Abstract
B cell antigen receptor signals development, activation, proliferation, or apoptosis of B cells depending on their condition, and its proper signaling is critical for activation and homeostasis of the immune system. The B cell-restricted adaptor protein BASH (also termed BLNK/SLP-65) is rapidly phosphorylated by the tyrosine kinase Syk after BCR ligation and binds to various signaling proteins. BASH structurally resembles SLP-76, which is essential for T cell development and T cell receptor signaling. To evaluate the role for BASH in B cell development and function in vivo, we disrupted BASH alleles in embryonic stem cells by means of homologous recombination and used these cells to complement lymphocyte-incompetent blastocysts from RAG2-deficient mice. In the resultant chimeric mice, T cell development was apparently normal, but B cell development was impaired, and a normally rare population of large preB cells expressing preB cell receptor dominated in the bone marrow in place of small preB cells, although they were mostly noncycling. In addition, the mature B cell populations in the periphery and the bone marrow profoundly decreased in size, as did B-1 cells in the peritoneal cavity, and serum Ig was severely reduced. The BASH-deficient B cells scarcely proliferated or up-regulated B7-2 in response to BCR ligation and poorly proliferated upon CD40 ligation or lipopolysaccharide stimulation. This phenotype indicates that BASH is critical for preB cell receptor signaling inducing proliferation of large preB cells and the following differentiation, for peripheral B cell maturation, and for BCR signaling inducing activation/proliferation of B cells.
Antigen receptor complex on B cells (BCR) is composed of a membrane-form of Ig H chain, Ig L chains, and Ig-α/Ig-β (CD79a/b) cytoplasmic components. Upon binding to antigen, BCR generates signals provoking various events on B cells, such as cytokinetic change to internalize the antigen, cell activation manifested as up-regulation of surface proteins including B7-2 (CD86), increase in size and cell-cycle entry, and survival or, in some condition, apoptosis. Such cellular events are prerequisites for collaboration with T cells, clonal expansion, and terminal differentiation to antibody-secreting or memory cells, or self-tolerance (1). Recently, it was shown that the continued presence of BCR, without obvious antigen binding, is required for the persistence of mature B cells (2), suggesting that BCR on the cell surface constitutively signals the survival and perhaps maturation of the cells. The preBCR has a similar structure to BCR, and is composed of Ig-α/Ig-β, μH chain (μH) and the surrogate L chains, λ5 and VpreB. The formation of the preBCR is critical for early B cell development because the mice lacking the components of the preBCR exhibit a developmental block at the proB cell stage (3–8) and loss of H chain allelic exclusion (9, 10). The developmental block presumably stems from the lack of preBCR-signaled proliferation and/or survival of preB cells and the following differentiation into small, resting preB cells, the major site of L chain gene rearrangement (11, 12).
Ig-α/Ig-β transduces the signal from BCR or preBCR primarily through the activation of tyrosine kinases such as Src-family kinases, Lyn, Fyn, Blk, or Syk and Btk kinases. These kinases phosphorylate various signaling intermediates, including enzymes such as phospholipase C (PLC),γ2, phosphatidylinositol 3-kinase, and Vav, or adaptor molecules such as Shc, Grb2, Nck, and HS1 (13–15). Deficiency of either one of the Src-family kinases did not cause serious impairment of B cell development or weakening of BCR signaling. On the other hand, Syk deficiency caused early block of B cell development at proB cell stage, indicating that Syk is primarily responsible for preBCR signaling (16, 17). Mutations in the btk gene were identified as being responsible for X-linked agammaglobulinemia (Bruton's disease) in humans and X-linked immunodeficiency (Xid) in mice. Xid mice, as well as btk gene knockout mice, are characterized by the impaired function of peripheral B cells, primarily because of deficiency of BCR signaling (ref. 18, and the references therein). Studies have evidenced that Syk and Btk are necessary for activation of PLCγ2 and the following intracellular calcium flux (19–22). However, it is unclear how these kinases interact with PLCγ2 or other substrates.
Recently, we isolated a molecule that we termed BASH (for B cell adaptor containing SH2 domain) as a molecule expressed selectively in B cells in the bursa of Fabricius in the chicken (23). BASH was structurally similar to the T cell adaptor protein SLP-76 (24), became tyrosine phosphorylated upon BCR crosslinking, and bound to Syk and Shc. We also isolated a mouse homologue of BASH whose amino acid sequence was identical to BLNK, with four amino acid differences, and to SLP-65. BLNK was purified as 70/68-kDa phosphoproteins bound to the SH2 domain of PLCγ1 (25, 26), whereas SLP-65 as a 65-kDa protein rapidly tyrosine-phosphorylated upon pervanadate treatment in the presence of surface BCR (27, 28). BASH/BLNK/SLP-65 transcripts and proteins were dominantly expressed in tissues containing B cells, although a weak expression was detected in other tissues, and restricted to B-lineage cell lines. BASH/BLNK/SLP-65 was phosphorylated primarily by Syk, translocated into the membrane fraction after BCR stimulation, and bound to PLCγ1/2, Grb2, Vav, and Nck (23, 26, 27). It has been proposed that phosphorylated BLNK recruits PLCγ1/2 to the proximity of Syk, thus facilitating tyrosine phosphorylation and activation of PLCγ1/2 by Syk, and elevation of intracellular calcium upon BCR stimulation (26). Analysis of a chicken B cell line deficient for BLNK confirmed this and further revealed that BLNK is necessary for activation of JNK and p38 upon BCR ligation (29). Recently, it has been shown that BLNK also binds to Btk and mediates the phosphorylation of PLCγ1/2 by Btk (30). Together, BASH/BLNK/SLP-65 is proposed to function as a scaffold protein to focus various signaling effectors at the plasma membrane for phosphorylation by Syk and/or Btk.
To elucidate the in vivo function of BASH/BLNK/SLP-65 in the development and function of lymphoid system, we used gene targeting in the mouse. Because BASH/BLNK/SLP-65 gene was weakly expressed in several nonlymphoid tissues, like liver, kidney, and ovary in the mouse (ref. 26, and our unpublished results), and its transcripts were identified even in fertilized eggs (DDBJ accession no. C87337), we decided to apply the RAG2-deficient blastocyst complementation assay (31) to avoid possible embryonic lethality or deficiency in tissues other than B or T lymphoid tissues. Here, we describe B cell development and function in the chimeric mice whose lymphoid system is derived from homozygous BASH-mutant embryonic stem (ES) cells.
Materials and Methods
Targeted Disruption of BASH Genes and Generation of Chimeric Mice.
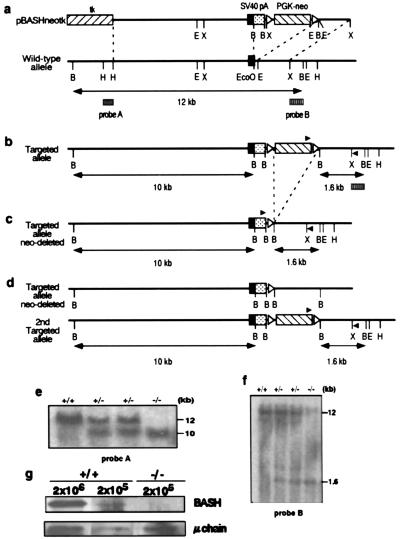
cDNA containing complete mouse BASH coding sequence (DDBJ/EMBL/GenBank accession no. AB015290) was obtained by standard PCR and 5′-rapid amplification of cDNA ends method (Marathon cDNA amplification kit; CLONTECH) based on an incomplete cDNA sequence identified as a homologue of chicken BASH in GenBank (BCA, accession no. AJ222814). Using the mouse BASH cDNA as a probe, a λEMBL4 genomic library from TT2 ES cell line (Lifetech Oriental, Tokyo) was screened, and a 17-kb genomic fragment containing an exon encoding amino acids 39–55 of BASH and the flanking introns was isolated. A HindIII–XbaI genomic fragment was used in the gene-targeting vector (pBASHneotk), in which a part of the exon and the following intron (EcoO109I–EcoRI) was replaced with two fragments: (i) BamHI–MluI fragment of pEGFP-C1 (CLONTECH) containing three-frame stop codons and an SV40 polyadenylation signal; (ii) Pgk-neor flanked with loxP sites derived from pKSTKNeoLoxP (a gift of W. Reith, University of Geneva Medical School; see Fig. 1a). The vector also contains HSV-tk gene for negative selection (32). The linearized vector was electroporated into B6III ES cells (33) and the cells were selected by G418 (0.2 mg/ml; Wako Biochemicals, Osaka) and Gancyclovir (0.5 μg/ml; Syntex, Palo Alto, CA). Drug-resistant colonies were screened for homologous recombination events by PCR with 5′-primer (5′-TGCTAAAGCGCATGCTCCAGACTG-3′) and 3′-primer (5′-ATGCTTGACAGTGTGGGCTTCTGT-3′) as described (3). Positive colonies were propagated and verified by Southern blot analysis for a precise targeted event (Fig. 1 b, e, and f). The heterozygously targeted clones were transfected with a vector expressing Cre recombinase and puromycin-resistant gene (Cre-Pac; ref. 34) and selected with puromycin (0.5 μg/ml) for 54 hr. The resistant colonies were tested by neomycin-sensitivity and PCR (Fig. 1c, and data not shown) for the deletion of Pgk-neo sequence by loxP-mediated recombination from the targeted allele. An ES cell clone that had undergone this recombination (BASH+/−) was transfected again with pBASHneotk and screened as above to derive a homozygously targeted (BASH−/−) clone (Fig. 1d). Southern blot analysis was performed with BamHI-digested genomic DNA from ES-cell clones and probes A and B (Fig. 1 e and f). BASH+/− and BASH−/− ES cell clones were injected into blastocysts obtained from RAG2−/− mice that had been backcrossed to BALB/c mouse strain (a gift of Y. Shinkai, Institute for Virus Research, Kyoto University), as described (31).
Figure 1.
Targeted disruption of the mouse BASH gene. (a) Structure of the targeting vector and a partial restriction map of BASH gene locus. Filled rectangles represent exons; open triangles represent loxP sites. Probes A and B used in Southern blot analysis are shown as boxes below a double-headed arrow, indicating a restriction fragment detected by these probes. tk, HSV-tk gene; pA, polyadenylation signal; E, EcoRI; X, XbaI; B, BamHI; H, HindIII; EcoO, EcoO109I. (b) Primary targeted allele. Filled triangles represent the locations of PCR primers used for the detection of homologous recombination events. (c) The primary targeted allele after Cre-mediated recombination of loxP sites, resulted in the deletion of PGK-neo cassette. (d) The primary and secondary targeted alleles. (e and f) Southern blot analysis of B6III cells (+/+), the cells with the primary targeted allele as in b (+/−, Left), the cells with the primary targeted allele undergone Cre-mediated recombination as in c (+/−, Right), and the cells with both targeted alleles as in d (−/−). The sizes of the BamHI-restriction fragments hybridized with the probe A (e) and probe B (f) are indicated. (g) Lysates of spleen cells containing indicated number of B cells from wild-type C57BL/6 mice (+/+) or BASH−/−RAG−/− chimera (−/−) were analyzed by Western blotting with rabbit anti-BASH antibody, and reprobed with anti-μH chain antibody .
Flow Cytometry.
Single-cell suspensions were prepared from lymphoid organs, and red blood cells were lysed in hypotonic buffer. After washing, cells were stained with anti-mouse antibodies conjugated with FITC, phycoerythrin (PE), or biotin, followed by streptavidin (SA)-RED670 (GIBCO) in the latter case, and analyzed on FACSort with cellquest software (Becton Dickinson). Monoclonal antibodies (mAbs), FITC-conjugated anti-CD3ɛ (145-2C11), CD4 (RM4–5), CD43 (S7), CD25 (7D4), CD5 (53-7.3), Mac-1 (M1/70), PE-conjugated anti-CD8α (53-6.7), B220/CD45R (RA3–6B2), and biotin-anti-Ly9.1 (30C7) were purchased from PharMingen; PE-anti-IgD mAb (SBA-1), FITC-goat-anti-κ and λ, and FITC- or PE-goat-anti-IgM (μH chain-specific) were from Southern Biotechnology Associates. Biotin-goat-anti-IgM (μH) was from Cappel. Biotin-SL156 (35) was provided by H. Karasuyama (The Tokyo Metropolitan Institute of Medical Science). For the analysis of B cell activation, spleen cells were depleted of T cells as described below, stimulated with 10 μg/ml goat anti-mouse IgM F(ab′)2 fragment (Jackson Laboratory) for 24 hr, then stained with PE-anti-B220 and FITC-anti-B7-2 (GL-1; a gift of R. Abe, Research Institute for Biological Sciences, Science University of Tokyo, Chiba). B220+ cells were electrically gated for the analysis of B7-2 expression.
Cell Cycle Analysis.
Cell sorting was performed by FACSvantage (Becton Dickinson). The sorted cells were fixed, treated with RNase A, stained with propidium iodide, and analyzed for DNA content by FACSort, as described (36).
Proliferation Assay.
B cells were enriched as follows: spleen cells were incubated on ice for 30 min in RPMI 1640 medium supplemented with 1% FCS, 25% culture supernatant of hybridoma T-24 (anti-Thy1 mAb), and 2.5 μg/ml biotin-anti-Ly9.1. After washing, T cells and Ly9.1+ cells were killed with rabbit complement (Cedarlane Laboratories) and further depleted with a mixture of anti-Thy1- and SA-coated magnetic beads (Dynal, Great Neck, NY), according to the supplier's instruction. Despite many trials, enrichment of B cells from the spleen of BASH−/−RAG2−/− chimera was inefficient (up to 15%) because of low quantity of B cells, compared with those of wild-type or BASH+/−RAG2−/− chimeric mice (>80%). Therefore, the enriched B cells from the latter were mixed with RAG2−/− spleen cells, treated as above, to reproduce the proportion of B cells in the samples from BASH−/−RAG2−/− chimera. In another assay, B220+ cells were sorted from spleen by FACSvantage. Thus prepared samples, each containing 1 × 104 B cells, were stimulated in 100 μl culture medium containing 10 μg/ml goat anti-mouse IgM F(ab′)2 fragment (The Jackson Laboratory), 10 μg/ml LPS (Sigma), 10 μg/ml anti-mouse CD40 mAb (LB429; ref. 37), or 2 ng/ml recombinant mouse IL-4 (PeproTech, Rocky Hill, NJ), alone or in combinations, as indicated in Fig. 4. Forty hours later, [3H]thymidine (0.5 μCi per well; Amersham Pharmacia) was added. Cells were harvested 8 hr later, and incorporated [3H]thymidine was counted in a BetaPlate scintillation counter (Wallac, Gaithersburg, MD).
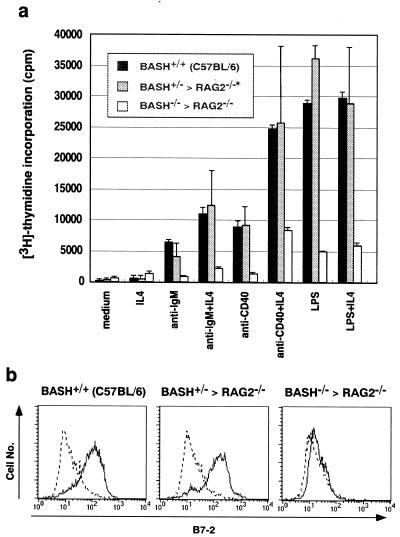
Figure 4.
Defective proliferative response and activation of BASH−/− B cells in vitro. (a) T cell-depleted cells from a spleen of wild-type C57BL/6 mouse, BASH+/−RAG2−/− chimera, or admixed two spleens of BASH−/−RAG2−/− chimeras were stimulated with the indicated reagents for 48 hr, and the incorporation of [3H]thymidine was measured. Proportion of B cells in the samples was made uniform to10%, as described in Materials and Methods, and that of T cells was <1%. Data are expressed as mean cpm (±SD) of duplicates. *, Separate data from two spleens of BASH+/−RAG2−/− chimeras were combined. (b) T cell-depleted spleen cells were stimulated with 10 μg/ml goat anti-IgM for 24 hr, then stained with PE-anti-B220 and FITC-anti-B7-2 mAb (GL-1). B220+ cells were gated for the analysis of B7-2 expression.
Serum Ig Titration.
Igs in serial dilutions of serum were measured by ELISA using antibody pairs specific for different mouse Ig isotypes (PharMingen). 2,2′-Azino-bis (3-ethylbenz-thiazolne-6-sulfonic acid) was used as a substrate for the horseradish peroxidase (HRP)-conjugated secondary antibodies, and the absorbance at 405 nm was measured in a microplate reader (Bio-Rad).
Western Blot Analysis.
Polyclonal anti-BASH antibody was produced by immunizing a rabbit with keyhole limpet hemocyanin-conjugated peptide corresponding to mouse BASH (amino acids 42–52). Whole-cell lysates were subjected to SDS/PAGE and blotted onto nitrocellulose filter. BASH protein was detected by the above antibody and HRP-conjugated goat-anti-rabbit or IgG (Zymed), and developed with ECL system (Amersham Pharmacia). The filter was stripped and reprobed with biotin-goat anti-mouse IgM (Cappel) and SA-HRP (Calbiochem).
Results and Discussion
Generation of Chimeric Mice with BASH-Deficient Lymphoid System.
The genes encoding BASH were disrupted sequentially in C57BL/6-derived ES cells (33) by insertion of translational stop codons and polyadenylation signal into an exon coding the N-terminal portion of BASH protein by means of homologous recombination (Fig. 1 a–d). The BASH alleles were precisely targeted as shown by Southern blot analysis (Fig. 1 e and f). Heterozygous (+/−) or homozygous (−/−) mutant ES cell clones were injected into blastocysts from lymphocyte-deficient RAG2−/− mice on the BALB/c genetic background, to generate chimeric mice in which any lymphocyte must be derived from the mutant ES cells (31). Indeed, analysis of the Ly9.1 allelic marker indicated that T and B lymphocytes in the chimeric offspring were all derived from ES cells (Ly9.1−), not from the recipient (Ly9.1+) (data not shown). No BASH protein was detected by Western blot analysis in B cells derived from the homozygous mutant ES cells (Fig. 1g). Eight- to 16-week-old chimeric mice were used for the analyses shown hereafter, unless otherwise noted.
Severe Reduction of Mature B Cells in the Spleen and B-1 Cells in the Peritoneal Cavity of BASH−/− Chimeras.
Chimeric mice derived from RAG2−/− blastocysts injected with BASH−/− ES cells (BASH−/−RAG2−/− chimera) and those with BASH+/− ES cells (BASH+/−RAG2−/− chimera) were analyzed for lymphocyte cellularity by flow cytometry. Population analyses shown herein were on the cells in the lymphocyte gate as defined by forward and side light scatters. C57BL/6 (B6) and RAG2−/− (on BALB/c background) mice were analyzed at the same time as normal and T/B lymphocyte-deficient controls, respectively. BASH−/−RAG2−/−, as well as BASH+/−RAG2−/−, chimera had normal numbers of thymocytes and no apparent defects in T cell development, as demonstrated by the expression of CD4 and CD8 on thymocytes (data not shown) and normal numbers of T cells in spleen (Fig. 2a). This result is consistent with the fact that BASH is not expressed in T cells (refs. 23, 26, and 27; and our unpublished results), and indicates that the generation of common lymphoid precursors from the mutant ES cells is intact.
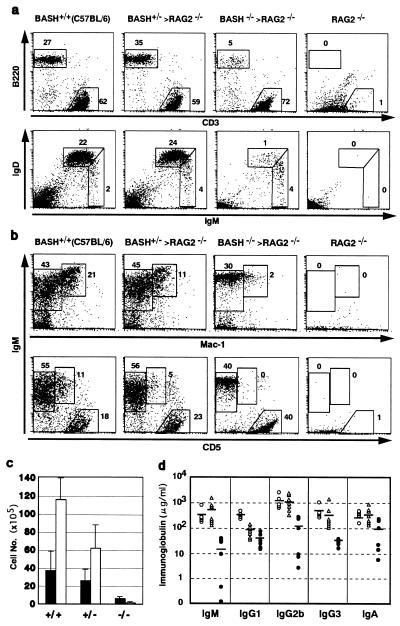
Figure 2.
Peripheral B cell deficiency in BASH−/−RAG2−/− chimera. Representative flow cytometric analyses of cells from spleens (a) or peritoneal cavities (b) of the indicated mice. Cell surface antigens stained by antibodies are indicated. The number at each window indicates the percentage of the total cells within the lymphocyte gate. Spleen cells were simultaneously stained for Ly9.1 antigen, and Ly9.1− cells (derived from ES cells) were plotted and counted for the population analyses for the chimeras. Peritoneal cells were not gated by Ly9.1 marker because of its highly nonspecific staining. (c) Actual numbers (mean ± SD) of immature (IgMhigh, IgDlow; filled bars) and mature (IgMlow, IgDhigh; open bars) B cells (as defined in a) in spleens from wild-type C57BL/6 mice (+/+, n = 3), BASH+/−RAG2−/−chimeras (+/−, n = 3), or BASH−/−RAG2−/− chimeras (−/−, n = 4). (d) Ig concentrations in the sera from wild-type C57BL/6 mice (○), BASH+/−RAG2−/−chimeras (▵), or BASH−/−RAG2−/− chimera (●). Mice were 6–8 wk old. Mean values of each isotype are indicated by horizontal bars.
Analysis of spleen cells from the chimeras for the Ly9.1 allelic marker showed that contribution of ES cell-derived (Ly9.1−) cells was relatively high (87 ± 3% in BASH+/−RAG2−/− chimeras, n = 3; 73 ± 11% in BASH−/−RAG2−/− chimeras, n = 4). However, B cell numbers were markedly reduced in the peripheral lymphoid organs of the BASH−/−RAG2−/− chimeras. The proportion of B220 (CD45R)+, IgM+ B cells among Ly9.1− lymphoid cells in the spleens of BASH−/−RAG2−/− chimeras was 4- to 9-fold smaller compared with BASH+/−RAG2−/− chimeras or wild-type B6 mice (Fig. 2a). The reduction of the B cell population in the former animals was mainly the result of the disappearance of mature B cells (IgMlow, IgDhigh), whereas immature (IgMhigh, IgDlow) B cells were less affected (Fig. 2 a and c). This was supported by Western blot analysis showing that BASH−/− B cells express more μH chain than wild-type B cells on a per-cell basis (Fig. 1g). An almost identical phenotype was observed with lymphocytes in the blood (data not shown). These results indicate that BASH is critical for maturation and expansion of B cells in the peripheral lymphoid system.
The B cells in the peritoneal cavity consist of two phenotypically distinct subpopulations: IgM+, B220high, CD5−, Mac-1− B-2 (conventional B) cells and IgM+, B220low, CD5+, Mac-1+ B-1 cells. The B-1 cell population was essentially absent in the peritoneal cavity of BASH−/−RAG2−/− chimeras and reduced in BASH+/−RAG2−/− chimeras compared with wild-type B6 mice (Fig. 2b), although the B-2 cell population was not reduced in BASH+/−RAG2−/−chimeras and only slightly in BASH−/−RAG2−/− chimeras. This indicates that expansion of B-1 cells requires BASH and may depend on the dosage of BASH. Interestingly, the B-2 cells of BASH−/−RAG2−/− chimeras expressed IgM uniformly at a high level in contrast to those of wild-type mice or BASH+/−RAG2−/− chimera expressing IgM heterogeneously. This, as well as the phenotype of spleen B cells, may indicate that BASH−/− B cells require a high amount of IgM on the cell surface to compensate for inefficient BCR signaling for expansion and/or survival (2). In accord with the reduction of B cells in the periphery, the levels of all Ig classes were markedly reduced in the sera of BASH−/−RAG2−/− chimeras (Fig. 2d). The level of IgG1, but not the other classes, was constantly reduced also in BASH+/−RAG2−/− chimeras, the reason for which is currently unknown.
Accumulation of Noncycling Large PreB Cells and the Lack of Small PreB Cells in the Bone Marrow of BASH−/− Chimeras.
B cells are generated from hematopoietic stem cells through proB, large preB, and small preB cell stages in the bone marrow (1). As reported previously (5), B cell development was completely blocked at the proB cell stage (B220low, μH−, CD43high, CD25−) in RAG2−/− bone marrow cells deficient for Ig gene rearrangements (Fig. 3a). A similar block had been observed in mice deficient for the components of preBCR, such as membrane-form μH and λ5 (3, 4, 38); thus, it has been established that preBCR signaling is essential for the proB to preB cell transition (1). In the bone marrow of BASH−/−RAG2−/− chimeras, μHhigh (IgM+), B220high mature B cells were nearly absent, and the μHhigh (IgM+), B220low immature B cell population was moderately reduced compared with wild-type and BASH+/−RAG2−/− chimeric mice (Fig. 3a, Left). This may reflect profoundly reduced numbers of circulating B cells and inefficient generation of B cells.
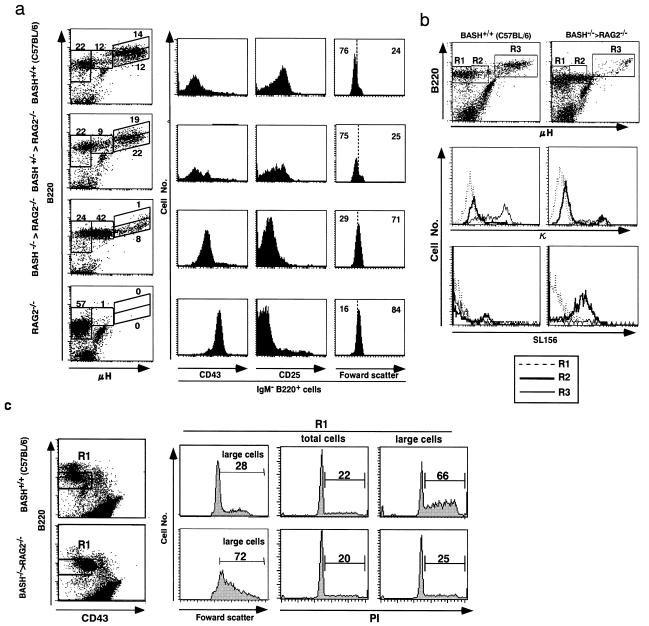
Figure 3.
Impairment of BASH−/− B cell development in the bone marrow. Shown are representative flow cytometric analyses of bone marrow cells from the indicated mice. Only the cells in the lymphocyte gate were analyzed. (a) Cells were stained for μH, B220, and Ly9.1, and Ly9.1− cells were gated for the analysis of the chimeras. B220 vs. μH expression is shown as dot plots (Left). The number at each window indicates the percentage of the total cells plotted. Cells were also stained simultaneously for either CD43 or CD25, and pro/preB cell fraction (the leftmost two windows set on the dot plots) were gated for the analyses of CD43 or CD25 expression, or cell size by the forward scatter. The analysis of Ly9.1 marker on proB cells (CD43high, B220low) indicated that, on average, about half of the proB cells in BASH+/−RAG2−/− or BASH−/−RAG2−/− chimeras were derived from ES cells. (b) Cells were stained for μH, B220, and either κL chain or preBCR (by SL156). B220 vs. μH expression are shown as dot plots (Top). μH−, μHlow, and μHhigh (IgM+) cells (in the windows R1, R2, and R3, respectively) were analyzed for κL chain (Middle) or preBCR (SL-156; Bottom) expression. The nature of the bright κL chain staining on a small fraction of the μHlow cells from BASH−/−RAG2−/− chimera (Middle Right, R2) remains to be elucidated. (c) Cells were stained for CD43, B220, and μH, then μHlow–μH− cells within the window (R1) set on each dot plot were sorted by flow cytometry. The sorted cells were fixed, stained with propidium iodide (PI), analyzed for cell size by forward scatter, and gated on large cells (Left). DNA contents of the total sorted cells (Middle) or the gated large cells (Right) were shown as histograms of PI staining, in which cells in S/G2/M phases of cell-cycle were indicated by bars. Numbers indicate the percentage of the total cells plotted in each histogram. The antibody-mediated fluorescence on the cell surface became negligible in the analysis of the strong PI fluorescence.
Most notably, B220low cells weakly expressing μH (μHlow) were dominant among B220+ BASH−/− bone marrow cells. Such cells were essentially absent in RAG2−/− mice and formed a minor population in wild-type and BASH+/−RAG2−/− chimeric mice (Fig. 3a, Left). Thus, in the pro/preB cell fraction (the leftmost two windows in the dot plots in Fig. 3a), small preB (CD43−, CD25high) and large proB (CD43high, CD25−) cells are the majority in normal control and RAG2−/− mice, respectively, whereas large cells with the transitional phenotype (CD43low, CD25low), which correspond to the μHlow cells (data not shown), were the majority in BASH−/−RAG2−/− chimeras (Fig. 3a, Right). Strikingly, most of the μHlow cells from BASH−/−RAG2−/− chimeras were positively stained by the mAb specific for preBCR complex (SL156; ref. 35) but only marginally stained for κL chain (Fig. 3b, Right, R2) as well as λL chain (data not shown). The same μHlow cell fraction in the wild-type mice contained a few SL156+ cells (Fig. 3b, Left, R2) and such cells increased after culture at 37°C for 1 hour (data not shown, refs. 35, 11). Neither μHhigh (IgM+) cells nor μH− cells were positive for SL156 (Fig. 3b, R3 and R1, respectively). These results indicate that CD43low, CD25low, large preB cells expressing preBCR on their surface were abnormally accumulated in the bone marrow of BASH−/−RAG2−/− chimeras.
Although large in cell size, most of the BASH−/− large preB cells were not in cell cycle, in contrast to the normal, large preB cells (Fig. 3c). Therefore, BASH appears to be necessary for preBCR signaling that promotes preB cells to enter into cell cycle, and this cycling is likely to be a prerequisite for down-regulation of preBCR expression and the subsequent differentiation into small preB cells. These results also indicate, however, that BASH is not essential for preBCR signaling for the development of large preB cells from proB cells.
Despite the lack of small preB cells, the production of B cells was not severely reduced in the bone marrow of BASH−/−RAG2−/− chimera. This may suggest an alternative pathway of B cell development that does not go through the small preB cell stage as suggested by the leaky B cell development in λ5-knockout mice (4, 39). In the latter mice, however, B cell generation is very inefficient because it depends on rare L chain gene rearrangements occurring on the proB cell stage (12, 38–40). Therefore, it is possible that the L chain gene rearrangement remains effective in the BASH−/− large preB cells because of impaired preBCR signaling, which normally down-regulates the expression of RAG genes in large preB cells (41). This possibility remains to be tested.
BCR-Signaled Activation and Proliferation Are Impaired in BASH−/− B Cells.
Proliferative responses to various stimuli of spleen B cells, either enriched by T cell depletion (Fig. 4a) or purified by cell sorting (data not shown), were examined. In contrast to the wild-type or BASH+/− cells, BASH−/− B cells negligibly responded to anti-IgM stimulation, and did poorly to anti-CD40 and lipopolysaccharide (LPS). IL-4 synergized with anti-IgM or anti-CD40 on the BASH−/− cells as on the wild-type or BASH+/− cells. BCR-mediated activation of spleen B cells was measured by up-regulation of B7-2 (CD86) antigen (Fig. 4b). In contrast to wild-type or BASH+/− B cells, B7-2 was not up-regulated on the surface of BASH−/− B cells after stimulation with anti-IgM antibody. These results indicate that BASH is essential for BCR-mediated activation and proliferation of B cells. Hyporesponsiveness of BASH−/− B cells to CD40 ligation or LPS stimulation may be the result of their immature phenotype, although a possibility that BASH might be involved in CD40 and LPS receptor signaling cannot be ruled out.
Although the in vivo immune responses have not been analyzed because they cannot be evaluated properly in chimeric mice with variable frequencies of T and B cells, so far, the observed phenotype of peripheral B cells in BASH−/−RAG−/− chimeras is similar to that of Btk-knockout mice or Xid mice (see ref. 18 and references therein). Both are characterized by the reduced number of mature B cells (with more severe reduction in BASH−/−RAG−/− chimera), the lack of the peritoneal B-1 cells, serum Ig deficiency, and defective proliferative responses to anti-IgM, anti-CD40, and LPS in vitro. This suggests a role for BASH in the function of Btk in the peripheral B cells. This is in line with the observations indicating that BASH/BLNK is necessary for BCR-mediated activation of PLCγ2 and the following intracellular calcium flux (29), for which Btk is also crucial (20, 21), and that BASH/BLNK binds to Btk in addition to PLC-γ2, thus mediating PLC-γ2 phosphorylation by Btk (30). Taken together, BASH may be critical for the BCR signal transduction mediated by Btk that induces maturation and activation of peripheral B cells.
During the submission of this manuscript, the phenotypes of two mutant strains of mice deficient for BASH/BLNK/SLP-65 have been published (42, 43). The phenotypes of these mutants were mostly equivalent to that of BASH−/− chimeric mice shown here, although the early B cell development was not analyzed in detail in the former. One marked difference is that Pappu et al. (43) have described that CD86 and CD69 were normally up-regulated on spleen B cells of their BLNK-deficient mice after BCR crosslinking, whereas we did not detect any significant up-regulation of CD86 (Fig. 4b) and CD69 (data not shown) by the same anti-IgM F(ab′)2 treatment. The reason for this discrepancy is unclear, but it might be because they stimulated whole spleen cells, whereas we used spleen cells depleted of T cells. The data indicating that the combination of T cell-mediated stimuli (i.e., anti-CD40 + IL-4) partially promoted the proliferation of BASH−/− B cells (Fig. 4a, and ref. 42) may support this possibility.
Acknowledgments
We thank Y. Shinkai for genomic library and RAG2−/− mice, W. Reith for pKSTKNeoLoxP, K. Rajewsky for B6III cells, T. Yagi for Cre-Pac, K. Miyake and N. Sakaguchi for anti-CD40 mAb, H. Karasuyama for SL156, and R. Abe for anti-B7-2 and reagents for ELISA. This work was supported in part by grants from the Ministry of Health and Welfare and the Science and Technology Agency in Japan.
Abbreviations
- BCR,B cell receptor
ES, embryonic stem
- PE
phycoerythrin
- SA
streptavidin
- HRP
horseradish peroxidase
- LPS
lipopolysaccharide
- PLC
phospholipase C
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040575697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040575697
References
- 1.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Lam K P, Kuhn R, Rajewsky K. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 3.Kitamura D, Roes J, Kuhn R, Rajewsky K. Nature (London) 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 4.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 5.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 7.Torres R M, Flaswinkel H, Reth M, Rajewsky K. Science. 1996;272:1804–1808. doi: 10.1126/science.272.5269.1804. [DOI] [PubMed] [Google Scholar]
- 8.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura D, Rajewsky K. Nature (London) 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 10.Loffert D, Ehlich A, Muller W, Rajewsky K. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- 11.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt F W, Melchers F. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 12.Li Y S, Hayakawa K, Hardy R R. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reth M, Wienands J. Annu Rev Immunol. 1997;15:453–479. doi: 10.1146/annurev.immunol.15.1.453. [DOI] [PubMed] [Google Scholar]
- 14.DeFranco A L. Curr Opin Immunol. 1997;9:296–308. doi: 10.1016/s0952-7915(97)80074-x. [DOI] [PubMed] [Google Scholar]
- 15.Kurosaki T. Curr Opin Immunol. 1997;9:309–318. doi: 10.1016/s0952-7915(97)80075-1. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A M, Rowley B, Pao W, Hayday A, Bolen J B, Pawson T. Nature (London) 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 17.Turner M, Mee P J, Costello P S, Williams O, Price A A, Duddy L P, Furlong M T, Geahlen R L, Tybulewicz V L. Nature (London) 1995;378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
- 18.Khan W N, Alt F W, Gerstein R M, Malynn B A, Larsson I, Rathbun G, Davidson L, Muller S, Kantor A B, Herzenberg L A, et al. Immunity. 1995;3:283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 19.Takata M, Sabe H, Hata A, Inazu T, Homma Y, Nukada T, Yamamura H, Kurosaki T. EMBO J. 1994;13:1341–1349. doi: 10.1002/j.1460-2075.1994.tb06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takata M, Kurosaki T. J Exp Med. 1996;184:31–40. doi: 10.1084/jem.184.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluckiger A C, Li Z, Kato R M, Wahl M I, Ochs H D, Longnecker R, Kinet J P, Witte O N, Scharenberg A M, Rawlings D J. EMBO J. 1998;17:1973–1985. doi: 10.1093/emboj/17.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharenberg A M, El-Hillal O, Fruman D A, Beitz L O, Li Z, Lin S, Gout I, Cantley L C, Rawlings D J, Kinet J P. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goitsuka R, Fujimura Y, Mamada H, Umeda A, Morimura T, Uetsuka K, Doi K, Tsuji S, Kitamura D. J Immunol. 1998;161:5804–5808. [PubMed] [Google Scholar]
- 24.Peterson E J, Clements J L, Fang N, Koretzky G A. Curr Opin Immunol. 1998;10:337–344. doi: 10.1016/s0952-7915(98)80173-8. [DOI] [PubMed] [Google Scholar]
- 25.Fu C, Chan A C. J Biol Chem. 1997;272:27362–27368. doi: 10.1074/jbc.272.43.27362. [DOI] [PubMed] [Google Scholar]
- 26.Fu C, Turck C W, Kurosaki T, Chan A C. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 27.Wienands J, Schweikert J, Wollscheid B, Jumaa H, Nielsen P J, Reth M. J Exp Med. 1998;188:791–795. doi: 10.1084/jem.188.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Wienands J, Zurn C, Reth M. EMBO J. 1998;17:7304–7310. doi: 10.1093/emboj/17.24.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishiai M, Kurosaki M, Pappu R, Okawa K, Ronko I, Fu C, Shibata M, Iwamatsu A, Chan A C, Kurosaki T. Immunity. 1999;10:117–125. doi: 10.1016/s1074-7613(00)80012-6. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto S, Iwamatsu A, Ishiai M, Okawa K, Yamadori T, Matsushita M, Baba Y, Kishimoto T, Kurosaki T, Tsukada S. Blood. 1999;94:2357–2364. [PubMed] [Google Scholar]
- 31.Chen J, Lansford R, Stewart V, Young F, Alt F W. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 33.Ledermann B, Burki K. Exp Cell Res. 1991;197:254–258. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi M, Sanbo M, Watanabe S, Naruse I, Mishina M, Yagi T. Nucleic Acids Res. 1998;26:679–680. doi: 10.1093/nar/26.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler T H, Rolink A, Melchers F, Karasuyama H. Eur J Immunol. 1995;25:446–450. doi: 10.1002/eji.1830250221. [DOI] [PubMed] [Google Scholar]
- 36.Fukuda T, Kitamura D, Taniuchi I, Maekawa Y, Benhamou L E, Sarthou P, Watanabe T. Proc Natl Acad Sci USA. 1995;92:7302–7306. doi: 10.1073/pnas.92.16.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomura J, Inui S, Yamasaki T, Kataoka S, Maeda K, Nakanishi K, Sakaguchi N. Immunol Lett. 1995;45:195–203. doi: 10.1016/0165-2478(95)00006-q. [DOI] [PubMed] [Google Scholar]
- 38.Ehlich A, Schaal S, Gu H, Kitamura D, Muller W, Rajewsky K. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- 39.Pelanda R, Schaal S, Torres R M, Rajewsky K. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 40.Novobrantseva T I, Martin V M, Pelanda R, Muller W, Rajewsky K, Ehlich A. J Exp Med. 1999;189:75–88. doi: 10.1084/jem.189.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grawunder U, Leu T M, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 42.Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen P J. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- 43.Pappu R, Cheng A M, Li B, Gong Q, Chiu C, Griffin N, White M, Sleckman B P, Chan A C. Science. 1999;286:1949–1954. doi: 10.1126/science.286.5446.1949. [DOI] [PubMed] [Google Scholar]