR, von Caemmerer S. Plant Physiol. 1996;110:339–346. doi: 10.1104/pp.110.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisler MJ, Sack FD. New Phytol. 2002;153:469–476. doi: 10.1046/j.0028-646X.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 20.Weber H, Borisjuk L, Wobus U. Annu Rev Plant Biol. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
- 21.Manz B, Müller K, Kucera B, Volke F, Leubner-Metzger G. Plant Physiol. 2005;138:1538–1551. doi: 10.1104/pp.105.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoddad WS, McNulty I, Trebes JE, Anderson EH, Lernque RA, Yang L. Science. 1994;2:1213–1215. doi: 10.1126/science.266.5188.1213. [DOI] [PubMed] [Google Scholar]
- 23.Cloetens P, Barrett R, Baruchel J., Guigay JP, Schlenker M. J Phys D Appl Phys. 1996;29:133–146. [Google Scholar]
- 24.Snigirev A, Snigireva I, Kohn V, Kuznetsov S, Schelokov I. Rev Sci Instrum. 1995;66:5486–5492. [Google Scholar]
- 25.Wilkins SW, Gureyev TE, Gao D, Pogany A, Stevenson AW. Nature. 1996;384:335–338. [Google Scholar]
- 26.Nugent KA, Gureyev TE, Cookson DF, Paganin D, Barnea Z. Phys Rev Lett. 1996;77:2961–2964. doi: 10.1103/PhysRevLett.77.2961. [DOI] [PubMed] [Google Scholar]
- 27.Cloetens P, Ludwig W, Baruchel J, Van Dyck D, Van Landuyt J, Guigay JP, Schlenker M. Appl Phys Lett. 1999;75:2912–2914. [Google Scholar]
- 28.Zabler S, Cloetens P, Guigay JP, Baruchel J, Schlenker M. Rev Sci Instrum. 2005;76:073705. [Google Scholar]
- 29.Coene W, Janssen G, Op de Beeck M, Van Dyck D. Phys Rev Lett. 1992;69:3743–3746. doi: 10.1103/PhysRevLett.69.3743. [DOI] [PubMed] [Google Scholar]
- 30.Labiche JC, Segura-Puchades J, Van Brussel D, Moy JP. ESRF Newslett. 1996;25:41–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.