Abstract
In mantle cell lymphoma (MCL), the translocation t(11;14) is considered the cytogenetic hallmark of the disease. Recently, however, deletion of the chromosomal region 11q22-q23 has been identified as a frequent event in this type of cancer, indicating the existence of a pathogenically relevant tumor suppressor gene in this region. The deleted segment contains the ATM (ataxia telangiectasia mutated) gene. ATM is an interesting candidate as a tumor suppressor gene because constitutive inactivation of the gene predisposes ataxia telangiectasia patients to lymphoid malignancies. To assess the potential involvement of the gene in MCL lymphomagenesis, we performed mutation analysis of ATM in 12 sporadic cases of MCL, 7 of them with a deletion of one ATM gene copy, by using single-strand conformation polymorphism analysis of reverse transcription–PCR-amplified mRNA and subsequent DNA sequencing. In all seven cases containing a deletion of one ATM allele, a point mutation in the remaining allele was detected, which resulted in aberrant transcript splicing, truncation, or alteration of the protein. In addition, biallelic ATM mutations were identified in two MCLs that did not contain 11q deletions. Interestingly, in three cases analyzed, the ATM mutations detected in the tumor cells were not present in nonmalignant cells, demonstrating their somatic rather than germ-line origin. The inactivation of both alleles of the ATM gene by deletion and deleterious point mutation in the majority of cases analyzed indicates that ATM plays a role in the initiation and/or progression of MCL.
Mantle cell lymphoma (MCL) is a specific subtype of non-Hodgkin's lymphoma derived from naive CD5+ B- cells residing in the primary follicles or in the mantle zones of secondary follicles (1, 2). In addition to distinctive morphological, immunophenotypic, and clinical features, MCL is characterized by the chromosomal translocation t(11;14)(q13;q32), which is present in the vast majority of cases and is, therefore, regarded as the cytogenetic hallmark of the disease (3, 4). This translocation results in the juxtaposition of the cyclin D1 (CCND1) gene to transcriptional control elements from the Ig heavy chain locus (5–7). The resulting overexpression of the CCND1 gene (6, 8, 9) is thought to play a crucial role in the pathogenesis of MCL, because the gene product is one of the key proteins regulating the G1-to-S phase transition of the cell cycle (10). The potential of cyclin D1 to induce lymphomas, however, is limited: transgenic mice overexpressing cyclin D1 do not develop spontaneous B-cell lymphomas, and lymphomagenesis in these animals requires cooperation with other oncogenic factors such as C-MYC (11, 12). Thus, other mechanisms, in addition to cyclin D1 overexpression, are necessary for the development and progression of MCL.
Additional genetic alterations involved in the pathogenesis of MCL are not yet well characterized. Recent studies indicate that mutations of TP53 (13, 14) and CDKN2A inactivation (15, 16) are associated with aggressive variants of MCL. Furthermore, the involvement of additional tumor suppressor genes is strongly suggested by the recurrent deletion of distinct chromosomal regions (17, 18). However, the disease-associated genes residing in two of the most frequently deleted regions in MCL, namely, 13q14 and 11q22-q23, have not yet been identified (19, 20).
In an attempt to isolate the putative tumor suppressor gene at 11q22-q23, we recently have characterized 11q deletions in a series of MCLs by interphase fluorescence in situ hybridization (FISH). Through these analyses, we have identified a commonly deleted region smaller than 1 Mbp in size that includes the ATM (ataxia telangiectasia mutated) gene locus (20). Mutations of the ATM gene are causative for the inherited recessive disease ataxia telangiectasia (A-T; MIM 208900). The 66 ATM exons, which encompass 146 kb of genomic DNA, encode a 370-kDa nuclear phosphoprotein that shares homology with phosphatidylinositol-3 (PI-3) kinase (21–25). PI-3 kinase-related proteins are known to function in DNA repair, DNA recombination, and cell-cycle control. Concordantly, constitutional inactivation of ATM in cells of A-T patients causes defective double-strand break repair, defective cell-cycle checkpoint control, and radiation sensitivity (for review see refs. 26 and 27). ATM recently has been shown to directly phosphorylate the oncogenic factors c-Abl and p53 in response to DNA damage (the ATM kinase and binding domains are schematically illustrated in Fig. 1) (28–31). ATM is an interesting candidate for a tumor suppressor gene with pathogenic function in MCL for the following reasons. (i) A-T patients show an increased predisposition to develop cancer, in particular, neoplasms of the lymphoid system including both B- and T-cell tumors (32). The risk of these patients developing leukemia is approximately 70 times higher than in the normal population (33). (ii) Mutational inactivation of the ATM gene recently has been demonstrated in T-prolymphocytic leukemia (T-PLL) and a subset of B-cell chronic lymphocytic leukemias (B-CLL) in patients without A-T history, indicating a tumor suppressor function of ATM in both sporadic leukemias (34–41).
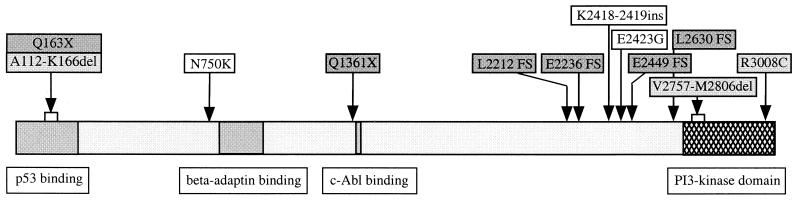
Figure 1.
Schematic representation of the ATM protein including the location of mutations detected in different MCLs with respect to the domains that are responsible for the kinase function (21, 44) and the binding of p53 (46), β-adaptin (47), and c-Abl (28, 29). The positions of the mutations are indicated by arrows. Dark-gray boxes represent truncating mutations (X, new stop codon; FS, frameshift leading to truncation). Mutations that correspond to A-T alleles and/or mutations associated with T-PLL are displayed in light gray: R3008C has been described as an A-T allele (48) and as a mutation in two T-PLL cases (34, 37); the splice-site mutations in MCL-J and MCL-A both resemble A-T mutations, which cause skipping of exons 7 and 59 from the transcript, respectively (42, 49–51). Note that the nucleotide change causative for exon 7 skipping and the deletion of 55 aa (A112-L166del) created a new stop codon (Q163X) in the low level of correctly spliced transcripts produced from this allele. Mutations in unshaded boxes are amino acid changes, the functional consequences of which are not yet known.
Two lines of evidence prompted us to analyze the ATM gene in MCL: the frequent deletion of one copy of the ATM gene in MCL and the intriguing properties of ATM that point to a tumor suppressor function of the gene product. Mutational inactivation of the gene in tumor cells would provide evidence for a pathogenic role of ATM in MCL.
Patients and Methods
Patients.
The study comprised samples from 12 MCLs that were diagnosed according to established morphological and immunophenotypical criteria (2). All MCLs were shown to have a t(11;14)(q13;q32). Deletions of chromosome bands 11q22-q23 were detected in seven cases by dual-color interphase FISH (20). None of the patients had clinical evidence for A-T or a family history of A-T. The tumor specimens were derived from peripheral blood (n = 11) or tonsil (n = 1). From three patients (MCL-G, MCL-H, and MCL-K), leukapheresis samples were obtained in remission, and from one patient (MCL-A), a skin biopsy was available. These samples were used for analysis of the ATM germ-line status. Samples were collected and used with the patient's consent.
RNA and DNA Preparation.
Cells from tonsillectomy samples were obtained by mechanical disaggregation. In leukemic and normal control cases, mononuclear cells from peripheral blood samples were obtained by Ficoll density gradient centrifugation. Total RNA and genomic DNA from tumor samples and from mononuclear cells of healthy controls were isolated with Trizol reagent (GIBCO/BRL). Genomic DNA from skin biopsy cells and leukapheresis cells was prepared by using the QIAamp kit according to the manufacturer's instructions (Qiagen, Chatsworth, CA).
Mutation Analyses.
Reverse-transcription–PCR (RT-PCR) and single-strand conformation polymorphism (SSCP) analyses were performed as described previously (34, 41, 42). Briefly, first-strand cDNA was synthesized from total RNA (1 μg) with murine leukemia virus reverse transcriptase and random hexamers (GeneAmp RNA PCR System; Perkin–Elmer). Primer sets 1A/B, 2A/B, 3A/B, 4A/B, 5A/B, 6A/B, 7A/B, and 8n1A/8n1B (Table 1) were used to amplify eight overlapping fragments covering the entire coding region of the ATM transcript. For SSCP analysis, RT-PCR products were digested with restriction endonucleases and end-labeled with T4 polynucleotide kinase in the presence of [γ-33P]ATP. Denatured fragments were analyzed by electrophoresis both at room temperature and at 4°C on nondenaturing 6% polyacrylamide gels containing 5% glycerol (exclusively for runs at room temperature). After electrophoresis at 8 W for at least 10 hr, the gels were dried and subjected to autoradiography.
Table 1.
Oligonucleotide primers for PCR amplifications and sequence analyses
| Primer | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| 1A (exon 36) | CTTCAGTGGACCTTCATAATGC |
| 1B (exon 43) | CCATACAAACTATCTGGCTCC |
| 2A (exon 29) | CAGAGATTGTGGTGGAGTTATTG |
| 2B (exon 36) | GCATTATGAAGGTCCACTGAAG |
| 3A (exon 42/43) | CTGGAATAAGTTTACAGGATCTTC |
| 3B (exon 51) | GATGATTTCATGTAGTTTTCAATTC |
| 3A1 (exon 45/46) | CATTGCACTTCCGTCAGC |
| 3A2 (exon 48) | CTATCATGGCTCTACGCAC |
| 3B3 (exon 49) | AATACTTGTGCTTCTTCCAG |
| 4A (exon 56) | AAGATGTTGTTGTCCCTACTATG |
| 4B (exon 65) | AAGGCTGAATGAAAGGGTAATTC |
| 4A2 (exon 64) | CCTTCTATATGATCCACTC |
| 4B3 (exon 61) | TCTGTACATGTCTATCACC |
| 5A (exon 51) | GATGGAGAAAGTAGTGATGAGC |
| 5B (exon 57) | AGTCACCAGATTTCCATATTCTC |
| 5A2 (exon 54/55) | GCTCTCAGCTTGATGAGG |
| 5B1 (exon 52) | TACCCACATATCATGTTC |
| 6A (exon 22) | CTAGGTCAAAGCAATATGGACTC |
| 6B (exon 30) | CATGCGATGGAAAATGAGGTG |
| 6A2 (exon 28) | ACATCTGGTGATTAGAAGTC |
| 7A (exon 36) | GAGGTCAAACCTAGAAAGCTCAC |
| 7B (exon 22/23) | CCTCTCCTTTGTTAGATGCC |
| 7A1 (exon 13) | TGCCTCCAATTCTTCACAG |
| 7B5 (exon 19) | ATGGTACTTTGGCTCTCTCC |
| 8n1A (exon 3) | AGGCATACATCACAATTTGG |
| 8n1B (exon 12) | TTGCTCAGAACTTATACCACG |
| 8A5 (exon 6) | CAGCCTCAACACAAGCCTC |
| 8B4 (exon 8) | GCATCCTTTGGTAACAGCAT |
| 29A (intron 28) | AGATTGTGGTGGAGTTATTGA |
| 29B (intron 29) | TTAAAAAGAGTGATGTCTATAA |
| 48A (intron 47) | CTCTTGCTTACATGAACTCTA |
| 48B (intron 48) | AAGAGGTAAGATGACATAGTT |
| 59A (intron 58) | AGGTCAACGGATCATCAAAT |
| 59B (intron 59) | TTAATTTTGGGTGTCACTC |
Primer sets 1A/B, 2A/B, 3A/B, 4A/B, 5A/B, 6A/B, and 7A/B and primer pairs 29A/B, 48A/B, and 59A/B have been described previously (42, 43). 8n1A, 8n1B, 3A1, 3A2, 3B3, 4A2, 4B3, 5A2, 5B1, 6A2, 7A1, 7B5, 8A5, and 8B4 were derived from cDNA sequence data from the ATM gene (GenBank accession no. U33841) (44).
Germ-Line Analyses.
For analyses of the germ-line status of the ATM gene, exons 29, 48, and 59 and their flanking intronic regions were amplified from genomic DNA as described previously by using primer pairs 29A/B, 48A/B, and 59A/B (Table 1) (43).
DNA Sequence Analyses.
Direct sequencing of PCR and RT-PCR products was performed by cycle sequencing with ABI PRISM BigDye Terminator chemistry (Perkin–Elmer) followed by electrophoresis on a Perkin–Elmer ABI-377 automated sequencer.
Results
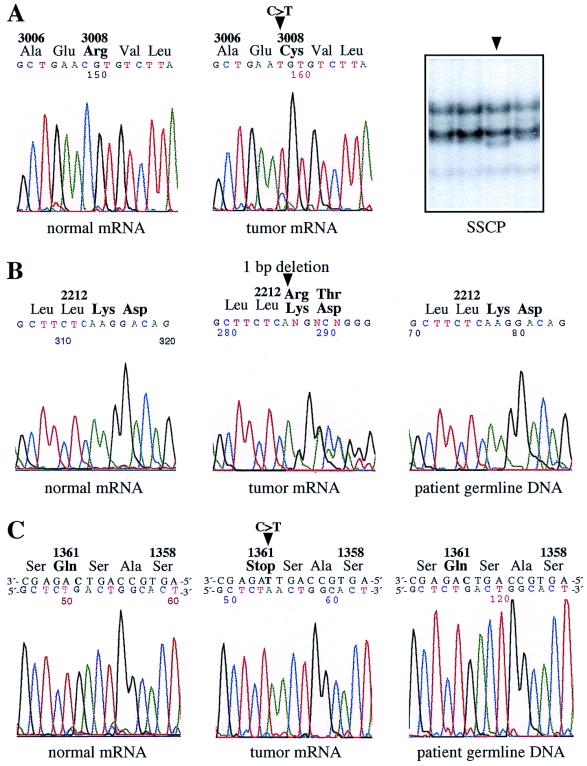
To address the question of whether the nondeleted allele of the ATM gene is mutated in MCLs with a monoallelic deletion of the gene, we selected seven MCLs for mutation analysis that were shown by FISH to exhibit deletions of the ATM region. The entire 9.2-kb coding region of the ATM transcript was subjected to SSCP analysis. Aberrant migration of single-stranded DNA fragments, because of mutation-associated conformation changes, was observed in six cases. Direct sequencing of the corresponding RT-PCR fragments identified two small deletions, one single-nucleotide insertion and three single-base substitutions (Table 2 and Fig. 1). In samples MCL-C (Fig. 2A) and MCL-D, the nucleotide substitutions led to amino acid changes in the PI-3 kinase domain and near the β-adaptin-binding domain. The C→T transition in sample MCL-G (Fig. 2C) created a new stop codon that removed the c-Abl and kinase domain from the translated product. Small insertions and deletions in samples MCL-E, MCL-F, and MCL-H (Fig. 2B) all caused frameshift changes and termination of translation not more than 22 codons downstream of the mutation site, thus resulting in truncated ATM protein lacking the PI-3 kinase domain.
Table 2.
Alterations affecting both ATM alleles in mantle cell lymphomas of non-A-T individuals*
| Mantle cell lymphoma | First
allele
|
Second allele
|
||
|---|---|---|---|---|
| DNA†‡ | Gene product | DNA‡ | Gene product | |
| MCL-A§¶ | del(11q22-q23) | — | IVS59 → T (intron 59) | Exon 59 skipped: Val2757-Met2806del |
| MCL-C | del(11q22-q23) | — | 9022C → T (exon 65) | Arg3008Cys |
| MCL-D | del(11q22-q23) | — | 2250C → G (exon 16) | Asn750Lys |
| MCL-E | del(11q22-q23) | — | 6709–6710delAA (exon 48) | Frameshift after Glu2236 + truncation |
| MCL-F | del(11q22-q23) | — | 7349–7350insT (exon 52) | Frameshift after Glu2449 + truncation |
| MCL-G§ | del(11q22-q23) | — | 4081C → T (exon 29) | Gln1361ter |
| MCL-H§ | del(11q22-q23) | — | 6638delA (exon 48) | Frameshift after Leu2212 + truncation |
| MCL-B¶ | 7268A → G (exon 51) | Glu2423Gly | 7253–7254insGAA (exon 51) | Lys2418–2419ins |
| MCL-J | 487C → T (exon 7) | Gln163ter | 7890–7891insTATTA (exon 55) | Frameshift after Leu2630 + truncation |
| Exon 7 skipped: Arg111Lys + Ala112-Lys166del | ||||
The polymorphic change Asp1853Asn detected in cases with (MCL-G) and without (MCL-K) monoallelic ATM deletion is not listed.
† The deletion mapping by interphase FISH is described in ref. 20; del(11q22-q23) indicates loss of one copy of the chromosomal region 11q22-q23 containing ATM.
‡ The exon numbering is according to refs. 22 and 23 (GenBank accession no. U82828). The nucleotide positions refer to the sequence of the ATM transcript (GenBank accession no. U33841); the first codon of the ORF was designated +1. The nomenclature of the mutations was used as suggested by Antonarakis and Group (45).
§ For those patients, the analysis of the germ-line status of ATM was possible.
¶ MCL-A and MCL-B were included in a previous study (41).
Figure 2.
Point mutations in the ATM gene in MCLs containing a deletion of one ATM copy. Arrowheads indicate the positions of the mutations. (A) MCL-C. SSCP analysis of a 196-bp restriction fragment of the RT-PCR product containing exons 56–65 and DNA sequence analysis of the corresponding RT-PCR products identified the missense mutation Arg3008Cys. This mutation has been described in two T-PLL cases (34, 37) and is known as an A-T-causing ATM alteration (48). (B) MCL-H. Using SSCP analysis, an aberrantly migrating restriction fragment of the RT-PCR product harboring exons 42–51 was detected. Sequence analysis of the respective RT-PCR fragments identified a single-nucleotide deletion (6638delA) that caused a frameshift and truncation. Note that the amount of cells with an 11q deletion was only 79% in the tumor sample, which is responsible for the unusually high signal intensity of the normal allele. The patient's germ-line DNA did not contain the mutation. (C) MCL-G. SSCP and subsequent DNA sequence analysis of RT-PCR products encompassing exons 22–30 identified the truncating mutation Gln1361ter. This mutation was not present in the patient's germ-line DNA.
In the seventh case with a monoallelic deletion analyzed (MCL-A), sequence analysis of the RT-PCR fragment encompassing exons 56–65 of ATM revealed the absence of exon 59 from the ATM transcript. A nucleotide substitution at the donor splice junction of exon 59/intron 59 (IVS59+1G→A) was detected by genomic DNA sequencing. This alteration led to aberrant transcript splicing, resulting in the in-frame loss of 50 aa from the kinase domain (41).
To determine whether ATM is also involved in MCLs without 11q deletion, five lymphomas without microscopically detectable deletion of the ATM locus were analyzed for mutations by using SSCP and sequence analyses. In two cases, both alleles of the gene were found to contain point mutations (Table 2 and Fig. 1). In sample MCL-J, we detected a heterozygous 5-bp insertion that expanded a TA-rich region containing two tandem repeats by a third one: TTA[TATTA][TATTA][TATTA]. As this alteration was confirmed in multiple independent PCR amplifications, the insertion is likely because of DNA polymerase slipping during replication. The insertion caused a frameshift and premature termination of translation, giving rise to a mutated ATM protein lacking the entire kinase domain. A second heterozygous mutation in this case was identified in exon 7: the nucleotide substitution 487C→T (near the exon 7/intron 7 splice junction) led to the skipping of exon 7 from most of the transcripts, which resulted in the deletion of 55 aa from the p53-binding domain. In the low levels of correctly spliced transcripts produced from this allele, the mutation created a new stop codon, causing premature termination of translation in the amino-terminal region of the protein. As estimated from the SSCP gels and the DNA sequence profiles, the correctly spliced transcripts amounted to some 10%. In a second sample (MCL-B), both alleles of ATM were altered by two independent mutations affecting the same region of the protein: a nucleotide substitution caused an amino acid change (Glu2423Gly), and a trinucleotide insertion induced the in-frame insertion of an additional lysine (Lys2419–2419ins) (41).
Cell samples for the analyses of the ATM germ-line status were available from three patients who had ATM mutations in their tumor cells. Sequence analyses of genomic DNA from skin cells (MCL-A) and from leukapheresis cells obtained from patients in remission (Fig. 2 C, MCL-G, and B, MCL-H) showed that the nucleotide changes in ATM detected in tumor cells were not present in nonmalignant cells. Thus, the mutations were of somatic rather than germ-line origin in all three cases.
Discussion
Deletion of the chromosomal region 11q22-q23 is one of the most frequent genetic alterations in MCL. In two recent studies using FISH analyses, loss of chromosome 11 material was observed in 46% and 49% of the MCLs analyzed (20, 52). These data emphasize the pivotal role of a putative tumor suppressor gene localized in this region. In the present study, a series of 12 MCLs, 7 of them with deletions of one copy of the chromosomal region 11q22-q23, were analyzed for mutations of the ATM gene. In all seven cases with monoallelic deletions, the nondeleted ATM allele was found to contain a sequence alteration. Furthermore, two MCLs without 11q deletion were found to contain mutations in both ATM alleles. Therefore, the involvement of genetic aberrations affecting the ATM gene in the pathogenesis of MCL is higher than originally anticipated based on deletion analyses. The frequent inactivation of both alleles of the gene by deletion and/or deleterious mutation is in accordance with the two-hit model of tumor suppressor gene inactivation (53) and, thus, supports a pathogenic function of ATM inactivation in MCL development.
The high proportion of MCLs with ATM inactivation is at variance with the data of another B-cell lymphoma, for which ATM mutations were described recently, namely, B-CLL (39–41): (i) the frequency of deleterious mutations of the ATM gene is considerably higher in MCL, whereas in B-CLL missense mutations are predominant; (ii) in all MCLs containing a monoallelic deletion of ATM, the remaining allele was subject to inactivation, whereas in a fraction of B-CLLs with monoallelic ATM deletion no mutation could be detected within the coding region (41). These data suggest a second gene located within 11q22.3-q23.1 that may have a pathogenic role in B-CLL, although it cannot be excluded that a mutation remained undetected. Hence, ATM inactivation is highly characteristic for MCL. According to our data, there is no evidence for a second gene within 11q22.3-q23.1 being associated with MCL. However, in a recent 11q deletion mapping study by Monni et al. (52), large deletions including the ATM gene were reported in all but one MCL, which had a small deletion 2–3 Mbp distal to the ATM gene locus.
Heterozygous germ-line mutations of ATM currently are discussed as predisposing factors for tumor development, because the frequency of heterozygotes amounts to 1% (54). In two recent studies, heterozygous ATM germ-line mutations were reported in patients with B-CLL (39, 40). In this study, the analysis of the ATM germ-line status was possible for three patients who contained ATM point mutations in their tumor cells. In all three cases, the ATM mutations were not present in the nonmalignant cells, indicating that they arose de novo during tumor development. Thus, by analogy to our previous study of B-CLL (41), the present data do not provide evidence for a genetic predisposition to MCL that would be based on heterozygous germ-line inactivation of the ATM gene.
The ATM mutations identified in this study were of deleterious character: the majority of the 11 mutations resulted either in premature termination of translation, resulting in a truncated protein lacking the PI-3 kinase domain (five cases, Fig. 1), or corresponded to disease mutations associated with T-PLL or causative for A-T (three cases, Fig. 1). All mutations directly affected either the p53 binding or the kinase domain of the protein, which is crucial for the phosphorylation of ATM substrates (30, 31), and therefore are predicted to cause substantial functional impairment. With respect to known aspects of the biological function of ATM there are several conceivable mechanisms by which ATM inactivation may act as a tumorigenic promoter in MCL lymphomagenesis.
(i) Recent studies indicate that ATM is a key regulator of the cellular response to DNA double-strand breaks induced by irradiation or physiological processes, such as V(D)J recombination (reviewed in refs. 27 and 55). One therefore may speculate that ATM inactivation may be involved in the rearrangement of the Ig gene locus with the cyclin D1 gene characteristic for MCL. In support of this hypothesis, lymphocytes and lymphoid tumor cells of A-T patients (32, 56) and Atm-deficient mice (57, 58) regularly present chromosomal translocations and inversions that typically involve the Ig and T cell receptor genes, suggesting illegitimate joining of these loci during V(D)J recombination as the causative mechanism. The resulting rearrangements with oncogenes are regarded as crucial for the development of lymphoid malignancies in A-T patients (32) and in Atm-deficient mice (57, 58). In addition, chromosome translocations and inversions that typically involve a break in the T-cell receptor gene locus TCRα/δ at 14q11 characteristically are associated with sporadic T-PLL (59, 60), which recently was shown to be associated with recurrent somatic ATM mutation (34–37).
(ii) ATM-mediated response to DNA breaks includes regulation of cell-cycle checkpoint control, induction of apoptosis, and DNA repair (see, for review, refs. 27 and 55). Because p53 (30, 31) and c-Abl (28, 29) have been identified as nuclear substrates for ATM, it is likely that defects in the ATM-dependent phosphorylation of these molecules account for the genetic instability and defective cell-cycle control observed in A-T cells. ATM inactivation in tumor cells thus may act synergistically with cyclin D1 overexpression to override cell-cycle checkpoint controls and thereby promote the accumulation of additional genomic aberrations during tumorigenic development.
Acknowledgments
The support by Ralf Klären, Stefan Wiemann, Martin Bentz, Elke Leupolt, and Brenda Stride is gratefully acknowledged. This study was supported by the Wilhelm-Sander-Stiftung (97.003.1), the Deutsche Krebshilfe (10-1289-StI), and the Tumorzentrum Heidelberg/Mannheim (I/I.1).
Abbreviations
- MCL
mantle cell lymphoma
- B-CLL
B-cell chronic lymphocytic leukemia
- ATM
ataxia telangiectasia mutated
- A-T
ataxia telangiectasia
- PI-3
phosphatidylinositol-3
- T-PLL
T-prolymphocytic leukemia
- FISH
fluorescence in situ hybridization
- RT-PCR
reverse transcription–PCR
- SSCP
single-strand conformation polymorphism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050400997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050400997
References
- 1.Tolksdorf G, Stein H, Lennert K. Br J Cancer. 1980;41:168–182. doi: 10.1038/bjc.1980.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weisenburger D D, Armitage J O. Blood. 1996;87:4483–4494. [PubMed] [Google Scholar]
- 3.Leroux D, Le Marc'Hadour F, Gressin R, Jacob M C, Keddari E, Monteil M, Caillot P, Jalbert P, Sotto J J. Br J Haematol. 1991;77:346–353. doi: 10.1111/j.1365-2141.1991.tb08582.x. [DOI] [PubMed] [Google Scholar]
- 4.Vandenberghe E, De Wolf Peeters C, Wlodarska I, Stul M, Louwagie A, Verhoef G, Thomas J, Criel A, Cassiman J J, Mecucci C. Br J Haematol. 1992;81:212–217. doi: 10.1111/j.1365-2141.1992.tb08209.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsujimoto Y, Yunis J, Onorato-Showe L, Erikson J, Nowell P C, Croce C M. Science. 1984;224:1403–1406. doi: 10.1126/science.6610211. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg C L, Wong E, Petty E M, Bale A E, Tsujimoto Y, Harris N L, Arnold A. Proc Natl Acad Sci USA. 1991;88:9638–9642. doi: 10.1073/pnas.88.21.9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Withers D A, Harvey R C, Faust J B, Melnyk O, Carey K, Meeker T C. Mol Cell Biol. 1991;11:4846–4853. doi: 10.1128/mcb.11.10.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimokh R, Berger F, Delsol G, Charrin C, Bertheas M F, Ffrench M, Garoscio M, Felman P, Coiffier B, Bryon P A. Blood. 1993;81:3063–3067. [PubMed] [Google Scholar]
- 9.Bosch F, Jares P, Campo E, Lopez-Guillermo A, Piris M A, Villamor N, Tassies D, Jaffe E S, Montserrat E, Rozman C. Blood. 1994;84:2726–2732. [PubMed] [Google Scholar]
- 10.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 11.Bodrug S E, Warner B J, Bath M L, Lindeman G J, Harris A W, Adams J M. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovec H, Grzeschiczek A, Kowalski M B, Moroy T. EMBO J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greiner T C, Moynihan M J, Chan W C, Lytle D M, Pedersen A, Anderson J R, Weisenburger D D. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- 14.Hernandez L, Fest T, Cazorla M, Teruya-Feldstein J, Bosch F, Peinado M A, Piris M A, Montserrat E, Cardesa A, Jaffe E S, et al. Blood. 1996;87:3351–3359. [PubMed] [Google Scholar]
- 15.Dreyling M H, Bullinger L, Ott G, Stilgenbauer S, Müller-Hermelink H K, Bentz M, Hiddemann W, Döhner H. Cancer Res. 1997;57:4608–4614. [PubMed] [Google Scholar]
- 16.Pinyol M, Hernandez L, Cazorla M, Balbin M, Jares P, Fernandez P L, Montserrat E, Cardesa A, Lopez-Otin C, Campo E. Blood. 1997;89:272–280. [PubMed] [Google Scholar]
- 17.Monni O, Oinonen R, Elonen E, Franssila K, Teerenhovi L, Joensuu H, Knuutila S. Genes Chromosomes Cancer. 1998;21:298–307. doi: 10.1002/(sici)1098-2264(199804)21:4<298::aid-gcc3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 18.Bea S, Ribas M, Hernandez J M, Bosch F, Pinyol M, Hernandez L, Garcia J L, Flores T, Gonzalez M, Lopez-Guillermo A, et al. Blood. 1999;93:4365–4374. [PubMed] [Google Scholar]
- 19.Stilgenbauer S, Nickolenko J, Wilhelm J, Wolf S, Weitz S, Döhner K, Boehm T, Döhner H, Lichter P. Oncogene. 1998;16:1891–1897. doi: 10.1038/sj.onc.1201764. [DOI] [PubMed] [Google Scholar]
- 20.Stilgenbauer S, Winkler D, Ott G, Schaffner C, Leupolt E, Bentz M, Möller P, Müller-Hermelink H-K, James M R, Lichter P, et al. Blood. 1999;94:3262–3264. [PubMed] [Google Scholar]
- 21.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 22.Uziel T, Savitsky K, Platzer M, Ziv Y, Helbitz T, Nehls M, Boehm T, Rosenthal A, Shiloh Y, Rotman G. Genomics. 1996;33:317–320. doi: 10.1006/geno.1996.0201. [DOI] [PubMed] [Google Scholar]
- 23.Platzer M, Rotman G, Bauer D, Uziel T, Savitsky K, Bar-Shira A, Gilad S, Shiloh Y, Rosenthal A. Genome Res. 1997;7:592–605. doi: 10.1101/gr.7.6.592. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Lee E Y-H P. J Biol Chem. 1996;271:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- 25.Brown K D, Ziv Y, Sadanandan S N, Chessa L, Collins F S, Shiloh Y, Tagle D A. Proc Natl Acad Sci USA. 1997;94:1840–1845. doi: 10.1073/pnas.94.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeggo P A, Carr A M, Lehmann A R. Trends Genet. 1998;14:312–316. doi: 10.1016/s0168-9525(98)01511-x. [DOI] [PubMed] [Google Scholar]
- 27.Rotman G, Shiloh Y. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 28.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, et al. Nature (London) 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 29.Shafman T, Khanna K K, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Nature (London) 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 30.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 31.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 32.Taylor A M R, Metcalfe J A, Thick J, Mak Y-F. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 33.Morrell D, Cromartie E, Swift M. J Natl Cancer Inst. 1986;77:89–92. [PubMed] [Google Scholar]
- 34.Stilgenbauer S, Schaffner C, Litterst A, Liebisch P, Gilad S, Bar-Shira A, James M R, Lichter P, Döhner H. Nat Med. 1997;3:1155–1159. doi: 10.1038/nm1097-1155. [DOI] [PubMed] [Google Scholar]
- 35.Vorechovsky I, Luo L, Dyer M J S, Catovsky D, Amlot P L, Yaxley J C, Foroni L, Hammarström L, Webster A D B, Yuille M A R. Nat Genet. 1997;17:96–99. doi: 10.1038/ng0997-96. [DOI] [PubMed] [Google Scholar]
- 36.Stoppa-Lyonnet D, Soulier J, Lauge A, Dastot H, Garand R, Sigaux F, Stern M H. Blood. 1998;91:3920–3926. [PubMed] [Google Scholar]
- 37.Yuille M A R, Coignet L J A, Abraham S M, Yaqub F, Luo L, Matutes E, Brito-Babapulle V, Vorechovsky I, Dyer M J S, Catovsky D. Oncogene. 1998;16:789–796. doi: 10.1038/sj.onc.1201603. [DOI] [PubMed] [Google Scholar]
- 38.Starostik P, Manshouri T, O'Brien S, Freireich E, Kantarjian H, Haidar M, Lerner S, Keating M, Albitar M. Cancer Res. 1998;58:4552–4557. [PubMed] [Google Scholar]
- 39.Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd P J, Moss P A H, Taylor A M R. Lancet. 1999;353:26–29. doi: 10.1016/S0140-6736(98)10117-4. [DOI] [PubMed] [Google Scholar]
- 40.Bullrich F, Rasio D, Kitada S, Starostik P, Kipps T, Keating M, Albitar M, Reed J C, Croce C M. Cancer Res. 1999;59:24–27. [PubMed] [Google Scholar]
- 41.Schaffner C, Stilgenbauer S, Rappold G A, Döhner H, Lichter P. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- 42.Gilad S, Khosravi R, Shkedy D, Uziel T, Ziv Y, Savitsky K, Rotman G, Smith S, Chessa L, Jorgensen T J, et al. Hum Mol Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 43.Vorechovsky I, Rasio D, Luo L, Monaco C, Hammarstrom L, Webster A D B, Zaloudik J, Barbanti-Brodani G, James M, Russo G, et al. Cancer Res. 1996;56:2726–2732. [PubMed] [Google Scholar]
- 44.Savitsky K, Sfez S, Tagle D A, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 45.Antonarakis S E, Group N W. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, et al. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 47.Lim D S, Kirsch D G, Canman C E, Ahn J H, Ziv Y, Newman L S, Darnell R B, Shiloh Y, Kastan M B. Proc Natl Acad Sci USA. 1998;95:10146–10151. doi: 10.1073/pnas.95.17.10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hacia J G, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor S P A, Tagle D A, Collins F S. Genome Res. 1998;8:1245–1258. doi: 10.1101/gr.8.12.1245. [DOI] [PubMed] [Google Scholar]
- 49.Ejima Y, Sasaki M S. Hum Genet. 1998;102:403–408. doi: 10.1007/s004390050712. [DOI] [PubMed] [Google Scholar]
- 50.Wright J, Teraoka S, Onengut S, Tolun A, Gatti R A, Ochs H D, Concannon P. Am J Hum Genet. 1996;59:839–846. [PMC free article] [PubMed] [Google Scholar]
- 51.Stankovic T, Kidd A M, Sutcliffe A, McGuire G M, Robinson P, Weber P, Bedenham T, Bradwell A R, Easton D F, Lennox G G, et al. Am J Hum Genet. 1998;62:334–345. doi: 10.1086/301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monni O, Zhu Y, Franssila K, Oinonen R, Hoglund P, Elonen E, Joensuu H, Knuutila S. Br J Haematol. 1999;104:665–671. doi: 10.1046/j.1365-2141.1999.01257.x. [DOI] [PubMed] [Google Scholar]
- 53.Knudson A G. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swift M, Morrell D, Massey R B, Chase C L. N Engl J Med. 1991;325:1831–1836. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 55.Brown K D, Chakravarti A. Am J Hum Genet. 1999;64:46–50. doi: 10.1086/302223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojis T L, Gatti R A, Sparkes R S. Cancer Genet Cytogenet. 1991;56:143–156. doi: 10.1016/0165-4608(91)90164-p. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Ashley T, Brainerd E E, Bronson R T, Meyn M S, Baltimore D. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 58.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J N, Ried T, Tagle D, et al. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 59.Brito-Babapulle V, Pomfret M, Matutes E, Catovsky D. Blood. 1987;70:926–931. [PubMed] [Google Scholar]
- 60.Zech L, Gahrton G, Hammarstrom L, Juliusson G, Mellstedt H, Robert K H, Smith C I. Nature (London) 1984;308:858–860. doi: 10.1038/308858a0. [DOI] [PubMed] [Google Scholar]