Abstract
Leukemias with MLL gene translocations are a complication of primary cancer treatment with DNA topoisomerase II inhibitors. How early translocations appear during primary cancer treatment has not been investigated. We tracked the leukemic clone with an MLL gene translocation during neuroblastoma therapy in a child who developed acute myeloid leukemia. The karyotype of the leukemic clone showed del(11)(q23). We used panhandle PCR-based methods to isolate the breakpoint junction involving MLL and an unknown partner gene. Marrow DNA from neuroblastoma diagnosis and DNA and RNA from serial preleukemic marrows were examined for the translocation. The karyotypic del(11)(q23) was a cryptic t(11;17). GAS7, a growth arrest-specific gene at chromosome band 17p13, was the partner gene of MLL. Two different MLL-GAS7 fusion transcripts were expressed. The translocation was already detectable by 1.5 months after the start of neuroblastoma treatment. The translocation was not detectable in the marrow at neuroblastoma diagnosis or in peripheral blood lymphocyte DNAs of six normal subjects. GAS7 is a new partner gene of MLL in treatment-related acute myeloid leukemia. MLL gene translocations can be present early during anticancer treatment at low cumulative doses of DNA topoisomerase II inhibitors. Although MLL has many partner genes and most have not been characterized, panhandle PCR strategies afford new means for detecting MLL gene translocations early during therapy when the partner gene is unknown.
There are two forms of treatment-related leukemia. The form associated with alkylating agents is characterized by chromosome 5 and 7 loss, presents as myelodysplasia, and has a latency of 5–7 years (reviewed in ref. 1). The form associated with DNA topoisomerase II inhibitors shows translocations of the MLL gene at chromosome band 11q23 and other translocations. Monoblastic acute myeloid leukemia (AML) is the most common presentation and the latency is short (reviewed in ref. 1). The incidence of leukemia is as high as 20% with intensive alkylating agent regimens for pediatric solid tumors (2, 3). The incidence of epipodophyllotoxin-related cases is 2–3% (4). There is a dose-response effect in alkylating agent-related, but not epipodophyllotoxin-related, cases (4, 5).
Presumably, the drugs play a role in pathogenesis, but the timing and mechanism of putative drug effects remain unknown. This study addresses when MLL gene translocations first appear during primary cancer treatment. MLL presents an extreme example of a gene involved in translocations with many different partner genes, most of which are uncharacterized. We developed panhandle PCR methods that do not require partner gene-specific primers and, therefore, are well suited to this situation. Using this approach, we followed the leukemic clone in a child who developed AML during neuroblastoma treatment. This led to discovery of the partner gene and detection of the translocation early in the therapy. The same approach also could be useful for other translocations.
Case Report
A 13-year-old boy presented with metastatic neuroblastoma. He was treated with modified N6 therapy including cyclophosphamide, doxorubicin, vincristine, cisplatin, etoposide, surgical resection, radiation therapy, and anti-GD2 (3F8) antibodies with granulocyte/macrophage colony-stimulating factor (Fig. 1A) (2, 6–8). Six months after starting therapy, peripheral monocytosis and cytopenias appeared. The G-banded marrow karyotype obtained 12 months from diagnosis as a routine test was 46,XY,del(11)(q23)[8]/46,XY[12] (3, 8). Seventeen months after starting therapy, the marrow was replaced by French-American-British M4 leukemic blasts. Four months later the patient died from treatment-related AML (t-AML).
Figure 1.
(A) Treatment and clinical events in patient diagnosed with neuroblastoma. CPM denotes cyclophosphamide; ADR, doxorubicin; VCR, vincristine; CPPD, cisplatin; VP16, etoposide; XRT, local radiation therapy; MoAb, anti-GD2 mAbs (3F8); NBL, neuroblastoma; FAB M4 AML, French-American-British AML; DOD, dead of disease. Scale indicates months (mos) from neuroblastoma diagnosis. (B) Identification of MLL rearrangements by Southern blot analysis of cryopreserved marrows obtained during neuroblastoma treatment. Months (mos) from neuroblastoma diagnosis are indicated. BamHI-digested DNAs were hybridized with B859 cDNA fragment of ALL-1 exons 5–11 (34). Control peripheral blood lymphocyte DNA of normal subject shows the germ-line band (dash). Arrows show rearrangements. (C) PCR amplification of der(11) breakpoint junction with clonotypic primers in DNAs prepared from cryopreserved marrows or marrow aspirates on glass slides during neuroblastoma treatment. The translocation was detected at 1.5 mos from neuroblastoma diagnosis and in all specimens thereafter. No specimen was available after the first course of cyclophosphamide, doxorubicin, and vincristine. (D) Absence of der(11) breakpoint junction by PCR analysis of DNA prepared from morphologically normal marrow at neuroblastoma diagnosis (0 mos) with clonotypic primers. Ten reactions were performed on marrow aspirate DNA from a glass slide, five examples of which are shown (Left). Marrow DNA from AML diagnosis at 17 mos was positive control. PCR with p53 exon 8-specific primers shows amplifiable DNA in marrow from neuroblastoma diagnosis (0 mos) (Right).
Methods
The Institutional Review Boards at The Children's Hospital and Medical Center of Seattle, Memorial Sloan-Kettering Cancer Center, and The Children's Hospital of Philadelphia approved this research.
Translocation Breakpoint Cloning.
Genomic DNA and total RNA were isolated from cryopreserved marrow cells of the patient and peripheral blood lymphocytes of normal adults (9). MLL gene rearrangement was examined (10), and panhandle variant PCR analysis of the der(11) genomic breakpoint junction was performed as described (11). Products were subcloned by recombination PCR and sequenced (11). To determine the chromosomal origin of the partner sequence, forward and reverse primers 5′-TTGAAACAACCGGAGAATCC-3′ and 5′-CCACAGTGGCCATTTTCTTC-3′ were used to screen a somatic cell hybrid panel (Bios, New Haven, CT) and the Stanford G3 radiation hybrid panel (Research Genetics, Huntsville, AL). Clonotypic primers 5′-AGGCCTCAGTTTGTCCATCT-3′ and 5′-CCCGACGTGGATTTTCTTTA-3′ were used in two-phase touchdown PCR (12) to isolate the predicted der(17) genomic breakpoint junction.
Tracking the der(11) Breakpoint.
DNA was isolated from marrows on glass slides by using PUREGENE reagents for DNA isolation from buccal cells (Gentra Systems). Clonotypic primers used in two-phase touchdown PCR (12) were 5′-TGTGGTTGGATTATGGGTGA-3′ and 5′-TGATCCCAATTGTGGAGAAAA-3′. Each 50-μl PCR contained 100 ng of DNA from cryopreserved marrows or 2 μl of DNA from glass slides, 5 pmols each primer, 1 unit AmpliTaq Gold DNA polymerase, 200 μM each dNTP, 4 mM MgCl2 and 1× buffer II (Perkin–Elmer). After initial denaturation at 95°C × 10 min, the first phase included 22 cycles of 95°C × 45 sec, 70°C × 1 min (decrease 0.7°C/cycle) to reach 55.3°C. The second phase included 38 cycles of 95°C × 45 sec, 55°C × 30 sec, and 72°C × 1 min, and was followed by elongation at 72°C × 5 min. p53 exon 8 primers (13) were used in control reactions. Serial 1:10 dilutions of marrow DNA into peripheral blood lymphocyte DNA from a normal subject were analyzed to determine the sensitivity of the assay. For analysis of specificity, peripheral blood lymphocyte DNAs from six normal adults were amplified, and p53 exon 8 primers (13) were used in control reactions.
cDNA Panhandle PCR Analysis of der(11) Transcripts.
First-strand cDNAs were synthesized from 1 μg of total RNA by using oligonucleotides with MLL exon 5 positions 92–123 at the 5′ ends and six random bases at the 3′ ends (5′-CCTGAATCCAAACAGGCCACCACTCCAGCTTCNNNNNN-3′). The Superscript Preamplification System was used (Life Technologies, Gaithersburg, MD). Two microliters of first-strand cDNA was added to a 45.5-μl mixture that was preheated to 80°C for 5 min and contained 1.75 units of Taq/Pwo DNA polymerase, 385 μM each dNTP, 1.1 × buffer (Expand Long Template System, Boehringer Mannheim), and 12.5 pmols of primer 1, corresponding to MLL exon 5 positions 34–55 (14–16). Primer 1 extension from the first-strand cDNAs was achieved by denaturing at 94°C for 1 min followed by 1 cycle at 94°C for 10 sec, 68°C for 7 min. The sample was heated to 80°C for 5 min before adding 2.5 μl (12.5 pmols) of primer 2, corresponding to MLL exon 5 positions 136–159 (14–16). The 50-μl reactions then contained 350 μM of each dNTP and 1× buffer. Intrastrand annealing of complementary MLL sequences in the second strands and polymerase extension of the recessed 3′ ends occurred during PCR, completing formation of stem-loop templates containing the fusion point of the chimeric transcript in the loop. PCR with primers 1 and 2 was as described (14, 15). One microliter of the products was used for nested PCR with primers 3 (5′-GGAAAAGAGTGAAGAAGGGAATGTCTCGG-3′) and 4 (5′-GTGGTCATCCCGCCTCAGCCAC-3′). As a positive control, 2 μl of the first-strand cDNA was amplified with β-actin primers (16).
HindIII-digested pUC19 was amplified with primers 5′-ACATTCCCTTCTTCACTCTTTTCCTGGCGTAATCA-TGGTCATAGC-3′ and 5′-GTGGCTGAGGCGGGATGACCACCATGCCTGCAGGTCGACTC-3′, and cDNA panhandle PCR products were subcloned by recombination PCR (11).
To confirm the der(11) transcripts, 2 μl of the same first-strand cDNA was amplified with primers 5′-ACCACCAGAATCAGGTCCAG-3′ and 5′-AGAGCTCCTGTGAGAGCCAG-3′ from MLL exons 5–6 and GAS7 (growth arrest-specific 7) exon 3. Similar reverse transcription–PCR conditions have been described (17). Two microliters of the product was used as template in a second round of PCR.
Results
MLL Gene Translocation Is Detected Early During Treatment.
Six-kilobase and 8-kb MLL gene rearrangements were detected by Southern blot analysis in marrows from 3, 7, 11, and 12 months after starting treatment (Fig. 1B). Two rearrangements were more consistent with a t(11;?)(q23;?) than with del(11)(q23). The translocation was detectable by Southern blot 14 months before leukemia was diagnosed (Fig. 1 A and B). At 10 months, after chest and abdomen radiation (Fig. 1A), only the germ-line fragment was detected, suggesting that the radiation may have temporarily reduced the population of cells with the translocation to a level below the sensitivity of the Southern blot.
The der(11) breakpoint junction was amplified by panhandle variant PCR analysis of genomic DNA from the marrow obtained 12 months after starting treatment. Three independent PCRs yielded 6-kb products (Fig. 2A), indicating that the 6-kb rearrangement observed on Southern blot analysis was from the der(11) chromosome. Twenty-five of 56 subclones contained a 6-kb insert; one was sequenced in its entirety (Fig. 2B). The MLL breakpoint was in intron 8. The 3′ sequence was partner DNA.
Figure 2.
(A) Panhandle variant PCR products from der(11) chromosome in marrow DNA at 12 mos from neuroblastoma diagnosis. (Right) Negative control reactions, designated (−) ligase and dH2O. (B) Summary of panhandle variant PCR products containing der(11) breakpoint junction. The 31-base sequence of primer 3 used in final round of PCR and its complement are at 5′ and 3′ ends, respectively. The 4643–4645 bases of bcr sequence, including primer 3, are 5′ of MLL breakpoint at position 4662, 4663, or 4664 in intron 8 (corkscrew arrow). Depending on MLL breakpoint location, 1034–1036 bases of 3′ sequence are GAS7 partner DNA (GenBank accession no. AC005747). MLL breakpoint is 3′ of an AluY. GAS7 breakpoint is 5′ of an AluY. Identical 5′-AT-3′ sequences in MLL and GAS7 (outlined) preclude more precise assignment of breakpoint positions. Short homologies between MLL and GAS7 are underlined. (C) Genomic organization of 167-kb human GAS7 gene derived from alignment of genomic (GenBank accession no. AC005747) and cDNA sequences (GenBank accession nos. AB007854 and AJ224876). Boxes show the 14 exons. GAS7 der(11) breakpoint (corkscrew arrow) corresponds to nucleotide 165462, 165461, or 165460 upstream of exon 1 in GenBank accession no. AC005747; but sequence in GenBank entry is reverse complement relative to transcriptional orientation of the ORF (GenBank accession nos. AB007854 and AJ224876) and breakpoint corresponds to position 1240, 1241, or 1242 if GenBank accession no. AC005747 were in sense orientation. BamHI site indicates 3′ end of GAS7 sequence in panhandle variant PCR products shown in B.
By PCR analysis of the der(11) breakpoint junction with clonotypic primers, cryopreserved marrows from 3, 7, 10, 11, and 12 months after neuroblastoma diagnosis all contained the translocation (Fig. 1C). Analysis of marrows on glass slides showed that the translocation was already present by 1.5 months from neuroblastoma diagnosis (Fig. 1C Right), after two courses of cyclophosphamide, doxorubicin, and vincristine, but before any etoposide and cisplatin (Fig. 1A).
The translocation was not PCR-detectable in 10 reactions performed on marrow aspirate DNA from a glass slide from the time of neuroblastoma diagnosis, microscopic examination of which showed normal, trilinear maturation and no evidence of tumor infiltration (Fig. 1D). Although focal metastatic neuroblastoma was present in a marrow biopsy obtained from a different site at the same time, no neuroblastoma was evident in the specimen used for PCR. In addition, no abnormalities were detected by cytogenetic analysis of neuroblastoma tissue, indicating that the primary tumor cells did not contain the translocation. Analysis of serial 1:10 dilutions of marrow DNA from 12 months after neuroblastoma diagnosis into peripheral blood lymphocyte DNA from a normal subject indicated a sensitivity of between 1 cell in 1,000 and 1 cell in 10,000 for detection of the translocation (data not shown). The same PCR did not yield products from peripheral blood lymphocyte DNAs of six normal subjects (data not shown). Reactions with p53 primers demonstrated that the sample from neuroblastoma diagnosis and from each normal subject was amplifiable.
t(11;?)(q23;?) Fuses MLL with GAS7.
Sequencing of the panhandle variant PCR products identified the der(11) breakpoint in MLL intron 8 and partial sequence of the partner DNA (Fig. 2B). Direct sequencing of the products of PCRs performed with clonotypic primers confirmed the breakpoint junction in marrows from 1.5, 3, 7, 10, 11, and 12 months after neuroblastoma diagnosis and at diagnosis of t-AML. The partner DNA corresponded to human GAS7 genomic sequence (GenBank accession no. AC005747).
Comparison of the cDNA (GenBank accession nos. AB007854 and AJ224876) and genomic sequences suggests that GAS7 contains 14 exons (Fig. 2C). On the der(11) chromosome, the MLL breakpoint corresponded to position 4662, 4663, or 4664 in intron 8; the GAS7 breakpoint corresponded to position 165462, 165461, or 165460, upstream of exon 1 (GenBank accession no. AC005747) (Fig. 2B). MLL positions 4663–4664 and GAS7 positions 165462–165461 are 5′-AT-3′, precluding more precise localization of the breakpoints. The GenBank entry of the 167-kb GAS7 genomic sequence (GenBank accession no. AC005747) is the reverse complement relative to the ORF in the cDNA (GenBank accession nos. AB007854 and AJ224876). The der(11) GAS7 breakpoint would correspond to positions 1240, 1241, or 1242 if the genomic sequence were in sense orientation.
The MLL breakpoint was 3′ of an AluY and an AT-rich region. The GAS7 breakpoint was between a mammalian interspersed repeat and a similar AluY (Fig. 2B). In addition to the 5′-AT-3′, there were other identical short sequences in both genes (Fig. 2B).
Consistent with the location of GAS7, the somatic hybrid screen indicated that the partner DNA was from chromosome 17; the radiation hybrid screen localized the sequence to band p13.
A 360-bp PCR product was expected from amplification of the der(17) genomic breakpoint junction, but a 307-bp product was amplified from marrow DNA from 12 months after starting treatment (Fig. 3A). The GAS7 breakpoint was nucleotide 165499 (1203) upstream of exon 1 (GenBank accession no. AC005747); the MLL breakpoint was nucleotide 4680 (Fig. 3B). The der(11) and der(17) genomic sequences indicate that 53 bases containing regions of MLL and GAS7 were lost during the translocation process (Fig. 3C). Depending on the exact positions of the der(11) MLL and GAS7 breakpoints, the deleted region from MLL was 15–17 bp and the deleted region from GAS7 was 36–38 bp. Short homologous sequences were present near the der(17) breakpoints in both genes (Fig. 3B).
Figure 3.
(A) Amplification of der(17) breakpoint junction in marrow DNA at 12 months from neuroblastoma diagnosis using clonotypic primers. A 360-bp product was expected but a 307-bp product was obtained. Peripheral blood lymphocyte DNAs from two normal subjects and dH2O were negative controls. (B) Sequencing of the 307-bp products from duplicate reactions showed GAS7 breakpoint at position 1203 upstream of exon 1 and MLL breakpoint at position 4680 in intron 8. Homologous sequences are underlined. GAS7 nucleotide position is for sense orientation of genomic sequence (GenBank accession no. AC005747). (C) Expected der(17) breakpoint junction based on der(11) sequence (Fig. 2B). A total of 53 bases, including the indicated 36–38 bases from GAS7 and 15–17 bases from MLL, were lost in the translocation process. It cannot be determined from where AT was lost (outlined).
cDNA Panhandle PCR Identifies Two Chimeric Transcripts.
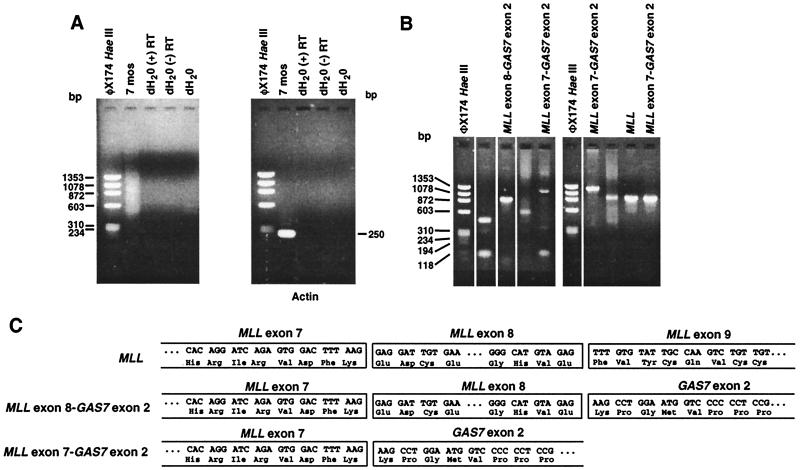
Total RNAs from cryopreserved marrows from 7 and 12 months after neuroblastoma diagnosis were studied. Fig. 4A shows the cDNA panhandle PCR products from 7 months. The products were subcloned by recombination PCR (Fig. 4B) and sequenced. Three subclones from 7 months and seven subclones from 12 months after neuroblastoma diagnosis ranging from 568 to 1,233 bp were from the normal MLL allele. Three subclones each from 7 months and 12 months after neuroblastoma diagnosis ranging from 701 to 1,263 bp contained an in-frame fusion of MLL exon 7 to GAS7 exon 2 at position 113 of the 7,979-bp full-length cDNA (GenBank accession no. AB007854). Because the MLL genomic breakpoint was in intron 8, this transcript indicates that MLL was alternatively spliced. One subclone each from 7 months and 12 months after neuroblastoma diagnosis that were 902 bp and 814 bp, respectively, contained a fusion of MLL exon 8 to GAS7 exon 2.
Figure 4.
(A) cDNA panhandle PCR analysis of marrow total RNA from 7 months after neuroblastoma diagnosis. As shown by smear in second lane of gel at left, a population of products of various sizes was obtained by reverse-transcribing first-strand cDNA from total RNA with 5′-MLL-NNNNNN-3′ oligonucleotides, generating second strands by primer 1 extension, forming stem-loop templates, and PCR with MLL-specific primers. As a positive control, a 2-μl aliquot of the same 5′-MLL-NNNNNN-3′-primed first-strand cDNAs was amplified with β-actin primers, which gave the expected 250-bp product (gel at right). (B) Representative recombination PCR-generated subclones of cDNA panhandle PCR products. One-eighth of the cDNA panhandle PCR products, represented by the smear in A, was used in each of three transformations. A total of 413 subclones were obtained, of which 14 were sequenced. (C) Sequences identified by cDNA panhandle PCR analysis. Three subclones contained normal MLL cDNA, three showed an in-frame fusion of MLL exon 7 to GAS7 exon 2, and one contained a fusion of MLL exon 8 to GAS7 exon 2. The other seven subclones contained empty vector sequence. The products from 12 months after neuroblastoma diagnosis were similar (see text).
Reverse transcription–PCR analysis using the first-strand cDNA from 7 months after neuroblastoma diagnosis corroborated these results (data not shown). A 409-bp product was obtained by using MLL and GAS7 primers; sequencing indicated that this product was from the alternatively spliced MLL exon 7-GAS7 exon 2 transcript. A less intense band of ≈523 bp also was detected. Although this was the predicted size of the product from the MLL exon 8-GAS7 exon 2 transcript, the quantity was insufficient for sequencing.
Discussion
We applied Southern blot analysis and panhandle PCR methods to track the MLL translocation during neuroblastoma treatment in a case of t-AML. This work contributes two elements to existing knowledge on leukemias that complicate anticancer treatment with DNA topoisomerase II inhibitors (1). The first pertains to natural history and the finding that the translocation can be present early during therapy. The second element is an approach to using MLL gene translocations as leukemia-associated biomarkers in patients undergoing treatment. MLL presents an extreme example of a translocation involving many partner genes; there may be up to 70 and most are uncharacterized. Southern blot analysis identifies MLL translocations regardless of the partner gene, but Southern blot analysis is not as sensitive as PCR. Previously we adapted panhandle PCR methods, which do not use partner gene-specific primers, to detect the translocation breakpoints in genomic DNA (11, 14–16). In the present study, we developed cDNA panhandle PCR to detect the fusion transcripts. This technology will facilitate detection of the leukemic clone in preleukemic samples.
When MLL gene translocations first appear during primary cancer treatment previously was unknown. In the patient we studied, Southern blot analysis detected MLL gene rearrangements in cryopreserved marrows available from 3 months after starting treatment, except for the marrow at 10 months that followed radiation. Although these findings demonstrate that the translocation was present early during treatment, after radiation the frequency of cells with the translocation was below the sensitivity of the Southern blot.
Panhandle variant PCR analysis of the marrow from 12 months after neuroblastoma diagnosis showed that the del(11)(q23) was a cryptic t(11;17)(q23;p13) and identified GAS7 as the partner gene of MLL. From the sequence of the panhandle variant PCR products, we designed clonotypic primers to track the translocation in sequential marrows. The translocation was not PCR-detectable at neuroblastoma diagnosis, but it was present in all marrows from 1.5 months after starting treatment forward, including the marrow at 10 months. The marrow at 1.5 months was after only two courses of cyclophosphamide, doxorubicin, and vincristine, before any cisplatin and etoposide. Monocytosis and cytopenias and then French-American-British M4 AML appeared 4.5 and 15.5 months after the translocation was PCR-detectable. cDNA panhandle PCR detected MLL fusion transcripts in the two marrows studied, from 7 and 12 months from the start of treatment, both several months before leukemia was diagnosed.
Panhandle PCR strategies are useful for leukemias with karyotypic del(11)(q23) because del(11)(q23) abnormalities often represent cryptic translocations of MLL and various unknown partner genes (18–21), as was true in this patient. GAS7 is the third partner gene of MLL to be described on chromosome 17. The AF17 gene at band 17q21 encodes a transcription factor (22). The second chromosome 17 partner gene, MSF, at band q25, also was discovered in t-AML (23). Both loci are distinct from GAS7.
Gas genes are expressed in quiescent cells (24). The human GAS7 cDNA (GenBank accession no. 007854) is 97% identical to its murine counterpart (24). Murine GAS7 is expressed in Purkinje cells and has putative roles in transcriptional regulation, neuronal differentiation, and neurotransmitter release (24).The murine protein contains an OCT2-like transcription factor structural motif, a region of homology to synapsins, a WW (Trp-Trp) module for protein-protein interactions, and a region resembling yeast ATP synthetase (24). MLL gene translocations are believed to be oncogenic by production of chimeric oncoproteins (25), but precisely how the putative MLL-GAS7 chimeric oncoprotein may affect the function of MLL remains unknown.
The der(11) MLL breakpoint in the t-AML was in intron 8 and the GAS7 breakpoint was upstream of exon 1. The MLL gene is known to be alternatively spliced (26). GAS7 cDNA sequences (GenBank accession nos. AB007854, U19860, and AJ003148) suggest that exon 2 contains the relevant translation initiation codon because exon 1 contains not only an ATG, but also a stop codon that would terminate the protein. cDNA panhandle PCR identified an MLL exon 8-GAS7 exon 2 transcript and a predominant, alternatively spliced MLL exon 7-GAS7 exon 2 transcript, but no transcript fusing MLL with GAS7 exon 1. No der(17) transcript would be predicted because the der(17) GAS7 genomic breakpoint was upstream of exon 1.
The leukemic clone emerged early in the course of treatment and the patient died from t-AML 4 months after leukemia diagnosis. Studies of additional patients will reveal whether leukemias with MLL-GAS7 translocations are especially aggressive. cDNA panhandle PCR provides an approach for detecting MLL fusion transcripts regardless of the partner gene and a useful tool to investigate the presence and natural history of the translocations in other patients who are undergoing treatment.
The der(11) and der(17) genomic breakpoints indicate loss of material from MLL and GAS7 during the translocation. There were identical 2- to 5-base sequences in MLL and GAS7 at both breakpoint junctions. This phenomenon suggests joining of homologous DNA ends during the translocation process (11, 14, 16, 27–30). Nearby AluY repeats also may have played a role in joining MLL with the involved genomic region of GAS7.
That we were able to detect the translocation very early in the course of treatment after the DNA topoisomerase II inhibitor, doxorubicin, and the alkylating agent, cyclophosphamide, suggests two possibilities. The first is that the chemotherapy caused the translocation. Use of anthracyclines in combination with alkylating agents and radiation for pediatric solid tumors is associated with t-AML characterized by translocations of chromosome band 11q23 (31). The potential role of chromosomal breakage brought about by DNA topoisomerase II inhibitors in forming translocations has been described (1). That the translocation was not detected in the marrow at neuroblastoma diagnosis nor in peripheral blood lymphocytes of normal subjects supports this possibility. Alternatively, if the frequency of cells with the translocation was below the sensitivity of the PCR, the possibility remains that the chemotherapy selected for a translocation that was already present. Indeed, MLL tandem duplications, a form of translocation, have been identified in peripheral blood and marrow of healthy individuals (32). Chemotherapy has profound effects on the kinetics of the marrow. Chemotherapy causes cell death, forcing many marrow stem cells to divide, which might select for the rare stem cell with a translocation (33). Favoring the former possibility is the specificity of the association of DNA topoisomerase II inhibitors, but not other chemotherapies that cause cell death in the marrow, with leukemias characterized by translocations. cDNA panhandle PCR will augment future studies of occult translocations. Regardless of which possibility proves to be correct, the early presence of the translocation is additional information on the natural history of t-AML. Most children who develop leukemia after dose-intensive therapy for primary pediatric cancers die from the disease (3). Panhandle PCR methods now render MLL breakpoint sequences biomarkers for early detection of leukemic clones in t-AML regardless of the partner gene and are generally applicable for analysis of unknown juxtaposed sequences and new gene discovery.
Acknowledgments
We thank Brian Lovett for helpful comments and critical review, and Julie Park, Douglas Hawkins, and Kim Kramer for providing samples from the patient and clinical information. C.A.F. was supported by National Institutes of Health Grants CA66140, CA77683, and CA80175, Leukemia Society of America Scholar Award, and The Children's Hospital of Philadelphia High Risk High Impact Grant.
Abbreviations
- AML
acute myeloid leukemia
- GAS
growth arrest-specific
- t-AML
treatment-related AML
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF231993, AF231994, AF231995, AF231996, AF231997, AF231998, AF231999, AF232000, AF232001, and AF232002).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050397097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050397097
References
- 1.Felix C A. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 2.Kushner B H, Cheung N K, Kramer K, Heller G, Jhanwar S C. J Clin Oncol. 1998;16:3880–3889. doi: 10.1200/JCO.1998.16.12.3880. [DOI] [PubMed] [Google Scholar]
- 3.Kushner B H, Heller G, Cheung N-K V, Wollner N, Kramer K, Bajorin D, Polyak T, Meyers P A. J Clin Oncol. 1998;16:3016–3020. doi: 10.1200/JCO.1998.16.9.3016. [DOI] [PubMed] [Google Scholar]
- 4.Smith M, Rubenstein L, Anderson J, Arthur D, Catalano P, Freidlin B, Heyn R, Khayat A, Krailo M, Land V, et al. J Clin Oncol. 1999;17:569–577. doi: 10.1200/JCO.1999.17.2.569. [DOI] [PubMed] [Google Scholar]
- 5.Meadows A T, Obringer A C, Marrero O, Oberlin O, Robison L, Fossati-Bellani F, Green D, Voute P A, Morris-Jones P, Greenburg M, et al. Med Pediatr Oncol. 1989;17:477–484. [PubMed] [Google Scholar]
- 6.Kushner B H, LaQuaglia M P, Bonilla M A, Lindsley K, Rosenfield N, Yeh S, Eddy J, Gerald W L, Heller G, Cheung N K. J Clin Oncol. 1994;12:2607–2613. doi: 10.1200/JCO.1994.12.12.2607. [DOI] [PubMed] [Google Scholar]
- 7.Felix C A, Walker A H, Lange B J, Williams T M, Winick N J, Cheung N-K V, Lovett B D, Nowell P C, Blair I A, Rebbeck T R. Proc Natl Acad Sci USA. 1998;95:13176–13181. doi: 10.1073/pnas.95.22.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung N-K V, Kushner B H, Cheung I Y, Kramer K, Canete A, Gerald W, Bonilla M A, Finn R, Yeh S J, Larson S M. J Clin Oncol. 1998;16:3053–3060. doi: 10.1200/JCO.1998.16.9.3053. [DOI] [PubMed] [Google Scholar]
- 9.Felix C A, Poplack D G, Reaman G H, Steinberg S M, Cole D E, Taylor B J, Begley C G, Kirsch I R. J Clin Oncol. 1990;8:431–442. doi: 10.1200/JCO.1990.8.3.431. [DOI] [PubMed] [Google Scholar]
- 10.Felix C A, Hosler M R, Winick N J, Masterson M, Wilson A E, Lange B J. Blood. 1995;85:3250–3256. [PubMed] [Google Scholar]
- 11.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felix C A, Megonigal M D, Chervinsky D S, Leonard D G B, Tsuchida N, Kakati S, Block A M W, Fisher J, Grossi M, Salhany K E, et al. Blood. 1998;91:4451–4456. [PubMed] [Google Scholar]
- 14.Felix C A, Kim C S, Megonigal M D, Slater D J, Jones D H, Spinner N B, Stump T, Hosler M R, Nowell P C, Lange B J, Rappaport E F. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- 15.Felix C A, Jones D H. Leukemia. 1998;12:976–981. doi: 10.1038/sj.leu.2401026. [DOI] [PubMed] [Google Scholar]
- 16.Megonigal M D, Rappaport E F, Jones D H, Kim C S, Nowell P C, Lange B J, Felix C A. Proc Natl Acad Sci USA. 1997;94:11583–11588. doi: 10.1073/pnas.94.21.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Megonigal M D, Rappaport E F, Nowell P C, Lange B J, Felix C A. Oncogene. 1998;16:1351–1356. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi H, Espinosa R, Thirman M J, Fernald A A, Shannon K, Diaz M O, LeBeau M M, Rowley J D. Genes Chromosomes Cancer. 1993;7:204–208. doi: 10.1002/gcc.2870070404. [DOI] [PubMed] [Google Scholar]
- 19.Leblanc T, Le Coniat M, Flexor M, Baruchel A, Daniel M T, Berger R. Leukemia. 1996;10:1844–1846. [PubMed] [Google Scholar]
- 20.Harbott J, Mancini M, Verellen-Dumoulin C, Moorman A V, Secker-Walker L M. Leukemia. 1998;12:823–827. doi: 10.1038/sj.leu.2401018. [DOI] [PubMed] [Google Scholar]
- 21.Behm F G, Raimondi S C, Frestedt J L, Liu Q, Crist W M, Downing J R, Rivera G K, Kersey J H, Pui C-H. Blood. 1996;87:2870–2877. [PubMed] [Google Scholar]
- 22.Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H, Huebner K, Berger R, Croce C, Canaani E. Proc Natl Acad Sci USA. 1994;91:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osaka M, Rowley J D, Zeleznik-Le N J. Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju Y-T, Chang A C Y, She B R, Tsaur M-L, Hwang H-M, Chao C C-K, Cohen S N, Lin-Chao S. Proc Natl Acad Sci USA. 1998;95:11423–11428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schichman S A, Canaani E, Croce C M. J Am Med Assoc. 1995;273:571–576. [PubMed] [Google Scholar]
- 26.Mbangkollo D, Burnett R, McCabe N, Thirman M, Gill H, Yu H, Rowley J D, Diaz M O. DNA Cell Biol. 1995;14:475–483. doi: 10.1089/dna.1995.14.475. [DOI] [PubMed] [Google Scholar]
- 27.Felix C A, Lange B J, Hosler M R, Fertala J, Bjornsti M-A. Cancer Res. 1995;55:4287–4292. [PubMed] [Google Scholar]
- 28.Felix C A, Hosler M R, Slater D J, Megonigal M D, Lovett B D, Williams T M, Nowell P C, Spinner N B, Owens N L, Hoxie J, et al. Mol Diagn. 1999;4:269–283. doi: 10.1016/s1084-8592(99)80002-2. [DOI] [PubMed] [Google Scholar]
- 29.Reichel M, Gillert E, Nilson I, Siegler G, Greil J, Fey G H, Marschalek R. Oncogene. 1998;17:3035–3044. doi: 10.1038/sj.onc.1202229. [DOI] [PubMed] [Google Scholar]
- 30.Atlas M, Head D, Behm F, Schmidt E, Zeleznik-Le N J, Roe B A, Burian D, Domer P H. Leukemia. 1998;12:1895–1902. doi: 10.1038/sj.leu.2401223. [DOI] [PubMed] [Google Scholar]
- 31.Sandoval C, Pui C-H, Bowman L C, Heaton D, Hurwitz C A, Raimondi S C, Behm F G, Head D R. J Clin Oncol. 1993;11:1039–1045. doi: 10.1200/JCO.1993.11.6.1039. [DOI] [PubMed] [Google Scholar]
- 32.Schnittger S, Wormann B, Hiddemann W, Griesinger F. Blood. 1998;92:1728–1734. [PubMed] [Google Scholar]
- 33.Knudson A G. Semin Cancer Biol. 1992;3:99–106. [PubMed] [Google Scholar]
- 34.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]