Abstract
Homeobox protein HOXA5 functions as a transcriptional factor for genes that are not only involved in segmentation identity but also in cell differentiation. Although HOXA5 has been shown to regulate the expression of the tumor-suppressor protein p53, its role in breast tumorigenesis is not well understood. Using yeast as a model system, we now demonstrate that overexpression of HOXA5 in yeast can be used to identify downstream target genes that are homologous in humans. One such identified gene was that of the mismatch repair pathway component MutL homolog 1. Analysis of the promoter region of the gene for human MutL homolog 1 (hMLH1) displayed several putative HOXA5-binding sites. In transient transfection experiments, the overexpression of HOXA5 transactivated the hMLH1 promoter-reporter construct. In addition, chromatin immunoprecipitation assay using a human breast cancer cell line MCF-7 demonstrated that HOXA5 binds to the hMLH1 promoter in vivo. Furthermore, we demonstrate that, in the presence of HOXA5, there is an increase in in vivo repair activity in MCF-7 cells. Taken together, our results indicate that HOXA5 is a transcriptional regulator of hMLH1 in breast cancer cells.
Keywords: Breast cancer, mismatch repair, homeotic gene, yeast, promoter analyses
Abbreviations: HOX, homeotic; hMLH1, human MutL homolog 1; ChIP, chromatin immunoprecipitation; MMR, mismatch repair; Luc, luciferase
Introduction
The homeobox protein HOXA5 has been shown to participate in the developmental regulation of the lung, gastrointestinal tract, spleen, kidney, and vertebrae in eukaryotic embryos [1]. Its function as a transcriptional factor is to modulate the expression of various proteins during the ontogeny of normal development. Besides its role in development, it has been recently demonstrated that dysregulation of HOXA5 in breast epithelium can contribute to breast cancer biogenesis primarily by regulating the expression of the tumor-suppressor protein p53 and the progesterone receptor [2,3]. Homeotic genes encode master regulatory transactivating proteins that likely regulate other genes within the mammary epithelium. To identify downstream target genes of HOXA5, we used yeast (Saccharomyces cerevisiae) as a model system. Overexpressing human HOXA5 in yeast cells resulted in a library of genes whose transcription was increased by HOXA5. One of the genes found to be upregulated was the human MutL homolog 1 (hMLH1) [4], a component of the mismatch repair (MMR) system.
The mammalian MMR system consists of multiple protein components, which detect and correct base mismatch abnormalities and insertion/deletion loops occurring within doublestranded DNA [4]. In addition to normal replication errors, environmental factors such as chemical toxins and ultraviolet radiation contribute to base modifications, which are also corrected by the MMR pathway [5]. Any disruption of cellular pathways that compromises genomic integrity is disastrous, giving rise to cell death or propagation of abnormal genetic codes. Thus, the absence of a complete set of MMR proteins results in replication errors, which contribute to genomic instability.
It is now established that many cancer types are deficient in MMR, which gives rise to genomic instability in these cancers. For example, hereditary nonpolyposis colorectal cancer (HNPCC) has germline mutations in human MutS homolog 2 (hMSH2) and hMLH1 in 98% of cases studied [6]. In breast cancer cell lines, it was shown that the hMLH1 gene can mutate [7]. Moreover, hMLH1 has been shown to be mutated in a subset of patients with lobular carcinoma in situ of the breast, resulting in truncated protein [8]. In addition, immunohisto-chemistry has shown that hMLH1 can be lacking or reduced in approximately 28% to 38% of human sporadic breast cancer cases [9]. Hypermethylation of hMLH1 promoter was shown to account for most of this loss of expression. However, in approximately 15% of these cases, the loss of hMLH1 was not explained. Thus, alternative mechanisms contributing to the loss of hMLH1 function in breast cancer exist.
Here we report that the putative HOXA5 DNA-binding sites found within the distal promoter of the human MLH1 gene bind HOXA5, but not other homeotic (HOX) proteins tested. In vivo binding of HOXA5 to the hMLH1 promoter was determined in breast cancer cells using chromatin immunoprecipitation (ChIP) assay and was also shown in luciferase (Luc) reporter analyses. In addition, overexpression of HOXA5 in the presence of a basepair mismatch within a reporter plasmid indicated that HOXA5 can be functionally relevant to the MMR in breast cancer cells, as repair of the mismatch was greatly increased during the overexpression of HOXA5. Taken together, we conclude that HOXA5 can regulate hMLH1 gene in breast cancer cells.
Materials and Methods
Generation of Full-Length cDNA Clone Encoding HOXA5 in a Yeast Shuttle Vector
The coding region of HOXA5 was cloned into the eukaryotic expression vector pcDNA3 (a gift of Dr. Judy Gasson, University of California at Los Angeles, Los Angeles, CA) and sequenced to rule out possible errors introduced during cloning. As pcDNA3 contains the T7 promoter at the 5′ end of the gene, in vitro transcription and translation, along with sodium dodecyl sulfate polyacrylamide gel electrophoresis, were performed to confirm that the protein product obtained from this clone was of the appropriate size. An EcoRI fragment containing the full-length HOXA5 clone and the inframe FLAG epitope at the N-terminus were then cloned into pYES2.1, a Gal1 yeast-inducible vector from Invitrogen (Carlsbad, CA). Following transformation into Escherichia coli DH5α cells, plasmid DNA was purified using a kit (Qiagen, Valencia, CA) and sequenced again.
Yeast Transformation
Yeast cells were transformed using the alkali cation method, as described by Ito et al. [10].
Growth and Induction of HOXA5 in Yeast Strain 334
Yeast strains used in this study were grown in synthetic media minus uracil (URA selection marker) plus 2% glucose for 12 to 14 hours at 30°C. Cells were then washed and seeded during logarithmic growth (optical density, OD = 0.6–0.9) in synthetic media minus URA plus 2% raffinose for an additional 12 to 14 hours. Induction was initiated by the addition of 1% galactose to raffinose-containing media, and HOXA5 expression was monitored in aliquots taken at regular intervals for the next 24 hours.
Affymetrix GeneChip Arrays
Within 2 hours following the induction of HOXA5 by galactose, RNA was extracted using a hot phenol method and purified using the oligotex mRNA kit (Qiagen). The target was prepared, processed, and hybridized to the Yeast Genome S98 Array (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol. Experimental parameters for scanning the chip are defined by the GeneChip software (Affymetrix) installed on a PC workstation with a Windows NT operating system (Redmond, WA). Probe array, sample description, and hybridization conditions were entered into the software and saved. The probes were scanned for accuracy at least three to four times using the HP Gene Array scanner (Affymetrix). The mean intensity of each probe was calculated and analyzed using the GeneChip software. The entire hybridization, scanning, and data analysis were performed at the Affymetrix core station located at the Johns Hopkins University School of Public Health and Hygiene (Baltimore, MD).
Microarray Data Analysis
Scanned output files were analyzed with the Affymetrix Microarray Suite 5.0 and independently normalized to an average intensity of 500 before comparison. To identify differentially expressed transcripts, pairwise comparison analysis was carried out with Data Mining Tool 3.0 (Affymetrix). The analysis compared differences in perfect match-to-mismatch values of each probe pair on baseline array to its matching probe pair on experimental array. P value was determined by Wilcoxon signed rank test and expressed as an increase, decrease, or no change. Data Mining Tool analysis also provided signal log ratio, which estimates the magnitude and direction of change of a transcript when two arrays are compared (experimental versus baseline). For convenience, we have converted the signal log ratio output into fold change using the formula recommended by Affymetrix: Fold change = 2a (a > 0) or (-) 2-(a) (a < 0), where a is the signal log ratio.
For this study, four pairwise comparisons were performed for each group [experimental (n = 2) versus baseline (n = 2)]. Only those altered genes that appeared in four of four comparisons were selected. This conservative analytic approach limited the number of false positives. In addition, a Mann-Whitney pairwise comparison test was performed in Data Mining Tool to rank the results by concordance as a calculation of the significance (P) of each identified change in gene expression.
Transfections
MCF-7 breast cancer cells were maintained in Eagle's minimal essential medium supplemented with 10% fetal bovine serum. Transfections were conducted using the Trans-IT transfection kit (Mirus Bio Corp. Madison, WI) according to the manufacturer's protocol. All transfections were allowed to proceed for 24 hours before cell harvest and analysis.
ChIP
ChIP was carried out following established protocols [11] using MCF-7 cells transfected with pcDNA3-HOXA5. Ten 10-cm plates, each with 2 x 106 MCF-7 cells (20 x 106 cells), were each transfected with 3 µg of pcDNA3-HOXA5. After 24 hours, DNA was crosslinked to bound proteins by the addition of formaldehyde (1% vol/vol, final concentration) to the culture medium with 5-minute incubation at room temperature. Glycine was then added to a final concentration of 0.125 M to quench the reaction. Cells were harvested, nuclei were isolated and lysed, and all 10 chromatin-protein preparations were combined. Samples were sonicated using a microtip with 1-second pulses for a total of 210 seconds, which sheared DNA-protein fragments to between 600 and 1000 bp of DNA. The sonicator was set at the 50% duty cycle and at the “microtip limit.” An aliquot (100 µl) of sonicated chromatin was designated as “total input chromatin.” Chromatin-protein complexes were immuno-precipitated using rabbit anti-HOXA5, anti-histone deacetylase, or anti-snail antibodies. Anti-snail samples, along with samples prepared without antibodies, served as negative controls. Sample crosslinking was reversed in 0.3 M NaCl. The DNA was extracted from the proteins using phenol/chloroform/isoamyl alcohol (25:24:1), precipitated with absolute alcohol, resuspended in water, and subsequently analyzed by polymerase chain reaction (PCR) using hMLH1 promoter-specific primers.
hMLH1 promoter primers used in PCR flanked HOXA5 cis elements and are listed in Table 1 as ChIP S1/ChIP AS1 and ChIP S2/ChIP AS2. PCR contained 0.025 mM deoxynucleoside triphosphates, 0.25 µM of each primer, 2 mM Mg2+, and 5 µl of each ChIP sample. PCR began with a 2-minute denaturation at 94°C and was then cycled 35 times with a melting temperature of 94°C (0 second), an annealing temperature of 54°C (0 second), and an extension at 74°C (15 seconds) in a Rapid Cycler (Idaho Technologies, Salt Lake City, UT).
Table 1.
PCR Primer and MMR Assay Oligonucleotide Sequences.
| Primer Designation | Sequence |
| hMLH1p S | 5′-TTCTCTGAGGGCAGGAAAGTCTGTTAG-3′ |
| hMLH1p AS | 5′-ACGTCTAGATGCTCAACGGAAGTGC-3′ |
| ChIP S1 | 5′-GCAAAGACTTTACTAACTCG-3′ |
| ChIP AS1 | 5′-AATCTGAATCAGAGCATGG-3′ |
| ChIP S2 | 5′-AATTCAGGTCGCCTAACG-3′ |
| ChIP AS2 | 5′-TGTAGGCCCTGAGTTGG-3′ |
| MMR S2 | 5′-TCGACGGTACCGCGGGATCCACCGGTCGCCACCATGGTGAGCAATCG-3′ |
| MMR AS2 | 5′-CGATTGCTCACGATGGTGGCGACCGGTGGATCCCGCGGTACCG-3′ |
Boldface indicates the G-G mispair that was introduced into the third nucleotide of the translation start site of the GFP cDNA.
Cloning the hMLH1 Promoter
Specific PCR primers (hMLH1p S and hMLH1p AS in Table 1) to the hMLH1 promoter were designed from its sequence (PubMed accession no. AB017806) using Primer-Select (Lasergene, DNAStar, Madison, WI). PCR was performed using a gradient annealing temperature scale (annealing range, 50–60°C). A 1.75-kb amplified DNA was purified from an agarose gel, ligated into pCR2.1 (Invitrogen), and sequenced. The promoter was then cloned into the pGL2-Basic vector (Promega, Madison, WI) upstream of the firefly Luc coding sequence.
Eight putative HOXA5-binding sites were identified in the hMLH1 promoter based on the reported HOXA5 corebinding consensus site of TAAT [12].
pGL2-Basic-hMLH1 Promoter Deletion Constructs
The 3′ proximal promoter region [13] of hMLH1 was excised by digestion with ApaI and HindIII. All further promoter deletion constructs were prepared from this 3′ truncation product. All plasmids were sequenced to verify the deletions generated.
Dual-Reporter Luc Assay
Transcriptional activation of pGL2-Basic-hMLH1 promoter deletion constructs was assayed using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions. MCF-7 breast cancer cells (5 x 105) were transfected with a combination of 1 µg of a pGL2-Basic-hMLH1 promoter vector construct plus 0.5 µg of pcDNA3-HOXA5 or 0.5 µg of pcDNA3-HOXB4, pcDNA3-HOXB5, pcDNA3-HOXB7, or pCR3.1 empty vector plus 5 ng of Renilla plasm id as transfection control. Samples were cultured overnight and then lysed using Passive Lysis Buffer (Promega, Madison, WI).
Luminosity measurements were obtained using a Berthold Sirius (Oak Ridge, TN) luminometer. Each Luc measurement was divided by its corresponding Renilla measurement, which normalized transfection efficiencies. To investigate fold activation due to HOXA5 and HOX variants, the ratios of HOX samples were divided by the ratios obtained for pCR3.1 cotransfected samples.
MMR Assay
A modified pEGFP-N1 vector (Clontech Mountain View, CA) p95-1 was a generous gift from Dr. Sun (University of Texas at San Antonio, San Antonio, TX). The p95-1 construct lacks the mammalian SV40ori site, which eliminates replication in mammalian cells [14]. p95-1 (30 µg) was digested with SalI and NruI, which excised the start codon of enhanced green fluorescent protein (EGFP) cDNA. Purified digested p95-1 was combined with annealed 5′-phosphorylated MMR S2/MMR AS2 oligonucleotides (see Table 1) at a plasmid/oligonucleotide molar ratio of 1:1.5 and ligated. This introduced a G-G mispair in the EGFP start codon (ATG/TAG), generating Δp95-1. MCF-7 cells (3 x 105) were transfected with 0.25 µg of Δp95-1 and 0.5 µg of pCR3.1-HOXA5 or pCR3.1-hMLH1 cDNA. Transfections with 0.25 µg of p95-1, in lieu of Δp95-1, constitute positive controls. All transfected cells were cotransfected with 0.25 µg of pDsRed2-N1 (Clontech) as internal control of transfection efficiencies.
Results
Identifying Downstream Effector Genes of HOXA5 in S. cerevisiae
The question arises as to whether HOX genes are present in single-cell eukaryotes. In support of this, it is been reported that the mating type A1 gene of yeast shares functional similarity with HOX genes [15]. To follow up on this line of reasoning, we did a protein BLAST search within the yeast genome database of the homeodomain region of HOX genes. We found that there is a striking conservation between homeobox-binding domains of yeast homeodomain proteins: Yox1 (AVQIWFQNKR), Pho1 (RVQVWFQNRR), and Pho2 (NVRIWFQNRR) and the same region of Antennapedia (QIKIWFQNRR), a homeodomain protein from Drosophila Moreover, the DNA sequence (TCTAATCC) that binds the Drosophila bicoid gene is nearly identical to a potential Yox1 DNA-binding sequence [TC(T/C)AATA(C/A)] [16]. This indicates the rationale for attempting to use yeast as a model system to study HOX gene functions, which would be in line with numerous similar model yeast systems where findings such as cyclin genes were extended and found to be ubiquitous to eukaryotes in general.
Effect of the Overexpression of HOXA5 in Yeast Strain 334
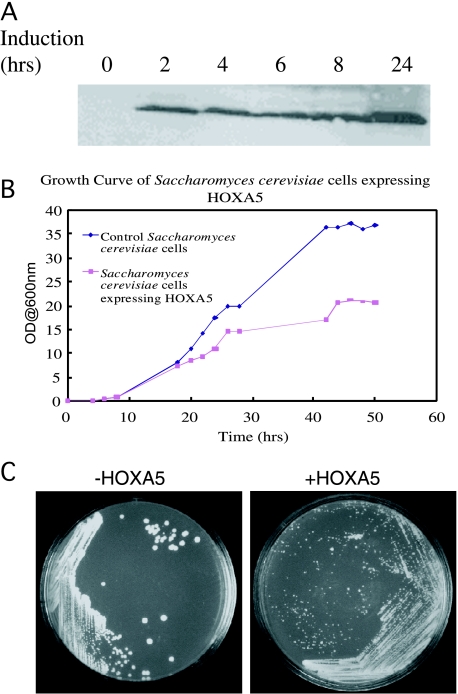
We explored the effect of HOXA5 expression on the growth of several yeast strains. Following determination of optimal induction conditions, we were able to achieve a robust expression of HOXA5 within 2 hours of galactose induction, as determined by immunoblotting (Figure 1A).
Figure 1.
Effects of HOXA5 expression in S. cerevisiae. (A) Time course evaluation of HOXA5 induction by galactose in S. cerevisiae strain 334. Following induction of HOXA5 gene by galactose in 334 cells, representative aliquots of cells were collected at the time points indicated and then lysed, and HOXA5 protein amount was estimated by Western blot assay with an affinity-purified polyclonal HOXA5 antibody (custom-made). (B) HOXA5 expression slows the growth of S. cerevisiae. Control and HOXA5-expressing S. cerevisiae cells were grown for 3 days in synthetic media minus URA plus 2% galactose. OD measurements were taken at different intervals during the course of 3 days. (C) Expression of HOXA5 in S. cerevisiae results in formation of petite colonies.
At established optimum induction conditions, we performed a growth curve experiment to determine whether these yeast cells would mimic the effect of HOXA5 expression in MCF-7 cells (i.e., show signs of hindered growth). This was the case as we observed a rapid decrease in cell growth, as measured by cell density, over a period of 36 hours (Figure 1B). To further confirm this finding, HOXA5 was induced in yeast cells, which exhibited a petite phenotype (Figure 1C). The change in morphology was observed in several yeast strains used in this study. Thus, the expression of HOXA5 in yeast cells appears to interfere with cell cycle progression in a way that is phenotypically similar to that observed in breast cancer cell lines.
A petite phenotype for yeast cells is thought to be primarily due to loss of normally functioning mitochondria [17]. Thus, before performing gene expression studies, we wanted to confirm that the expression of HOXA5 in yeast cells did not disrupt mitochondrial activity. To address this question, we expressed HOXA5 under the control of a constitutive alcohol dehydrogenase promoter in strain Y190. Following plating onto a synthetic media minus URA and with glycerol as the sole source of carbon, we observed no difference in the colony morphology of cells expressing HOXA5 vs control (data not shown). This indicated that the effects of HOXA5 protein on cell size and growth rate were not directly due to compromised mitochondrial function.
Gene Expression Profiles of Strain 334 in the Presence and Absence of HOXA5
Following procurement of Affymetrix data, gene homology searches were performed to identify the mammalian counterpart of each of the upregulated yeast genes. Once potential candidate genes had been chosen, we next sought to verify whether there were any putative HOX core-binding sites (TAAT) in their promoter regions. The finding of putative HOX-binding sites within the promoter region of human gene homologs provided strong support for the significance and relevance of using yeast as a model system to study HOX gene functions.
From the panel of genes that were upregulated by HOXA5 within 2 hours of induction in yeast, MutL had an average fold activation of 4.58 (SE ± 0.125). The hMLH1 gene was one of the promoters that contained putative HOX-binding sites. Because of its importance in MMR and the known functional significance of MMR in carcinogenesis, the hMLH1 promoter was selected for detailed analysis.
HOXA5 Transactivates hMLH1 Promoter-Reporter Constructs
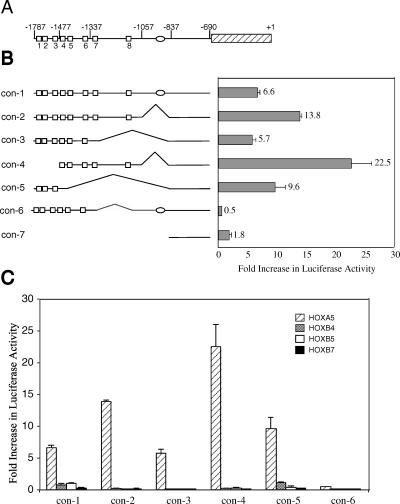
To assess whether HOXA5 transcriptionally modulates hMLH1 expression, cotransfection experiments were performed in breast cancer cell lines using effector and reporter plasmids. A schematic representation of different promoter deletion constructs indicating putative HOX-binding sites (open box), a possible p53-binding element (oval), and the proximal promoter (hatched box) is depicted in Figure 2A Our preliminary results showed that inclusion of the proximal and core promoter [13] into reporter vector constructs produced confounding results with unaltered high-Luc activity regardless of any upstream deletions. Thus, all promoter deletion constructs were prepared from a starting vector in which the proximal (basal) promoter had been excised. Following excision of these sites, the promoter-reporter construct (con-1) gave a 6.6 ± 0.4-fold increase in Luc activity in the presence of HOXA5. A similar strategy was used for the excision of a possible p53 cis element (oval) along with surrounding sequences. The absence of the p53 cis element (con-2) from con-1 resulted in a 13.8 ± 0.3-fold increase of Luc activity. Thus, either p53 or any other transcriptional factor that binds with the excised region represses promoter activity. When the two 3′ most putative HOX-binding sites (i.e., sites 7 and 8, Figure 2; con-3) were also excised, reporter activity dropped to 5.7 ± 0.6-fold activation. In contrast, excision of the p53 site and the first three 5′ HOX-binding sites (i.e., 1, 2, and 3, Figure 2; con-4) increased reporter activity to 22.5 ± 3.5-fold activation. However, excision of the p53 site and the five 3′ HOX-binding sites (i.e., 4, 5, 6, 7, and 8, Figure 2; con-5) gave a reporter activity of 9.6 ± 1.7-fold activation. An evaluation of these last three results appeared to indicate that the promoter sequence that contains the last two 3′ HOX-binding sites contributes the most to reporter activity in MCF-7 cells. To test whether this is the case, we excised sites 7 and 8 while maintaining the p53 site (con-6). This construct, in the presence of HOXA5, was completely repressed (0.5 ± 0.001-fold activation). Thus, all promoter constructs that lacked HOXA5-binding sites 7 and 8 had activation activity lower than those that included these sites. Furthermore, the inclusion of the p53 site in the absence of sites 7 and 8 (con-6) eliminated the activation activity of the first six sites (cf. con-3 with con-6, Figure 2). Finally, removal of all putative HOX cis elements (con-7) resulted in essentially no increase in fold activation (1.8 ± 0.4). Taken together, these results indicate that the eight HOX-binding sites modulate reporter activity in MCF-7 cells, with sites 7 and 8 contributing the most to the activation and apparently being capable of partially overcoming the repressor activity of the p53 region of the promoter. Similar results were obtained using SK-BR-3 and Hs 578T breast cancer cells (data not shown).
Figure 2.
HOXA5 transactivates hMLH1 promoter-reporter activity in MCF-7 cells. MCF-7 breast cells were transfected with a combination of 1 fig of one of the pGL2-Basic-hMLH1 promoter vector constructs plus 0.5 µg of pcDNA3-HOXA5 or 0.5 µg of pcDNA3-HOXB4, pcDNA3-HOXB5, pcDNA3-HOXB7, or pCR3.1 empty vector plus 5 ng of Renilla plasmid as transfection control. (A) Schematic representation of the 1.75-kb hMLH1 promoter fragment studied. Open boxes labeled 1 to 8 represent the relative positions of putative HOX cis elements; the open oval indicates the position of the p53-binding site; the proximal promoter is represented by a hatched rectangle; and +1 indicates the translational start site. (B) Left: hMLH1 promoter-reporter deletion constructs depicting the different HOX-binding sites (open boxes). Right: The corresponding bar graph, which indicates fold activation of Luc activity in the presence of HOXA5 in MCF-7 cells. (C) Effect of HOXB4, HOXB5, and HOXB7 on the reporter activity of different hMLH1 promoter-reporter constructs in MCF-7 cells.
In addition, we have demonstrated that the transactivation of the hMLH1 promoter-reporter construct is specific for HOXA5 and is not affected by other HOX proteins, as shown in Figure 2C. HOXB4, HOXB5, and HOXB7 showed little or no activation of Luc transcription compared with samples transfected with HOXA5. This indicates that transcriptional activation of the hMLH1 gene can occur through specific interaction with HOXA5 protein and not by other tested HOX proteins.
In Vivo Binding of HOXA5 to Its Cognate Sequence in the hMLH1 Promoter
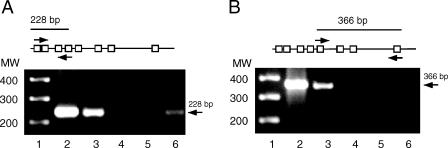
ChIP assays were used to assess whether HOXA5 binds in vivo to the hMLH1 promoter in MCF-7 cells overexpressing HOXA5. Two sets of PCR primers were designed for specific amplification of the first four or the last four HOX-binding sites in the hMLH1 promoter, respectively, as schematically shown in Figure 3. As shown in Figure 3, A and B (lane 3), the use of either primer pair in the PCR of ChIP DNA generated from anti-HOXA5 precipitations resulted in amplified products of 228 and 386 bp. The same amplification products were seen in positive control experiments. That is, hMLH1 promoter sequences were amplified from unprocessed chromatin and from the ChIP DNA generated from anti-acetylhistone H3 precipitations (Figure 3, A and B, lanes 2 and 6, respectively). In contrast, nonspecific antibody-mediated precipitations resulted in DNA templates that were not amplified (Figure 3, A and B, lane 4). In addition, no amplification was seen in the samples that were processed in the absence of precipitating antibodies (Figure 3, A and B, lane 5). These results indicate that HOXA5 binds directly or as part of a complex to the endogenous hMLH1 promoter in vivo. As ChIP analysis is dependent on the degree of shearing of chromosomal DNA, it is essential to perform additional PCR for a sequence distal to the promoter that does not contain any consensus HOX-binding sites. Based on PCR of the coding region of hMLH1, we did not observe any amplified products from the anti-HOXA5 immuoprecipitated sample, demonstrating the specificity of immunoprecipitation (data not shown).
Figure 3.
In vivo binding of HOXA5 protein to the hMLH 1 promoter sequence. (A) Top: Schematic representation of the portion of the hMLH1 promoter that contains putative HOX-binding sites represented as open boxes (see Figure 2). Small arrows above and below the promoter represent sense (ChIP S1: 5′-GCAAAGACTTTACTAACTCG-3′) and anti-sense (ChIP AS1: 5′-AATCTGAATCAGAGCATGG-3′) primer sites, respectively. Primers amplify a 228-bp fragment. Bottom: Agarose gel showing the ChIP S1/ChIP AS1-directed PCR product. (B) Top: Similar schematic presentation, as described above. Sense (ChIP S2:5′-AATTCAGGTCGCCTAACG-3′) and anti-sense (ChIPAS2: 5′-TGTAGGCCCTGAGTTGG-3′) priming sites are depicted with a PCR product of 366 bp. Lane 1, DNA ladder. PCR samples: lane 2, total input chromatin; lane 3, anti-HOXA5 precipitation; lane 4, nonspecific antibody precipitation; lane 5, no antibody; lane 6, anti-acetyl-histone H3 precipitation. Identical volumes from the final precipitate were used for PCR. Molecular weights indicated to the left of the gels are given in basepairs.
Overexpression of HOXA5 Enhances MMR Assay in MCF-7 Cells
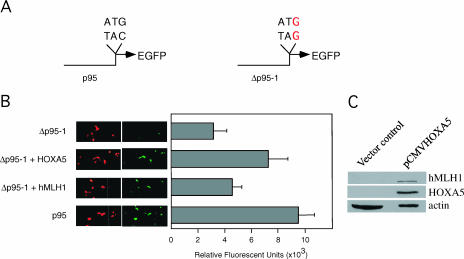
As HOXA5 could bind and transactivate the hMLH1 promoter, we wanted to determine if transient overexpression of HOXA5 in MCF-7 cells would increase the MMR capabilities of these breast cancer cells. To evaluate the in vivo functional role of HOXA5 in MMR, we used the method recently described by Lei et al. [14]. A vector encoding the red fluorescent protein, which served as transfection control, was included. A schematic representation of the native (p95-1) versus the mismatched (Δp95-1) portion of the vectors used in the assay is shown in Figure 4A Figure 4B shows the four transfections tested (Δp95-1, Δp95-1 + HOXA5, Δp95-1 + hMLH1, and p95-1), along with representative photomicrographs of red or green fluorescence and a corresponding bar graph of fluorescence with a scale of average relative fluorescence units (RFU). The positive transfection (p95-1) gave 9452 ± 1276 RFU. In contrast, Δp95-1 transfections alone averaged only 3124 ± 974 RFU. However, transfecting Δp95-1 with HOXA5 or hMLH1 resulted in 7218 ± 1479 and 4528 ± 747 RFU, respectively. No significant difference in red fluorescence intensity was observed across all transfected wells. The results from this assay provide evidence that HOXA5 induces MMR in MCF-7 cells. In addition, overexpression of HOXA5 in MCF-7 cells increases hMLH1 synthesis (Figure 4C). Taken together, the results indicate that HOXA5 is a transcriptional regulator of hMLH1 and provides an alternative mechanism to the loss of hMLH1 expression in some breast carcinomas.
Figure 4.
HOXA5 expression increases repair activity in MCF-7 cells. (A) Schematic representation of native (p 95-1) and mismatched (Δp95-1) regions of reporter plasmids used in the assay. (B) Cotransfections of reporter plasmids and effector plasmids (HOXA5 and hMLH1) in MCF-7 cells. MCF-7 cells were transfected with 0.25 fig of Δp95-1 and 0.5 µg of pCR3.1-HOXA5 or pCR3.1-hMLH1 cDNA. Transfections with p95-1 constitute positive controls. Plasmid pDsRed2-1 was used as internal control for transfection efficiencies. The graph represents relative fluorescent units. (C) Western blot analysis showing the activation of hMLH1 by HOXA5 in MCF-7 cells.
Discussion
Historically, HOX proteins have been considered as master transcriptional regulators of coordinated gene expression during development. HOX genes are essential for the proper control of anterior-posterior segmentation identity during development and are evolutionarily conserved over a broad range of metazoan species [18–20] (e.g., homologs have been identified in sea urchins, Drosophila, mice, and humans). However, in recent years, it has been shown that HOX proteins have important regulatory functions in normal adult tissues [21]. For example, Hoxa9, Hoxb9, and Hoxd9 have been shown to modulate adult mouse mammary gland development during pregnancy [22]. Dysregulation of HOX protein expression is now known to occur in many human cancers [21,23]. Examples include HOXC8 in prostate cancer [24], HOXD3 in lung cancer [25], and HOXA1, HOXB3, HOXB4, HOXB7, and HOXC6 in breast cancer [26–28]. Along these lines, we have previously demonstrated that HOXA5 can regulate both p53 and progesterone receptor expression in breast cancer [2,3]. As loss of HOXA5 expression is correlated to advanced breast carcinoma, we sought to decipher the possible multifaceted functions of HOXA5 in regulating cellular transformation. Although analyzing gene expression patterns in cancer cell lines overexpressing HOXA5 was an option, our interest was to use a homogenous system with little or no chromosomal variations. Based on the precedent of exploring humanized yeast systems [29], we used an inducible strategy in yeast to overexpress HOXA5 and to determine downstream effector genes.
To determine whether yeast could serve as a model system for studying HOX functions in eukaryotes, we did a protein BLAST search within the yeast genome database of the homeodomain region of HOX genes to first confirm that potential HOX-like proteins, along with their cis elements, are present in yeast. The findings showed that homeodomain motifs of yeast Yox1 (AVQIWFQNKR), Pho1 (ARVQVWFQNRR), and Pho2 (NVRIWFQNRR) are strikingly similar to the same region of Antennapedia (QIKIWFQNRR), a homeodomain protein from Drosophila. Moreover, the cis element (TCTAATCCC) that binds Drosophila bicoid gene appeared to be nearly identical to a potential Yox1 DNA-binding sequence (TCGAATCCC), as initially reported by Kaufmann [30], and fine mapping of the core-binding sequence [(T/C)AATA(C/A)] confirmed that report [16]. Although the phylogenetic distance between yeast and Homo sapiens is very large, yeast has historically been used as a general eukaryotic model system. For example, cell cycle-regulatory genes were initially identified in yeast, subsequent to their identification in mammalian cells [31]. In addition, yeast has been used as a model in which to identify human genes whose products function in DNA repair [32]. Furthermore, detailed genetic information is now available for yeast cells, which provides a well-characterized model in which to study the effector functions of HOX genes. Thus, the use of the yeast model as a screen for identifying the downstream effector genes of HOXA5 has been effective, and we anticipate that it will provide much useful information about potential, as yet unidentified, targets of HOXA5 regulation.
In this report, we focused on one of the genes that were identified using the yeast system, human homolog of bacterial MutL[4] or hMLH1, a component of the MMR system. The mammalian MMR system consists of multiple protein components, which detect and correct base mismatch abnormalities and insertion/deletion loops occurring within double-stranded DNA [4]. In addition to normal replication errors, environmental factors such as chemical toxins and ultraviolet radiation contribute to base modifications, which are also corrected by the MMR pathway [5]. The components of the MMR pathway (hMutLα and hMutLβ) are dimers composed of hMLH1 and its partners human postmeiotic segregation 2 (hPMS2) and hPMS1, respectively [4,31]. hMutLα is involved in the repair of mismatched basepairs [5], whereas hMutLβ participates in apoptosis signaling [32]. Thus, a fully functional MMR system is involved in maintaining cellular genetic integrity. In fact, an absence or a decrease in the expression levels of a complete set of MMR proteins results in replication errors, which contribute to genomic instability.
Analysis of the distal portion of the hMLH1 promoter identified eight putative HOX-binding (cis) elements. Systematic deletion of these potential HOXA5-binding sites, in the absence of the proximal promoter, affected the expression of Luc in an in vivo reporter assay. In all studies, Luc activity increased when promoter constructs containing HOX-binding sites were cotransfected with a HOXA5-encoding vector in MCF-7 breast cancer cells. Deletion of the region of the promoter containing a putative p53-binding site further increased reporter activation. In addition, the sequence context of HOX-binding sites affects the optimal activation of the reporter gene. That is, within the context of all eight sites, the first three 5′ HOX-binding sequences appear to be associated with some repressor activity, as their removal resulted in a large increase in reporter activity. However, when these first three 5′ sites were used alone, half of the maximal reporter activation returned. Nevertheless, removal of the last two 3′ HOX-binding sequences resulted in the largest decrease in reporter activity, indicating that one or both of these sites are essential for optimal activation process. When the last two 3′ HOXA5-binding sites were absent and the p53 region was present, Luc expression was abolished. This finding suggests that several HOXA5-related regulatory mechanisms could affect hMLH1 expression. For example, repressor activity at or near the p53-binding site could attenuate hMLH1 expression in circumstances of diminished HOXA5 expression, or in loss of HOXA5 expression, or through mutation, epigenetic silencing, or blocking of these two 3′ most HOXA5 cis elements. The possible repression by p53 is in agreement with recent reports [33–35] indicating that p53 can be a repressor of gene activity in certain cases where its two cis element half sites are inversions of each other. The p53 site found in the hMLH1 promoter has a topology that partially matches this description. Importantly, other HOX family members (HOXB4, HOXB5, and HOXB7) had little or no affect on reporter activity, which means that the observed increases in Luc activity in vivo in MCF-7 breast cancer cells were specific to binding and activation by HOXA5. These data confirmed the utility of the yeast system and helped identify hMLH1 as a potential target for HOXA5 regulation.
Previous reports aimed at characterizing the hMLH1 promoter have focused on the transcriptional regulation of basal activity or epigenetic regulation through methylation of CpG islands [36–39]. Kane et al. [36] initially described that methylation within the -670 to -67 region, relative to the transcription start site, gave rise to the loss of hMLH1 expression. This report also mentioned the presence of a p53 cis element, but it did not note its exact location. A refined mapping study narrowed a region of methylation, linked to relevant loss of expression apparently, to the -248 to -178 region [37]. However, none of these analyses has described the inactivation of the HOXA5 region of the promoter (i.e., roughly from -1690 to -1090) by epigenetic processes. The core or the proximal promoter for hMLH1 has been reported to be within the first 300 bp upstream of the transcription start site [10,37,38]. In addition, a -577 promoter-Luc construct generated similar levels of Luc activity as a -914 construct [39]. Both results are consistent with our analyses of the characterization of the hMLH1 promoter. Moreover, it was also reported that truncation of the promoter from -1781 to -914 resulted in loss of approximately one third of the activation of Luc [39], which supports the conclusion that the region of the promoter studied by the authors is important for full activation of hMLH1. It is possible that regulation of hMLH1 expression by HOXA5 is cell type-specific [39]. At this time, it is not yet fully understood under what circumstances HOXA5 exerts its possible enhancing affects on hMLH1 expression, and there have been no reports as to the developmental regulation of hMLH1 expression with respect to specific transcriptional factors.
In addition to transient cotransfection Luc reporter experiments, which are used to identify putative target promoters, an evaluation of putative endogenous regulation of gene activity by a transcriptional factor such as HOXA5 can be obtained through the use of ChIP assay. Of course, the extent of shearing of chromosomal DNA may have resulted in both primer regions being precipitated together. Thus, appropriate controls, including PCR within the coding region, were performed to verify the specificity of immunoprecipitation (i.e., no PCR products were observed when distal portions of the hMLH1 gene were targeted for amplification). Thus, our ChIP data demonstrated that, within the intact nucleus of MCF-7 cells, HOXA5 can specifically interact with regions of the hMLH1 promoter that contain HOXA5-binding sites. Finally, we sought to determine if binding of HOXA5 to the hMLH1 promoter could affect MMR activity in MCF-7 cells. Recently, Lei et al. [14] described an elegant assay for assessing MMR activity in vivo. Using their approach, we were able to show that HOXA5 expression increased the MMR capabilities of MCF-7 cells in vivo. Furthermore, we demonstrated that overexpression of HOXA5 in MCF-7 cells increased hMLH1 protein levels, thus underscoring the importance of HOXA5 in regulating MMR-related activities.
Lack of MMR activity can initiate carcinogenesis by augmenting genomic instability. One cause of this instability is loss of MMR proteins and, thus, reduced DNA damage signaling, which contributes to reduced apoptosis [4,32]. For example, in HNPCC, germline mutations in hMSH2 and hMLH1 have been observed in 98% of cases studied [6]. Moreover, loss or diminished function of hMLH 1 in breast cancer cell lines has been demonstrated to be due to missense mutation [7], which brings about microsatellite instability. Furthermore, truncated hMLH1 protein can be detected in a subset of patients with lobular carcinoma in situ of the breast [8]. Other studies have shown that hMLH1 gene expression can be silenced by promoter hypermethylation in approximately 28% to 38% of sporadic breast cancer cases [9], again resulting in microsatellite instability. However, in approximately 15% of the cases, the loss of hMLH1 expression remains unexplained. Thus, alternative mechanisms contributing to the loss of hMLH1 function in breast cancer exist. The evidence presented here indicates that HOXA5 can contribute to the regulation of hMLH1 expression in breast cancer cells and, by increasing or maintaining normal expression levels, may abrogate the deleterious effects that loss of MMR has on genomic instability. The present study describes the use of a yeast system for the identification of HOXA5 downstream effector targets. This approach has provided a list of potential genes that are regulated by HOXA5. This screen identified the MMR component hMLH1 as being potentially modulated by HOXA5. The results presented here indicate that HOXA5 may regulate hMLH1 in human breast cancer cells and thus have an indirect effect on the integrity of genetic information.
Acknowledgement
We thank James Kimble for technical assistance.
Footnotes
This work was supported by National Institutes of Health grant 1RO1 CA097226 to V.R.
References
- 1.Larochelle C, Tremblay M, Bernier D, Aubin J, Jeannotte L. Multiple cis-acting regulatory regions are required for restricted spatiotemporal Hoxa5 gene expression. Dev Dyn. 1999;214:127–140. doi: 10.1002/(SICI)1097-0177(199902)214:2<127::AID-AJA3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 3.Raman V, Tamori A, Vali M, Zeller K, Korz D, Sukumar S. HOXA5 regulates expression of the progesterone receptor. J Biol Chem. 2000;275:26551–26555. doi: 10.1074/jbc.C000324200. [DOI] [PubMed] [Google Scholar]
- 4.Li GM. DNA mismatch repair and cancer. Front Biosci. 2003;8:d997–d1017. doi: 10.2741/1121. [DOI] [PubMed] [Google Scholar]
- 5.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amsterdam) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Mitchel RJ, Farrington SM, Dunlop MG, Campbell H. Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am J Epidemiol. 2002;156:885–902. doi: 10.1093/aje/kwf139. [DOI] [PubMed] [Google Scholar]
- 7.Seitz S, Wassmuth P, Plaschke J, Schackert HK, Karsten U, Santibanez-Koref MF, Schlag PM, Scherneck S. Identification of microsatellite instability and mismatch repair gene mutations in breast cancer cell lines. Genes Chromosomes Cancer. 2003;37:29–35. doi: 10.1002/gcc.10196. [DOI] [PubMed] [Google Scholar]
- 8.Stone JG, Coleman G, Gusterson B, Marossy A, Lakhani SR, Ward A, Nash A, McKinna A, A'Hern R, Stratton MR, et al. Contribution of germline MLH1 and MSH2 mutations to lobular carcinoma in situ of the breast. Cancer Lett. 2001;167:171–174. doi: 10.1016/s0304-3835(01)00448-7. [DOI] [PubMed] [Google Scholar]
- 9.Murata H, Khattar NH, Gu L, Li GM. Roles of mismatch repair proteins hMSH2 and hMLH1 in the development of sporadic breast cancer. Cancer Lett. 2005;223:143–150. doi: 10.1016/j.canlet.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 12.Odenwald WF, Garbern J, Arnheiter H, Tournier-Lasserve E, Lazzarini RA. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989;3:158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- 13.Ito E, Yanagisawa Y, Iwahashi Y, Suzuki Y, Nagasaki H, Akiyama Y, Sugano S, Yuasa Y, Maruyama K. A core promoter and a frequent single-nucleotide polymorphism of the mismatch repair gene hMLH1. Biochem Biophys Res Commun. 1999;256:488–494. doi: 10.1006/bbrc.1999.0368. [DOI] [PubMed] [Google Scholar]
- 14.Lei X, Zhu Y, Tomkinson A, Sun L. Measurement of DNA mismatch repair activity in live cells. Nucleic Acids Res. 2004;32:e100. doi: 10.1093/nar/gnh098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong H, Vershon AK. The yeast homeodomain protein MATalpha2 shows extended DNA binding specificity in complex with Mcm1. J Biol Chem. 1997;272:8402–8409. doi: 10.1074/jbc.272.13.8402. [DOI] [PubMed] [Google Scholar]
- 16.Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth-Ben Arie Z, Altboum Z, Berdicevsky I, Segal E. Isolation of a petite mutant from a histidine auxotroph of Candida albicans and its characterization. Mycopathologia. 1998;141:127–135. doi: 10.1023/a:1006988119891. [DOI] [PubMed] [Google Scholar]
- 18.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 19.Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT, Pendleton JW. Evolution of HOX genes. Annu Rev Genet. 1994;28:423–442. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 20.Kappen C, Schughart K, Ruddle FH. Early evolutionary origin of major homeodomain sequence classes. Genomics. 1993;18:54–70. doi: 10.1006/geno.1993.1426. [DOI] [PubMed] [Google Scholar]
- 21.Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000;2:158–169. doi: 10.1186/bcr49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cillo C, Faiella A, Cantile M, Boncinelli E. Homeobox genes and cancer. Exp Cell Res. 1999;248:1–9. doi: 10.1006/excr.1999.4451. [DOI] [PubMed] [Google Scholar]
- 24.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 25.Hamada J-I, Omatsu T, Okada F, Furuuchi K, Okubo Y, Takahashi Y, Tada M, Miyazaki YJ, Taniguchi Y, Shirato H, et al. Overexpression of homeobox gene HOXD3 induces coordinate expression of metastasis-related genes in human lung cancer cells. Int J Cancer. 2001;93:516–525. doi: 10.1002/ijc.1357. [DOI] [PubMed] [Google Scholar]
- 26.Chario A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Commun. 1996;222:292–297. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- 27.Bodey B, Bodey B, Jr, Siegel S, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in breast carcinomas. Anticancer Res. 2000;20:3281–3286. [PubMed] [Google Scholar]
- 28.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringer M, Sauter G, Monni O, Elkahloun A, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 29.Nigro JM, Sikorski R, Reed SI, Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1357–1365. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufmann E. In vitro binding to the leucine tRNA gene identifies a novel yeast homeobox gene. Chromosoma. 1993;102:174–179. doi: 10.1007/BF00387732. [DOI] [PubMed] [Google Scholar]
- 31.Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol Cell Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett CB, Lewis LK, Karthikeyan G, Lobachev K, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Khalpey Z, Cascalho M. Cellular physiology of mismatch repair. Curr Pharm Des. 2004;10:4121–4126. doi: 10.2174/1381612043382468. [DOI] [PubMed] [Google Scholar]
- 34.Yanamadala S, Ljungman M. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol Cancer Res. 2003;1:747–754. [PubMed] [Google Scholar]
- 35.Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem. 2001;276:27716–27720. doi: 10.1074/jbc.C100121200. [DOI] [PubMed] [Google Scholar]
- 36.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 37.Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 38.Quaresima B, Faniello MC, Baudi F, Cuda G, Grandinetti C, Tassone P, Costanzo F, Venuta S. Transcriptional regulation of the mismatch repair gene hMLH1. Gene. 2001;275:261–265. doi: 10.1016/s0378-1119(01)00656-4. [DOI] [PubMed] [Google Scholar]
- 39.Arita M, Zhong X, Min Z, Hemmi H, Shimatake H. Multiple sites required for expression in 5′-flanking region of the hMLH1 gene. Gene. 2003;306:57–65. doi: 10.1016/s0378-1119(03)00385-8. [DOI] [PubMed] [Google Scholar]