Abstract
The aim of the current study was to evaluate the expression of ADAM15 disintegrin (ADAM15) in a broad spectrum of human tumors. The transcript for ADAM15 was found to be highly upregulated in a variety of tumor cDNA expression arrays. ADAM15 protein expression was examined in tissue microarrays (TMAs) consisting of 638 tissue cores. TMA analysis revealed that ADAM15 protein was significantly increased in multiple types of adenocarcinoma, specifically in prostate and breast cancer specimens. Statistical association was observed with disease progression within clinical parameters of predictive outcome for both prostate and breast cancers, pertaining to Gleason sum and angioinvasion, respectively. In this report, we also present data from a cDNA microarray of prostate cancer (PCa), where we compared transfected LNCaP cells that overexpress ADAM15 to vector control cells. In these experiments, we found that ADAM15 expression was associated with the induction of specific proteases and protease inhibitors, particularly tissue inhibitor of metalloproteinase 2, as validated in a separate PCa TMA. These results suggest that ADAM15 is generally overexpressed in adenocarcinoma and is highly associated with metastatic progression of prostate and breast cancers.
Keywords: ADAM15 disintegrin, breast cancer, cDNA microarray, prostate cancer, tissue microarray
Abbreviations: ADAM15, ADAM15 disintegrin; ECM, extracellular matrix; MMP, matrix metalloproteinase; PCa, prostate cancer; ER, estrogen receptor; PR, progesterone receptor; TIMP2, tissue inhibitor of metalloproteinase 2; TMA, tissue microarray; CI, confidence interval; RR, relative risk; SD, standard deviation; SE, standard error
Introduction
Due to advances in tumor detection, early diagnosis of human cancer has improved dramatically. Unfortunately, despite these improvements, some patients who are diagnosed early in their disease may still have micrometastasis, which leads to secondary tumor growth. Thus, the identification of early-stage aggressive tumors that are at high risk for recurrence remains a priority.
The progression of solid tumors to metastatic diseases includes essential steps that support the detachment of cells from the surrounding extracellular matrix (ECM), tumor cell survival, and tissue invasion [1]. One family of proteins supporting malignant progression is zinc-binding matrix metalloproteinases (MMPs). The role of MMPs in metastatic progression has been extensively studied in tumor biology. Specific MMPs have been implicated in the induction of tumor cell migration, invasion, and angiogenesis. These enzymes contain an active metalloproteinase domain that catalyzes the degradation of ECM proteins, such as collagen, laminin, and fibronectin [2]. This activity is believed to mediate the disruption of basement membrane integrity and to allow cancer cells to invade the surrounding tissues and vasculature.
Another aspect of proteolytic processing during tumor progression affects a diverse set of physiologic cell surface proteins, such as membrane-anchored growth factors and their receptors, ectoenzymes, and many cell adhesion molecules, including cadherins [3]. An intriguing family of membrane metalloproteinases has been identified in the extracellular processing of these cell surface molecules. This family of membrane-associated disintegrins, referred to as ADAMs (A Disintegrin that contains A Metalloproteinase), contains modular metalloproteinase motifs, an integrin-binding domain (disintegrin), and a cysteine-rich epidermal growth factor-like domain in the extracellular region of the molecule. One of the best characterized ADAMs is tumor necrosis factor convertase (ADAM 17), which processes pro-TNF-α, TNF receptors, interleukin-6 receptor, and amphiregulin [4,5]. ADAMs have been implicated in many biologic processes, including oocyte fertilization, neurogenesis, myogenesis, and growth factor shedding [6,7]. Additionally, this family of disintegrin metalloproteinases has also been shown to be involved in cancer progression. ADAM10 is overexpressed in pheochromocytomas and neuroblastomas, whereas ADAM12 is overexpressed in both breast and colon cancers [8,9]. ADAM10, ADAM12, and ADAM17 are implicated in gastrointestinal carcinoma [10]. The multiple domains of these proteinases impart several physiological features, including ECM degradation and shedding of transmembrane growth factors. ADAM9, ADAM10, ADAM12, and ADAM 17 have been shown to cleave the epidermal growth factor receptor (EGFR) ligand, heparin-binding (EGF), leading to transactivation of EGFR. This process has been implicated in many physiological responses, such as cell survival, proliferation, and migration, further supporting their role in neoplastic progression [11].
One understudied member of the ADAM family in human cancer is ADAM15 disintegrin (ADAM15), which is located on chromosome 1 at 1q21.3. This region is known to be amplified in many cancers, including metastatic prostate cancer (PCa), breast cancer, and melanoma [12,13]. Specifically, the region of chromosome 1 from 1q21 to 1q23 is increased in several types of adenocarcinoma and sarcomas, and thus appears to be a common aberration in many human cancers. Reports show that amplification of this region is higher in advanced metastatic cancers compared to tissues isolated from primary diseases [14–16].
ADAM 15 is thought to serve a dual function in metastasis by detaching cells from the ECM through its disintegrin domain and by degrading the ECM through the metalloproteinase domain. ADAM15 has been shown to degrade collagens I and IV and to cleave the inflammatory cytokine CD23 [17,18]. ADAM 15 has also been shown to influence ECM remodeling within rheumatoid synovial tissues and in atherosclerosis [19,20]. Additionally, ADAM15 is believed to interact with integrins αvβ3, α5β1, and α9β1 through its disintegrin domain. Uniquely, ADAM15 is the only family member to encode an RGD (Arg-Gly-Asp) binding motif, suggesting an important role in cell attachment and invasion [6,7]. In fact, ADAM15 induces glomerular mesangial cell migration within the kidney [7] and transactivates EGFR by cleaving the EGFR ligands amphiregulin and TGF-α in bladder cancer cell lines [21].
Knowledge concerning ADAM15 expression in human cancer is limited. We initiated the current study to compile the first comprehensive expression profile of ADAM15 by using multitumor tissue microarray (TMA) technology. Our preliminary findings demonstrated a significantly increased expression of ADAM15 in multiple adenocarcinomas. A thorough analysis of ADAM15 staining in regard to PCa and breast cancer, using TMA staining, revealed that the highest levels of expression correlated with advanced metastatic diseases, and a clear association with clinical parameters was determined.
Materials and Methods
Study Population and Tissue Collection
Formalin-fixed paraffin-embedded tissue blocks with tumor samples were previously set up for multitumor TMA. PCa and breast cancer TMAs were identified as part of the Institutional Review Board-approved project from a tissue bank.
Prostate tissue samples were taken from the radical prostatectomy series and the Rapid Autopsy Program [22] at the University of Michigan Prostate Cancer Specialized Program of Research Excellence (SPORE) Tissue Core (Ann Arbor, MI). Clinically localized PCa cases were taken from a cohort of men who underwent radical retropubic prostatectomy as monotherapy for clinically localized PCa between January 1995 and December 2001. Tumors were staged using the tumor-node-metastasis system [23] and were graded according to the system originally described by Gleason [24,25]. Snap-frozen prostate samples used for expression analysis were all evaluated histologically by a study pathologist (M.A.R.). All samples were trimmed to ensure that >95% of the sample represented the desired lesion. Breast cancer tissue samples were selected by a dedicated pathologist (C.G.K.) and categorized according to histology (i.e., carcinoma in situ, invasive breast cancer, or metastatic breast cancer).
cDNA Expression Profiles in PCa cDNA
RNA was isolated from 55 prostate tissue samples. Construction of cDNA microarrays was described in detail elsewhere [28]. In brief, plasmid templates for a maximum of 20,000 transcripts were isolated from bacterial clones, and inserts were amplified by polymerase chain reaction (PCR). Purified PCR fragments were printed on glass slides and cross-linked with DNA targets. cDNA generated from 55 PCa samples included 34 localized PCa and 21 metastatic tissue samples. These cDNA were labeled with distinguishable fluorescent dye and hybridized to a cDNA microarray. The cDNA microarray was analyzed using a scanner, and fluorescence ratios were determined for each gene. Color intensities were converted into ratios of gene expression. These ratios were inputted into a database for analysis.
TMAs
As preparation for the construction of the TMAs, all glass slides were examined to identify areas of benign or neoplastic lesions. To optimize transfer to the arrays, target areas were encircled on a glass slide template. TMAs were assembled using manual tissue array (Beecher Instruments, Silver Spring, MD), as previously described [26,27]. Tissue cores from circled areas were targeted for transfer to recipient array blocks. Several replicate tissue cores were sampled from each of the selected tissue types. TMA cores 0.6 mm in diameter were each spaced 0.8 mm from core center to core center. After construction, 4-µm sections were cut, and hematoxylin and eosin staining was performed on the initial slide to verify the histologic diagnosis. TMA images were acquired using the BLISS Imaging System (Bacus Laboratories, Lombard, IL). ADAM15 protein expression was evaluated in a blinded manner. All images were scored for ADAM 15 protein expression intensity by dedicated pathologists (M.A.R. and C.G.K.).
The tissue specimens on this array were derived from 22 different types of neoplastic tissue (i.e., lung, breast, and colon cancers). Thus, this array represents a unique source for evaluating and comparing different tumor types, under the same conditions, in a very efficient manner. The array consisted of tissue samples of 188 patients. The array was primarily set up with cores taken only from neoplastic areas. With several sections having been cut for experiments, 40 samples of the stained array were found to be benign. One hundred forty-eight samples were confirmed as neoplastic.
PCa TMAs. Two TMAs containing tissues from the prostatectomy series (benign versus localized disease) and from hormone-refractory PCa (benign versus localized versus metastatic) from the Rapid Autopsy Program (University of Michigan, SPORE) were constructed for this study. All TMA samples were assigned a diagnosis: benign, atrophic, and high-grade prostatic intraepithelial neoplasia, and PCa. One hundred sixty-four benign tissue cores, 85 localized PCa, and 82 tissue cores of far advanced metastatic diseases were suitable for analysis. Of the cases with localized PCa, the average age at surgery was 61.9 years (minimum = 47 years, maximum = 76 years). Eleven of 29 had a Gleason score of 5 or 6; the other cases had Gleason scores ≥7. Mean preoperative prostate-specific antigen (PSA) was 15 ng/ml (2.6–85.7 ng/ml). PSA failure as endpoint was defined as two consecutive PSA rises after having reached the nadir of <0.01 ng/ml after surgery. Mean follow-up time was 1354 days (median = 1669 days, minimum = 66 days, maximum = 2332 days).
Breast cancer TMA. The total number of patients represented on the TMA was 119. The following are the categories according to tissue type: stroma (n = 80), normal (n = 37), carcinoma in situ (ductal or lobular, n = 17), invasive carcinoma (ductal or lobular, n = 66), and metastatic carcinoma (n = 12). The total number of cases for evaluation after calculating the mean staining intensity for each tissue category was 212. Of the breast cancer cases, the average age at the time of diagnosis was 53.9 years (minimum = 29.9 years, maximum = 83.3 years). The mean tumor size was 2.6 cm (minimum = 1.0 cm, maximum = 8.0 cm). The mean number of total nodes removed was 15.7 (maximum = 29); of those, the mean number of positive nodes was 3.5 (minimum = 0, maximum = 25). Receptor status was available for the majority of cases. In this TMA, estrogen receptor (ER)-negative patients numbered 22, and the ER-positive patients, numbered 43. Additionally, 31 cases were progesterone receptor (PR)-negative and 33 cases were PR-positive. HER-2/neu was not overexpressed in 46 cases and was overexpressed only in 14 of these cases. Twenty of 112 cases had angioinvasion. The mean follow-up time was 1213 days (median = 719 days, minimum = 29 days, maximum = 7217 days). Of patients with known status on follow-up (n = 92), 55 had no evidence of disease, 25 were alive with recurrence, 6 had died due to breast cancer, and 6 died of other causes.
Immunohistochemistry (IHC) of ADAM15 and tissue inhibitor of metalloproteinase 2 (TIMP2)
Standard biotin-avidin complex IHC was performed to evaluate ADAM15 protein expression using a polyclonal anti-ADAM15 antibody (no. AB19035; Chemicon, Temecula, CA). Appropriate horseradish peroxidase-conjugated secondary antibody was obtained from BioRad (Hercules, CA). The antibody received microwave pretreatment (30 minutes at 100°C in 10 mM citrate buffer) for antigen retrieval. The reaction time for primary antibody incubation was 120 minutes at ambient temperature. This maybe needed for human samples, as this antibody was raised against the mouse peptide and was 85% homologous with human antibodies. The chromogen 3,3-diaminobenzidine tetrahydrochloride was used to visualize ADAM15-positive staining. Based on previous works, protein expression was scored as 1 = negative, 2 = weak, 3 = moderate, and 4 = strong [29,30]. Standard IHC was equally performed for the evaluation of TIMP2 protein expression using a monoclonal anti-TIMP2 antibody (no. MAB3310; Chemicon). Visualization of positive staining for TIMP2 was performed using the chromogen 3,3-diamino-benzidine tetrahydrochloride, and protein expression was scored the same as for ADAM 15 staining, as described here.
Cloning and Construction of the ADAM15-Tagged Vector
The cDNA for human ADAM 15 and the pCDNA3 vector containing a hemagglutinin (HA) tag were kind gifts of Carl Blobel and Gabrielle Nunez, respectively. The cDNA plasmid containing the hADAM15 sequence was used as template for PCR reactions. DNA primers that contained BamHI and XhoI sequences were used to create the PCR product BamHI/ADAM15/XhoI (5′ to 3′). This sequence did not contain a stop codon [forward “primer A (BamHI)” sequence: AATACGACTCACTATAGGGAGACCC; reverse “primer B (XhoI)” sequence: CTCGAGGAGGTAGAGCGAGGACACTGTC]. This BamHI/ADAM 15/XhoI DNA was then cloned into a shuttle vector from the TA Cloning Kit system (Invitrogen, Carlsbad, CA) prior to ligation into the pCDNA3 vector that contained an HA tag (3′). Next, GFP sequence was generated as a PCR product that contained XbaI sites at both the 5 prime and 3 prime ends using the pGFPN-1 vector as template for this reaction (InVitrogen). The GFP DNA sequence was inserted in frame into the pCDNA vector at the 3 prime end of the hemaglutanin tagged ADAM 15 (ending with a stop codon), as a second step cloning. This plasmid DNA was transformed and purified from bacteria. Purified plasmid DNA encoding the ADAM15-HA-GFP (designated LNCaP M142) was finally sequenced for verification.
Cell Culture and Transfection of ADAM15 (M142 cDNA) into LNCaP Cells
The PCa cell line LNCaP was obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained in RPMI1640 with 8% fetal bovine serum (FBS), 0.1% glutamine, and 0.1% penicillin/streptomycin (BioWhittaker, Walkersville, MD). LNCaP cells were grown to 70% confluence in serum containing a medium for transfection. TFX50 reagent (Promega, Madison, WI) was used for transfection as described by the manufacturer, with the following modifications: 15 µg of M142 DNA and 45 µl of TFX50 reagent were added to 5 ml of serum-free media for each dish of cells (grown in 100-mm culture dishes). This mixture was incubated for 4 hours at 37°C in a cell culture incubator. Following this incubation of TFX50 and DNA under serum-free conditions, 10 ml of 15% FBS medium was then added to the dishes, and transfection in the cell incubator continued for a total of 72 hours. On completion of this procedure, the cells were allowed to recover in the normal growth medium prior to selection into media containing 500 µg/ml G418. Stable cell lines were selected from pools of G418-resistant LNCaP cells. Additionally, a final round of selection using FACS sorting was used to select cells with high fluorescence expression of the GFP tag. Overexpression of ADAM15-tagged protein was evaluated by Western blot analysis using a polyclonal antibody (no. 19036; Chemicon, Temecula, CA). Results showed a high expression of exogenous ADAM 15 protein and no changes in growth rate, compared to vector controls (data not shown).
cDNA Microarray Analysis of PCa LNCap M142 Cells
RNA was prepared from the vector control LNCap cells and from the ADAM 15 overexpressing cell line, LNCaP M142. Because ADAM 15 is endogenously expressed in LNCaP cells, the vector control cell line was used as baseline for expression in comparison to the LNCap M142 cell line. RNA from both cell lines were prepared from log-phase cultures growing in 8% serum-containing culture medium. Microarray analysis was performed at the University of Ulm (Ulm, Germany). This microarray procedure has been previously reported [30].
Statistical Analyses
Statistical analysis was performed using SPSS (SPSS, Chicago, IL). Primary analysis of cDNA expression data was performed with Genepix software and Cluster analysis (Molecular Devices Corp., Downingtown, PA), was performed with the program Cluster, as described earlier [28,30]. ADAM15 expression was statistically evaluated using mean score results from each case for each tissue type (i.e., localized PCa and hormone-refractory PCa). To test for significant differences in mean ADAM15 protein expression between all tissue types, Mann-Whitney U test was performed. Mean expression scores for all examined cases were presented in a graphic format using error bars with 95% confidence interval (CI). For clinically localized PCa samples, ADAM15 protein expression was evaluated for association with pathology parameters (i.e., tumor stage) using t-test and Mann-Whitney U test. Univariable Cox regression analysis was used to evaluate the risk of PSA failure following prostatectomy (postsurgical PSA progression >0.2 ng/ml) and the risk of local or distant recurrence in breast cancer patients. Multivariable analysis using Cox hazards regression, with a stepwise backward selection (Wald), was used to simultaneously evaluate clinical, pathological, and molecular parameters. P < .05 was considered statistically significant.
Results
ADAM15 Expression on Multitumor TMA
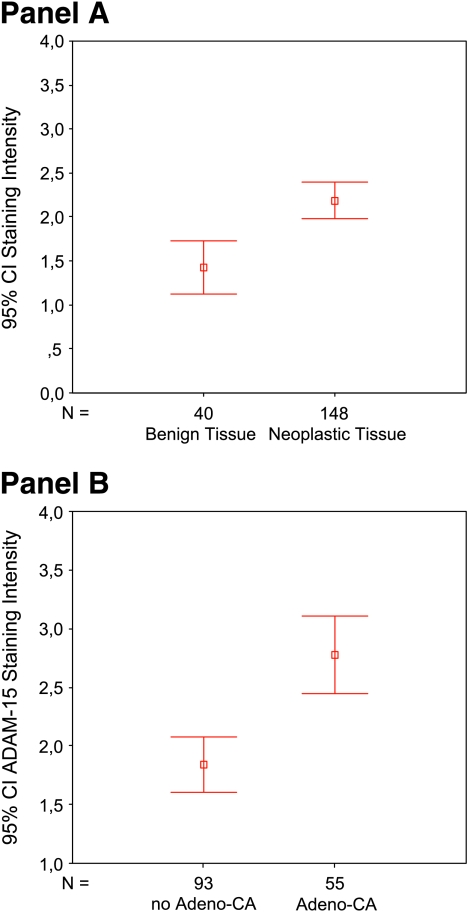
To explore the expression of ADAM15 in cancer, we used the Oncomine database created at the University of Michigan in the laboratory of A.M. Chinnaiyan [31]. In this database, we looked at the cDNA expression arrays of normal versus tumor cells. We found that ADAM15 was upregulated in many cancer arrays (see Figure W1). This was intriguing and led the authors to look at the protein expression of this ADAM protease in a broad spectrum of solid tumors. Thus, ADAM15 staining was evaluated on a multitumor TMA. Of 22 different tumor types represented in this TMA, the strongest staining was observed in tissue samples derived from ovary, breast, colon, and lung cancers. The lowest staining was observed in samples derived from pheochromocytoma, sarcoma, thyroid cancer, and testicular cancer (data not shown). All tumor tissues had higher expression levels compared to benign tissue samples (Figure 1A). The mean expression level for benign tissue was 1.43 [median = 1.00, standard error (SE) = 0.15, standard deviation (SD) = 0.96] compared to 2.19 (median = 2.00, SE = 0.10, SD = 1.26) for tumor tissue samples. The difference in expression levels was highly significant (Mann-Whitney U test, P < .001). Interestingly, when performing analysis between adenocarcinomas and non-adenocarcinomas (Figure 1B), we found that ADAM15 expression was significantly higher in all adenocarcinomas (mean = 1.78, median = 3.00, SE = 0.17, SD = 1.23) compared to other types of neoplastic tissue (mean = 1.84, median = 1.00, SE = 0.12, SD = 1.15) (Mann-Whitney U test, P < .001). Therefore, these data demonstrate that the highest staining occurred in cancers derived from epithelial lineages.
Figure 1.
(A) Comparison of benign tissue samples with neoplastic tissue samples of a multitumor TMA independent of organ site. There is a highly significant difference (Mann-Whitney U test, P < .001) in the expression values of ADAM15 between normal and neoplastic tissue samples, with an overexpression of ADAM15 in neoplastic tissue. (B) Comparison of non-adenocarcinomas and adenocarcinomas of different origins. There is a highly significant difference in the protein expression of ADAM 15 within these two categories. In adenocarcinomas, ADAM15 is significantly higher compared to other types of neoplastic tissue (P < .001).
ADAM15 was found to be overexpressed in adenocarcinomas compared to nonadenocarcinomas. Based on this observation, we then sought to further investigate ADAM15 expression in two of the most common hormone-regulated adenocarcinomas: PCa and breast cancer.
Expression in PCa cDNA Array
To explore the expression of ADAM15 in PCa progression, we first examined a PCa cDNA expression array derived form 103 benign and PCa specimens [28,30]. When we examined this array for dysregulated proteases, we found ADAM15 to be significantly overexpressed in PCa (Figure 2). Tissues were grouped under the following classifications: benign (normal adjacent prostate tissue), clinically localized PCa, and metastatic PCa. In relation to benign prostate tissues, localized PCa was significantly upregulated (Mann-Whitney U test, P < .005; localized PCa mean = 1.15, median = 1.14, SE = 0.03, SD = 0.24). In addition, there was significant upregulation in the expression of ADAM 15 in metastatic PCa compared to that in localized PCa cases (Mann-Whitney U test, P < .003; metastatic PCa mean = 1.37, median = 1.33, SE = 0.06, SD = 0.28).
Figure 2.
(A) ADAM15 is significantly overexpressed at the RNA level in neoplastic prostate tissue. Its expression is significantly higher in localized PCa than in benign prostatic tissue, and expression is highest in patients in the hormone-refractory metastatic group. (B) Three hundred twenty-one tissue samples were evaluated in this TMA. Staining intensity was significantly stronger in metastatic tissue samples than in localized PCa. Normal prostate tissue showed very low levels of ADAM15 expression (P < .001). (C) There is a strong association between ADAM15 expression in the TMA and Gleason sum categorized as <7 and ≥7 (P = .014).
ADAM15 Protein Expression in TMA of PCa
To validate the protein expression of ADAM15 in situ, an additional cohort of prostate samples from those used in the cDNA expression array analysis was used. A remarkable contrast in the levels of ADAM15 protein expression in malignant epithelia relative to adjacent benign epithelia was readily apparent. Many of the metastatic PCa samples demonstrated very strong ADAM15 expression. To assess ADAM15 protein expression in hundreds of prostate specimens (n = 313), we quantitated TMA data. From benign PCa (n = 164), localized PCa (n = 85), and metastatic PCa (n = 82), the mean ADAM15 protein staining intensity was determined to be 1.15 (median = 1.0, SE = 0.03, SD = 0.39), 2.16 (median = 2.0, SE = 0.11, SD = 1.00), and 2.68 (median = 3.00, SE = 0.1, SD = 0.94), respectively. Pairwise comparisons demonstrated dramatic differences in staining intensity between clinically localized PCa with respect to benign prostate tissues and metastatic hormone-refractory PCa samples (Mann-Whitney U test, P < .001). These data are graphically summarized using error bars representing the 95% CI for each tissue category (Figure 2B).
Staining of ADAM15 was then assessed for localized PCa and for potential associations with clinical and pathological parameters. The parameters, including age, predictor of PSA failure, preoperative PSA (and categorized PSA), gland weight, pathological tumor stage, Gleason score, extraprostatic extension, multifocality, tumor size, seminal vesicle involvement, and margin status, were examined for their ability to predict biochemical failure following surgical treatment. Using univariable Cox regression analysis of ADAM15 expression, we did not find a strong association with outcome [P = .26, relative risk (RR) = 1.43]. But when clinical parameters, including tumor size (P = .05, RR = 2.2), tumor stage (P = .05, RR = 1.7), seminal vesicle involvement (P = .006, RR = 2.6), and extraprostatic extension (P = .013, RR = 5.4), were evaluated, we saw an association with PSA failure.
We then looked for potential associations between clinical parameters and ADAM15 protein expression in advanced PCa disease. Interestingly, Gleason sum (categorized as <7 and ≥7) was significantly associated with ADAM15 expression (Mann-Whitney U test, P = .014). This is graphically demonstrated in Figure 2C. Extraprostatic extension and tumor stage also showed an association with ADAM15 expression (P < .001 and P = .004), whereas all other clinical parameters did not (P > .05).
The highest expression of ADAM15 in prostate tissue samples was observed in hormone-refractory metastatic PCa when analyzed at the RNA and protein levels. Our results also indicate that, in localized PCa cases, there is a strong association with Gleason sum, which is known to correlate with poor disease outcome.
ADAM15 Expression in TMA of Breast Cancer
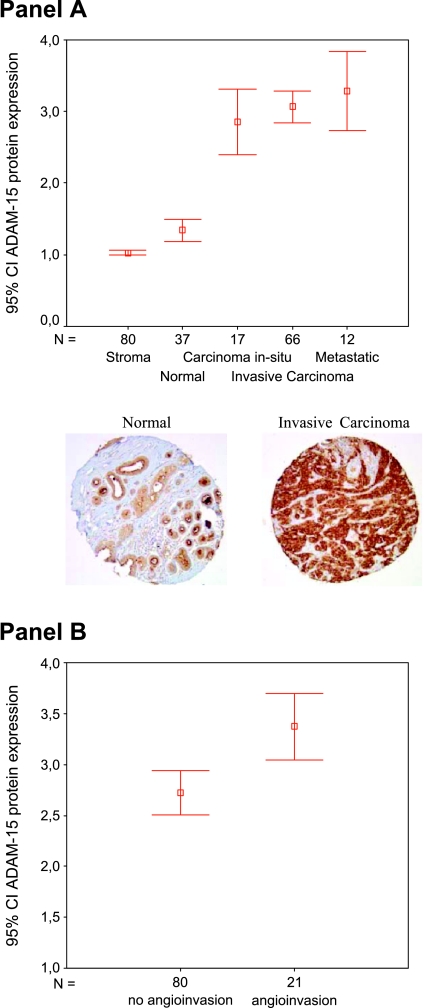
To validate the protein expression of ADAM15 in breast cancer, we analyzed a breast cancer TMA consisting of stroma (n = 80), normal breast tissue (n = 37), and neoplastic tissue [carcinoma in situ (n = 17), invasive carcinoma (n = 66), and metastatic deposit (n = 12)]. Staining intensities were compared between malignant epithelia and benign epithelia or stroma. Once quantified, the expression levels demonstrated the mean ADAM15 protein staining intensity for stroma (1.0; median = 1.0, SE = 0.02, SD = 0.16), normal tissue (1.34; median = 1.00, SE = 0.07, SD = 0.45), carcinoma in situ (2.85; median = 3.00, SE = 0.21, SD = 0.9), invasive breast cancer (3.07; median = 3.15, SE = 0.11, SD = 0.91), and metastatic disease (3.28; median = 3.5, SE = 0.25, SD = 0.87). Here, pairwise comparisons demonstrated highly significant differences in staining intensity between malignant and benign breast tissues (Mann-Whitney U test, P < .001). Although metastatic tissue samples had the highest mean expression levels, the difference between carcinoma in situ and invasive carcinoma was not significant (i.e., comparison of invasive ductal or lobular carcinoma versus metastases; P = .3). When the cutoff for staining intensity was set at 3.0, 42 of 66 (64%) cases of invasive carcinomas screened positive compared to 10 of 12 (83%) positive cases of metastatic samples. These results demonstrate that, although there is no statistically detectable difference in staining intensity between invasive samples and metastatic samples, the highest staining was observed in metastatic cases. Therefore, the trend demonstrates an increased expression in ADAM15, as the disease progresses from low-grade, to advanced, to metastatic disease. The data are graphically summarized for each tissue category in Figure 3A.
Figure 3.
(A) Protein expression of ADAM15 in different subtypes of breast cancer from breast TMA. A significantly higher expression of ADAM15 was seen in neoplastic samples (P < .001). Within different categories of malignant subtypes, the differences are not significant. The highest levels of ADAM15 were present in metastatic breast cancer tissue samples. Examples of two tissue cores (one from a normal tissue and the other from an invasive breast carcinoma) are depicted. (B) Breast cancer with angioinvasion has a significantly higher expression of ADAM15 protein than tissues without angioinvasion (P = .006).
To parallel our work with prostate array, we then assessed ADAM15 protein expression in breast tissues for association with clinical and pathological parameters and as predictor of disease recurrence. Using univariable Cox regression analysis of ADAM15 expression, we did not find a strong association with outcome, defined as local or distant recurrence (P = .517, RR = 1.075, CI = 0.865–1.335). Clinical parameters were as follows: tumor stage (P = .877, RR = 1.034, CI = 0.680–1.571), number of positive nodes (P = .686, RR = 1.01, CI = 0.963–1.059), ER (P = .416, RR = 0.775, CI = 0.419–1.432) or PR (P = .907, RR = 0.963, CI = 0.51–1.82), and HER-2/neu status (P = .549, RR = 0.913, CI = 0.679–1.229). In addition, the increase in ADAM15 staining showed no association with failure during follow-up. At the univariate level, histologic grade (P = .041, RR = 0.512, CI = 0.27–0.973), age at diagnosis (P < .000, RR = 1.044, CI = 1.021–1.068), and tumor size (P = .032, RR = 0.712, CI = 0.522–0.972) were associated with outcome. In our patient group, we found a strong association between angioinvasion and disease-free survival (P = .001, RR = 2.665, CI = 1.522–4.668), which remained significant in a multivariate analysis (backward stepwise selection; Wald, P = .002).
Additionally, we looked for potential associations between clinical parameters and ADAM15 protein expression in the cohort of neoplastic cases. There was no association between ADAM15 expression levels and parameters such as tumor stage, size of tumor, grade, number of positive lymph nodes, ER, PR, and HER-2/neu status (P > .05) compared within samples of advanced-stage cases. Interestingly, we found that angioinvasion was associated with high expression of ADAM15 protein (Mann-Whitney U test, P = .006). Therefore, patients with angioinvasive breast cancer had a significantly higher expression of ADAM15. This is graphically demonstrated in Figure 3B. The mean staining intensity of cases with angioinvasion was 3.37 (median = 3.5, SE = 0.16, SD = 0.72) compared to those without angioinvasion (mean = 2.72, median = 3.0, SE = 0.11, SD = 0.99).
ADAM15 is not expressed in stromal cells and is expressed at low levels in breast epithelial tissues. In normal glandular breast tissues, ADAM15 is expressed at the surface of the lumen in a highly polarized fashion. As the disease progresses to a more advanced stage, this polarized staining pattern is lost and ADAM15 expression becomes upregulated and diffused throughout the cytoplasm. Quantification of breast cancer TMA staining shows that ADAM15 protein expression is highest in cases of invasive breast cancer and metastatic breast cancer.
Evaluating the Expression of Proteases and Protease Inhibitors in the Overexpression of ADAM15 by cDNA Microarray
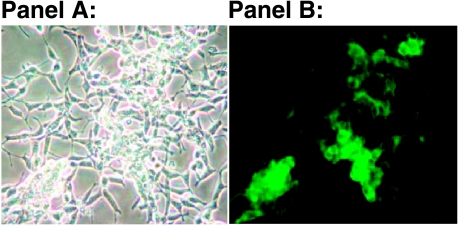
To determine the gene expression profile of ADAM15 overexpression in a PCa system, we created a stable LNCaP PCa cell line expressing a tagged ADAM15 protein (M142, described here). Figure 4, Panel B shows the protein expression of the ADAM15/GFP fusion protein in the LNCaP M142 cells under fluorescence microscopy. Figure 4 Panel A, shows the morphology of these same cells under bright field. By western analysis, we detected the exogenous ADAM15 protein in less than 2ug of total cell lysate per lane, (data not shown). This analysis proved that the ADAM15 fusion protein was very highly expressed in these cells in culture. Once a stable, highly expressing cell line of LNCaP M142 was established, RNA was isolated, and gene expression profile was generated on the same cDNA chip as the PCa profiles, as described in Materials and Methods section. In this experiment, we compared vector control to M142 cells in the presence of growth factors. Results showed that ADAM15 was 1.5-fold overexpressed in stable transfectants compared to vector control. Another 376 genes or expressed sequence tags were at least 1.5-fold overexpressed compared to vector control. Two hundred ninety-six genes were at least 0.5-fold down regulated. It has been previously published and is an accepted theme in the protease field that a change in the expression level of a single protease will then alter the expression of its inhibitor(s), and this change can additionally influence the expression of other proteases expressed by that cell [32,33]. Therefore, we focus on and report here the results of our array data, which show significant changes in the expression levels of protease inhibitors and other proteases affected specifically by the overexpression of ADAM15 in our LNCaP PCa model. A selection of these upregulated and dysregulated protease and protease inhibitor genes is given in Table 1. Interestingly, TIMP2 was overexpressed in M142 (ADAM15) LNCaP cells (1.7-fold compared to vector control). To validate this finding, we stained our tissue samples in the PCa TMA described here with an antibody against TIMP2.
Figure 4.
(A) Over-expression of ADAM15 protein in LNCaP cells in culture where, LNCaP M142 cells are shown under bright field microscopy and (B) the same field of cells under fluorescence, (magnification for both is 20X). Note that ADAM15/GFP fusion protein is localized to the cell membranes and the cell/cell junctions.
Table 1.
cDNA Microarray Experiment Showing Genes That Were Upregulated or Downregulated on the cDNA Chip (Comparing cDNA from LNCaP M142 ADAM15-Overexpressing Cells with cDNA from Control Cells).
| GENE | ADAM15 Ratio | |
| NOMANCLATURE | (Control LNCaP) | |
| Upregulated Proteases and Inhibitors: | ||
| membrane metallo-endopeptidase | MME | 2.47 |
| enolase 2 (gamma, neuronal) | ENO2 | 2.42 |
| matrix metalloproteinase 24 | MMP24 | 1.79 |
| tissue inhibitor of metalloproteinase 2 | TIMP2 | 1.69 |
| aconitase 1, soluble | ACO1 | 1.68 |
| a disintegrin and metalloproteinase domain15 | ADAM15 | 1.52 |
| Downregulated Proteases: | ||
| caspase 3, apoptosis-related | CASP3 | 0.462 |
| cysteine protease | ||
| disintegrin-like (reprolysin type) thrombospondin type 1, motif 3 | ADAMTS3 | 0.417 |
Given is the ratio of ADAM15 transcripts overexpressed in LNCaP M142 cells to ADAM15 transcripts overexpressed in vector control LNCaP cells.
TIMP2 Staining of PCa TMA
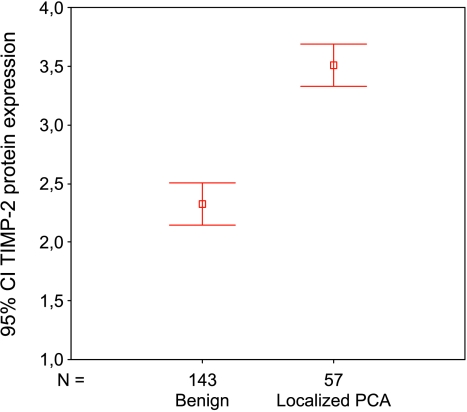
Acknowledging the fact that there are tremendous interactions between ADAMs and metalloproteinases and their inhibitors, we wanted to evaluate further the overexpression of TIMP2 in LNCaP M142 cells from a microarray cDNA experiment. To begin to validate this finding, we analyzed the protein expression of TIMP2 in a separate PCa TMA. The expression levels in normal prostatic tissue were compared to those of localized PCa cases. The result is graphically given in Figure 5, demonstrating a significant overexpression (Mann-Whitney U test, P < .001) of TIMP2 in PCa (mean = 3.51, median = 4.0, SD = 0.68) compared to normal tissue (mean = 2.32, median = 2.0, SD = 1.09), is graphically given in Figure 4. Based on the results shown in Table 1 and Figure 5, we hypothesize that there might be an important interaction between ADAM15 expression and TIMP2 expression levels in PCa. This interaction may affect a common protease pathway involved in the progression of advanced metastatic disease.
Figure 5.
Protein expression of TIMP2 in prostate tissues. TIMP2 expression is significantly higher in PCa compared to normal or benign prostate tissue (Mann-Whitney U test, P < .001).
Discussion
The ADAM family of proteases was first reported in 1995 [34]. Over the past 10 years, these proteins have been associated with distinct biologic processes, including reproduction (sperm/egg fusion), wound healing (migration and adhesion), inflammation (release of TGF-α), and tissue organogenesis [34]. More recently, members of the ADAM family have been implicated in tumorigenesis. This does not seem unreasonable, as this family of molecules has been associated with proteolytic activation, adhesion, and release of cytokines, which promote growth through signal transduction pathways.
Specifically, reports of ADAM15 have implied an association with tumorigenesis, but the functional role that ADAM15 plays in this process is not known. Recent reports, primarily utilizing DNA and RNA platforms, demonstrate increased expression of ADAM15 in ovary, gastric, lung, and breast cancer tissues [35–40]. Samples of these same tumors are represented in TMAs reported in this study and confirm increased ADAM15 protein expression in patients with ovary, gastric, lung, and breast cancers.
One of these reports analyzed the expression of ADAM15 in lung cancer cell lines and tissues and found that normal epithelial cells were negative for ADAM15 expression and that a higher expression was always found in tumor cells within the same tissue sections [39]. As observed in our study, adenocarcinoma of the lung showed the strongest staining. In comparison, small cell lung cancer showed only weak ADAM15 expression in our hands.
This report examined the expression of ADAM15 in multiple types of solid tumors, thus expanding the information base on ADAM15 expression in cancer. By using TMAs that included 22 different types of tumors, we can now report that the ADAM15 protein is upregulated in a variety of tumors but is most highly expressed in adenocarcinoma. From our TMA data, we observed that the highest levels of staining in tissue samples derived from ovary, breast, ileum, colon, and lung cancers. Lower values for protein expression were found in pheochromocytoma, sarcoma, carcinoma of the thyroid, and testicular cancer. In this study, all neoplastic tissue samples, compared to benign tissue samples, had higher expression levels, with adenocarcinomas exhibiting the highest level of ADAM15 staining.
Our previous work demonstrates that proteases play an important role in changing the homeostatic balance of both the prostate and breast epithelia [41–44]. In this study, we continued to focus on these two hormonally sensitive tissues and explored the expression of the ADAM15 protease in metastatic disease progression. We first found that ADAM15 was upregulated in many cancer cDNA arrays (see Table W1 generated on the Oncomine website Oncomine.org, including arrays of prostate and breast cancer). Now we have determined that the ADAM15 protein is upregulated in aggressive adenocarcinoma, and we suggest that it may play an essential regulatory role in the metastatic progression of both prostate and breast cancers.
For PCa, there is only very limited information available in the literature with respect to ADAM15. Of the metalloproteinases represented in our PCa cDNA expression array [28,30], only five metalloproteinases were significantly upregulated; this included three ADAM family proteases. The overexpression of ADAM15 mRNA in neoplastic prostate tissue was validated in two other independent cDNA array studies, thus providing strong evidence that ADAM15 levels are increased in cancer tissues compared to normal prostatic tissues [31,45,46]. In the present study, the highest expression of ADAM15 was observed in hormone-refractory and advanced metastatic PCa. In cell culture experiments using LNCaP cells, ADAM15 expression seemed to be independent of hormonal stimulus (data not shown). Related family members (ADAM9 and ADAM10) are thought to be sensitive to dihydrotestosterone levels, whereas ADAM17 seems to be repressed by androgens [7]. These results are based on studies in cell culture with the LNCaP cell line, which may represent an early stage of PCa disease. It cannot be excluded that, in the clinical setting and in advanced tumor stages, hormonal influences may still affect and influence the expression levels and activities of these disintegrins.
For cases specific to PCa, we demonstrate that, overall, increased expression of ADAM15 is associated with a more aggressive phenotype. This hypothesis is supported by the finding that, in localized PCa cases, there is a strong association with Gleason sum. Cases with a Gleason sum higher than 7 showed stronger ADAM15 expression compared to those with a Gleason sum lower than 7. The Gleason scoring system provides a well-described pathological parameter [24,25] in PCa disease and has proven to be a predictive value, and thus is considered as one of the strongest parameters for the prediction of outcome after specific therapy in clinical settings [37,38]. The results of this study suggest that the overexpression of ADAM15 may represent a more aggressive PCa phenotype.
In breast cancer, we found ADAM15 to be expressed at lower levels in the stroma and normal tissues, and at the highest levels in invasive and metastatic samples. This is in accordance with observations of an increased copy number of the ADAM15 gene in human breast cancer cell lines [38]. We also observed overexpression in the tumor aspect of breast tissues, where we found a very striking association with angioinvasion. This is interesting for three reasons. First, angioinvasion has been reported as a predictor of disease progression [49], local recurrence [50], and overall survival (RR = 4.26) [51]. Second, ADAM15 was recently described as a potential target for inhibitors of pathological neovascularization in ADAM15-null mice. In this model, neovascularization and melanoma tumor cell growth could be suppressed on this ADAM15 knockout background [52]. Third, human ADAM15 contains an RGD sequence and has been shown to interact with αvβ3 and αvβ1 integrins, which can drive an angiogenic response [53]. There are now several reports linking integrins to angiogenesis [54–56]. These data suggest that ADAM15 could be directly involved in the process of tumor cell growth through enhancement of angiogenesis. Taken together, these results suggest that ADAM15 could be a target for the development of inhibitors of angiogenesis, as mice carrying a null mutation in ADAM15 are viable and fertile without any severe pathological phenotypes [52].
In a separate set of experiments, we analyzed ADAM15 overexpression in the PCa cell line LNCaP using cDNA microarray technology. To uncover coregulators of disease progression, we examined gene expression profiles following the overexpression of ADAM15 in LNCaP M142 cells compared to vector control. We focused on proteases and protease inhibitors, which were associated with ADAM15 overexpression. This analysis revealed that the protease inhibitor TIMP2 was upregulated in LNCaP M142 cells. Our observation was verified in our study on protein level using a TMA of PCa. For technical reasons, TIMP2 staining was carried out on a separate TMA. Due to the result of the expression profile and the high number of cases stained for ADAM15 and TIMP2, we believe that the results are valid and representative of PCa. The observed overexpression of TIMP2 has to be interpreted with caution because it is known that TIMPs are not totally selective for MMPs [57]. So far, of these endogenous regulators of MMPs, four TIMPs are known. For example, TIMP3 is believed to inhibit ADAM17 and ADAM12. ADAM10 is also inhibited by TIMP3, in addition to TIMP1 [58,59]. However, not all MMPs seem to be sensitive to TIMPs. For example, ADAM8 and ADAM9 activity is not blocked by TIMPs [58]. Our data now suggest that TIMP2 may be an inhibitor of ADAM15; this observation warrants further study in the context of cancer progression. Additionally, we also report that ADAMTS3 and MMP24 cDNA expression is directly influenced by the overexpression of ADAM15, and we plan to verify these results on protein levels in future experiments.
Alternative splicing of primary RNA transcripts contributes to the functional regulation of many genes that impact normal and aberrant cellular functions. Spliced variants of ADAM gene family members have previously been found in several cancers, including grossly altered ADAM mRNA variants in breast cancer cells [38]. Although ADAM15 antibody recognition might be affected by some splicing events, ADAM15 splicing has not been investigated in PCa cells or tissues.
To summarize the present study, we can state that ADAM15 is overexpressed in a series of solid malignant tumors where the highest levels of expression are observed in adenocarcinomas of multiple tumor types. Specifically, we demonstrate that ADAM15 is highly upregulated in prostate and breast cancers, where the most significant staining was detected in advanced metastatic disease. For PCa, there is a strong association between ADAM15 expression and Gleason sum. In breast cancer, ADAM15 is significantly overexpressed in angioinvasive carcinoma; this observation may give us the best insight as to how the overexpression of ADAM15 directly impacts the growth and spread of more aggressive cancer cells. It is important to note here that Gleason sum and angioinvasion are two parameters associated with the worst outcome in a clinical course. In experiments wherein we analyze ADAM15 by cDNA expression in PCa cells that overexpress this disintegrin, we can report that ADAM15 directly influences the expression levels of specific protease inhibitors and proteases.
The results shown here demonstrate upregulation and change in the protein distribution of ADAM15 in many types of adenocarcinoma. This suggests that ADAM15 may play an essential role in the progression to metastatic disease, especially in cases of prostate and breast cancers. If this proves to be the case, then ADAM15 may serve as a drug target [13] for the treatment of patients who present with advanced stages of cancer and more aggressive types of cancer.
Supplementary Material
Acknowledgements
We thank Carl P. Blobel (Hospital for Special Surgery, Weill Medical College of Cornell University, Ithaca, NY) for providing us the cDNA for human ADAM15. Special thanks go to Gabrielle Nunez (Department of Pathology, University of Michigan) for the vectors used in cloning experiments.
Footnotes
This work was supported by grants from the National Institutes of Health (NIDDK R01 DK 56137-05 to M.L.D., R01CA107469 to C.G.K., and K08 CA090876 to C.G.K.) and the Department of Defense (PC030659).
This article refers to supplementary material, which is designated by “W” (ie, Table W1, Figure W1) and is available online at www.bcdecker.com.
References
- 1.Bogenrieder T, Herlyn M. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene. 2003;22(42):6524–6536. doi: 10.1038/sj.onc.1206757. [DOI] [PubMed] [Google Scholar]
- 2.Polette M, Nawrocki-Raby B, Gilles C, Clavel C, Birembaut P. Tumour invasion and matrix metalloproteinases. Crit Rev Oncol Hematol. 2004;49(3):179–186. doi: 10.1016/j.critrevonc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Wheelock MJ, Buck CA, Bechtol KB, Damsky CH. Soluble 80-kD fragment of cell-CAM 120/80 disrupts cell-cell adhesion. J Cell Biochem. 1987;34(3):187–202. doi: 10.1002/jcb.240340305. [DOI] [PubMed] [Google Scholar]
- 4.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15(5):598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282(5392):1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 6.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16(2):83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 7.McCulloch DR, Harvey M, Herington AC. The expression of the ADAMs proteases in prostate cancer cell lines and their regulation by dihydrotestosterone. Mol Cell Endocrinol. 2000;167(1–2):11–21. doi: 10.1016/s0303-7207(00)00305-1. [DOI] [PubMed] [Google Scholar]
- 8.Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997;235(2):437–442. doi: 10.1006/bbrc.1997.6714. [DOI] [PubMed] [Google Scholar]
- 9.Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM12 (meltrin alpha) supports tumor cell adhesion. Am J Pathol. 1999;154(5):1489–1501. doi: 10.1016/s0002-9440(10)65403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem. 2004;279(49):51323–51330. doi: 10.1074/jbc.M409565200. [DOI] [PubMed] [Google Scholar]
- 11.Fischer OM, Hart S, Gschwind A, Ullrich A. EGFR signal transactivation in cancer cells. Biochem Soc Trans. 2003;31(Part 6):1203–1208. doi: 10.1042/bst0311203. [DOI] [PubMed] [Google Scholar]
- 12.Alers JC, Rochat J, Krijtenburg PJ, Hop WC, Kranse R, Rosenberg C, Tanke HJ, Schroder FH, van Dekken H. Identification of genetic markers for prostatic cancer progression. Lab Invest. 2000;80(6):931–942. doi: 10.1038/labinvest.3780096. [DOI] [PubMed] [Google Scholar]
- 13.Glinsky GV, Krones-Herzig A, Glinskii AB. Malignancy-associated regions of transcriptional activation: gene expression profiling identifies common chromosomal regions of a recurrent transcriptional activation in human prostate, breast, ovarian, and colon cancers. Neoplasia. 2003;5(3):218–228. doi: 10.1016/S1476-5586(03)80054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goeze A, Schluns K, Wolf G, Thasler Z, Petersen S, Petersen I. Chromosomal imbalances of primary and metastatic lung adenocarcinomas. J Pathol. 2002;196(1):8–16. doi: 10.1002/path.1009. [DOI] [PubMed] [Google Scholar]
- 15.Qin LX. Chromosomal aberrations related to metastasis of human solid tumors. World J Gastroenterol. 2002;8(5):769–776. doi: 10.3748/wjg.v8.i5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson M, Meza-Zepeda LA, Mertens F, Forus A, Myklebost O, Mandahl N. Amplification of chromosome 1 sequences in lipomatous tumors and other sarcomas. Int J Cancer. 2004;109(3):363–369. doi: 10.1002/ijc.11716. [DOI] [PubMed] [Google Scholar]
- 17.Martin J, Eynstone LV, Davies M, Williams JD, Steadman R. The role of ADAM 15 in glomerular mesangial cell migration. J Biol Chem. 2002;277(37):33683–33689. doi: 10.1074/jbc.M200988200. [DOI] [PubMed] [Google Scholar]
- 18.Fourie AM, Coles F, Moreno V, Karlsson L. Catalytic activity of ADAM8, ADAM15, and MDC-L (ADAM28) on synthetic peptide substrates and in ectodomain cleavage of CD23. J Biol Chem. 2003;278(33):30469–30477. doi: 10.1074/jbc.M213157200. [DOI] [PubMed] [Google Scholar]
- 19.Bohm BB, Aigner T, Roy B, Brodie TA, Blobel CP, Burkhardt H. Homeostatic effects of the metalloproteinase disintegrin ADAM15 in degenerative cartilage remodeling. Arthritis Rheum. 2005;52(4):1100–1109. doi: 10.1002/art.20974. [DOI] [PubMed] [Google Scholar]
- 20.Al-Fakhri N, Wilhelm J, Hahn M, Heidt M, Hehrlein FW, Endisch AM, Hupp T, Cherian SM, Bobryshev YV, Lord RS, Katz N. Increased expression of disintegrin-metalloproteinases ADAM-15 and ADAM-9 following upregulation of integrins alpha5beta1 and alphavbeta3 in atherosclerosis. J Cell Biochem. 2003;89(4):808–823. doi: 10.1002/jcb.10550. [DOI] [PubMed] [Google Scholar]
- 21.Schafer B, Gschwind A, Ullrich A. Multiple G-protein-coupled receptor signals converge on the epidermal growth factor receptor to promote migration and invasion. Oncogene. 2004;23(4):991–999. doi: 10.1038/sj.onc.1207278. [DOI] [PubMed] [Google Scholar]
- 22.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6(3):1038–1045. [PubMed] [Google Scholar]
- 23.Greene FL. AJCC Cancer Staging Manual. 6th ed. New York: Springer-Verlag; 2002. AJCC cancer staging manual of the American Joint Committee on Cancer and American Cancer Society; p. 421. [Google Scholar]
- 24.Gleason D. Classification of prostate carcinoma. Cancer Chemother Rep. 1966;50(3):125–128. [PubMed] [Google Scholar]
- 25.Gleason D . The Veterans Administration Cooperative Urological Research Group, author. Histologic grading and clinical staging of prostate carcinoma. In: Tannenbaum M, editor. Urologic Pathology: The Prostate. Philadelphia: Lea and Febiger; 1977. pp. 171–198. [Google Scholar]
- 26.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 27.Perrone EE, Theoharis C, Mucci NR, Hayasaka S, Taylor JM, Cooney KA, Rubin MA. Tissue microarray assessment of prostate cancer tumor proliferation in African-American and white men. J Natl Cancer Inst. 2000;92(11):937–939. doi: 10.1093/jnci/92.11.937. [DOI] [PubMed] [Google Scholar]
- 28.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6489):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 29.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26(3):312–319. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;10(419(6907)):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noel A, Maillard C, Rocks N, Jost M, Chabottaux V, Sounni NE, Maquoi E, Cataldo D, Foidart JM. Membrane associated proteases and their inhibitors in tumour angiogenesis. J Clin Pathol. 2004;57(6):577–584. doi: 10.1136/jcp.2003.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handsley MM, Edwards DR. Metalloproteinases and their inhibitors in tumor angiogenesis. Int J Cancer. 2005;115(6):849–860. doi: 10.1002/ijc.20945. [DOI] [PubMed] [Google Scholar]
- 34.Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131(2):275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck V, Herold H, Benge A, Luber B, Hutzler P, Tschesche H, Kessler H, Schmith M, Geppert HG, Reuning U. ADAM15 decreases integrin alphavbeta3/vitronectin-mediated ovarian cancer cell adhesion and motility in an RGD-dependent fashion. Int J Biochem Cell Biol. 2005;37(3):590–603. doi: 10.1016/j.biocel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rocken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol. 2005;26(1):17–24. [PubMed] [Google Scholar]
- 37.Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H, Rocken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol. 2005;131(1):41–48. doi: 10.1007/s00432-004-0619-y. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz RM, Karkkainen I, Huovila AP. Aberrant alternative exon use and increased copy number of human metalloprotease-disintegrin ADAM15 gene in breast cancer cells. Genes Chromosomes Cancer. 2004;41(4):366–378. doi: 10.1002/gcc.20102. [DOI] [PubMed] [Google Scholar]
- 39.Schutz A, Hartig W, Wobus M, Grosche J, Wittekind Ch, Aust G. Expression of ADAM15 in lung carcinomas. Virchows Arch. 2005;446(4):421–429. doi: 10.1007/s00428-004-1193-z. [DOI] [PubMed] [Google Scholar]
- 40.Hart S, Fischer OM, Prenzel N, Zwick-Wallasch E, Schneider M, Hennighausen L, Ullrich A. GPCR-induced migration of breast carcinoma cells depends on both EGFR signal transactivation and EGFR-independent pathways. Biol Chem. 2005;386(9):845–855. doi: 10.1515/BC.2005.099. [DOI] [PubMed] [Google Scholar]
- 41.Rashid MG, Sanda MG, Vallorosi CJ, Rios-Doria J, Rubin MA, Day ML. Posttranslational truncation and inactivation of human E-cadherin distinguishes prostate cancer from matched normal prostate. Cancer Res. 2001;61(2):489–492. [PubMed] [Google Scholar]
- 42.Rios-Doria J, Day KC, Kuefer R, Rashid MG, Chinnaiyan AM, Rubin MA, Day ML. The role of calpain in the proteolytic cleavage of E-cadherin in prostate and mammary epithelial cells. J Biol Chem. 2003;278(2):1372–1379. doi: 10.1074/jbc.M208772200. [DOI] [PubMed] [Google Scholar]
- 43.Rios-Doria J, Kuefer R, Ethier SP, Day ML. Cleavage of beta-catenin by calpain in prostate and mammary tumor cells. Cancer Res. 2004;64(20):7237–7240. doi: 10.1158/0008-5472.CAN-04-1048. [DOI] [PubMed] [Google Scholar]
- 44.Rios-Doria J, Day ML. Truncated E-cadherin potentiates cell death in prostate epithelial cells. Prostate. 2005;63(3):259–268. doi: 10.1002/pros.20179. [DOI] [PubMed] [Google Scholar]
- 45.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61(12):4683–4688. [PubMed] [Google Scholar]
- 46.Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61(15):5692–5696. [PubMed] [Google Scholar]
- 47.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90(10):766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 48.Kattan MW, Potters L, Blasko JC, Beyer DC, Fearn P, Cavanagh W, Leibel S, Scardino P. Pretreatment nomogram for predicting freedom from recurrence after permanent prostate brachytherapy in prostate cancer. Urology. 2001;58(3):393–399. doi: 10.1016/s0090-4295(01)01233-x. [DOI] [PubMed] [Google Scholar]
- 49.Martin SA, Perez-Reyes N, Mendelsohn G. Angioinvasion in breast carcinoma. An immunohistochemical study of factor VIII-related antigen. Cancer. 1987;59(11):1918–1922. doi: 10.1002/1097-0142(19870601)59:11<1918::aid-cncr2820591113>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 50.Sinn HP, Anton HW, Magener A, von Fournier D, Bastert G, Otto HF. Extensive and predominant in situ component in breast carcinoma: their influence on treatment results after breast-conserving therapy. Eur J Cancer. 1998;34(5):646–653. doi: 10.1016/s0959-8049(97)10106-x. [DOI] [PubMed] [Google Scholar]
- 51.Watermann D, Madjar H, Sauerbrei W, Hirt V, Prompeler H, Stickeler E. Assessment of breast cancer vascularisation by Doppler ultrasound as a prognostic factor of survival. Oncol Rep. 2004;11(4):905–910. [PubMed] [Google Scholar]
- 52.Horiuchi K, Weskamp G, Lum L, Hammes HP, Cai H, Brodie TA, Ludwig T, Chiusaroli R, Baron R, Preissner KT, Manova K, Blobel CP. Potential role for ADAM15 in pathological neovascularization in mice. Mol Cell Biol. 2003;23(16):5614–5624. doi: 10.1128/MCB.23.16.5614-5624.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nath D, Slocombe PM, Stephens PE, Warn A, Hutchinson GR, Yamada KM, Docherty AJ, Murphy G. Interaction of metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on different haemopoietic cells. J Cell Sci. 1999;112(Part 4):579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- 54.Eliceiri BP, Cheresh DA. The role of alphav integrins during angiogenesis: insights into potential mechanisms of action and clinical development. J Clin Invest. 1999;103(9):1227–1230. doi: 10.1172/JCI6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9(15):1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156(4):1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 58.Amour A, Knight CG, Webster A, Slocombe PM, Stephens PE, Knauper V, Docherty AJ, Murphy G. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473(3):275–279. doi: 10.1016/s0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- 59.Loechel F, Fox JW, Murphy G, Albrechtsen R, Wewer UM. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem Biophys Res Commun. 2000;278(3):511–515. doi: 10.1006/bbrc.2000.3835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.