Abstract
Cyclooxygenase-2 (COX-2) overexpression and mutations of p53 (a known COX-2 regulator) are inversely associated with microsatellite instability—high (MSI-H) and CpG island methylator phenotype (CIMP), characterized by extensive promoter methylation, is associated with MSI-H. However, no studies have comprehensively examined interrelations between COX-2, p53, MSI, and CIMP. Using MethyLight, we measured DNA methylation in five CIMP-specific gene promoters [CACNA1G, CDKN2A (p16/INK4A), CRABP1, MLH1, and NEUROG1] in relatively unbiased samples of 751 colorectal cancer cases obtained from two large prospective cohorts; 115 (15%) tumors were CIMP-high (≥ 4 of 5 methylated promoters), 251 (33%) were CIMP-low (1 to 3 methylated promoters), and the remaining 385 (51%) were CIMP-0 (no methylated promoters). CIMP-high tumors were much less frequent in COX-2+/p53+ tumors (4.6%) than in COX-2+/p53- tumors (19%; P < .0001), COX-2-/p53+ tumors (17%; P= .04), and COX-2-/p53- tumors (28%; P < .0001). In addition, COX-2+/p53+ tumors were significantly less common in MSI-H CIMP-high tumors (9.7%) than in non-MSI-H CIMP-low/CIMP-0 tumors (44–47%; P< .0001). In conclusion, COX-2 and p53 alterations were synergistically inversely correlated with both MSI-H and CIMP-high. Our data suggest that a combined analysis of COX-2 and p53 may be more useful for the molecular classification of colorectal cancer than either COX-2 or p53 analysis alone.
Keywords: Colon cancer, cyclooxygenase-2, DNA methylation, MethyLight, TP53
Introduction
Transcriptional inactivation by cytosine methylation at promoter CpG islands of tumor-suppressor genes is thought to be an important mechanism in human carcinogenesis [1]. A number of tumor-suppressor genes, including CDKN2A (cyclin-dependent kinase inhibitor 2A; p16/INK4A), MGMT, MLH1, and others, are silenced by promoter methylation in colorectal cancer [2]. A subset of colorectal cancer cases exhibits promoter methylation in multiple genes, referred to as CpG island methylator phenotype (CIMP) [2–4]. CIMP+ colorectal tumors appear to have a distinct clinical and molecular profile, including associations with the female sex, proximal tumor location, high frequencies of microsatellite instability (MSI), and BRAF mutations [4–12]. Promoter CpG island methylation has been shown to occur early in colorectal carcinogenesis [13–15].
Cyclooxygenase-2 (COX-2 or PTGS2, the HUGO-approved gene symbol) has been considered to be important in colorectal carcinogenesis [16] and has been shown to be expressed in approximately 70% to 80% of colorectal cancer cases [17–19]. COX-2 overexpression is considered to be important in the early stage of colorectal cancer development [20]. A study showed that COX-2 overexpression was associated with poor prognosis [18], whereas another study showed that COX-2 expression was not correlated with poor survival [17]. The COX inhibitor aspirin has been known to decrease the risk for colorectal cancer [21], and the new COX-2-specific inhibitor celecoxib has been shown to inhibit the growth of colorectal cancer cells in vitro [22,23]. Thus, COX-2 is an attractive chemopreventive target [16,24,25]. COX-2 overexpression has been shown to be inversely associated with MSI-high (MSI-H) [26,27] and CIMP [28]. Aspirin has been shown to act through COX-independent mechanisms by increasing protein expression and subsequent apoptosis in mismatch repair [29]. COX-2 expression has been shown to be regulated by wild-type p53, suggesting that TP53 mutation may cause deregulation of COX-2 expression [30]. p53 mutations have also been shown to be inversely associated with MSI-H [31] and CIMP [4]. Although both COX-2 and p53 are important molecules in colorectal carcinogenesis and both CIMP and MSI appear to be important molecular features, no study to date has comprehensively examined interrelations between COX-2, p53, CIMP, and MSI in colorectal cancer. One way of dissecting these interrelations is to correlate the combined status of COX-2 and p53 expressions with the combined CIMP and MSI status.
In this study, using a quantitative real-time polymerase chain reaction (PCR; MethyLight) assay [11,32–35] and relatively unbiased samples of colorectal cancer from two large prospective cohort studies, we quantified DNA methylation in five CIMP-specific gene promoters and assessed the interrelations between COX-2, p53, MSI, and CIMP in colorectal cancer. In contrast to methylation-specific PCR (MSP), which has widely been used in previous studies [4–10,36], MethyLight can reliably distinguish high levels from low levels of DNA methylation, with the latter likely having little or no biologic significance [33,34,37].
Materials and Methods
Study Group
To recruit patients into this study, we used the databases of two large prospective cohort studies: the Nurses′ Health Study (n = 121,700 women), which has been in progress since 1976 and is managed by the Channing Laboratory at the Brigham and Women's Hospital (Boston, MA) [38], and the Health Professional Follow-up Study (n = 51,500 men), which has been in progress since 1986 and is managed by the Harvard School of Public Health (Boston, MA) [39]. Informed consent was obtained from all participants before inclusion in the cohorts. All cohort participants were free of cancer (except for nonmelanoma skin cancer) at the time of study entry. A subset of the cohort participants developed colorectal cancer during prospective follow-up. We excluded cases if adequate paraffin-embedded tumor tissue was not available for quantitative DNA methylation analysis at the time of this study, or if there were not enough tissue sections for the evaluation of COX-2 and p53 expression. As a result, 751 colorectal cancer cases (328 from the men's cohort and 423 from the women's cohort) were included in this study. Tissue collection and analyses were approved by the Institutional Review Boards of the Dana-Farber Cancer Institute and the Brigham and Women's Hospital.
Genomic DNA Extraction
To enrich tumor DNA, areas composed entirely of tumor were encircled with a pen on the coverslip of a hematoxylin and eosin (H&E)-stained tumor slide. Using the marked H&E slide as a guide, the tumor tissue was dissected manually from additional tissue sections by a sterile needle. Normal tissue was obtained from normal colorectal tissues at the margins of resection specimens. Dissected tissue was placed in buffered proteinase K solution at 56°C for 3 hours. Genomic DNA was extracted using QIAmp DNA Mini Kit (Qiagen, Valencia, CA), according to the manufacturer's instructions.
Quantitative Real-Time PCR for DNA Methylation (MethyLight)
Sodium bisulfite treatment on genomic DNA was performed as previously described [37]. Real-time PCR was performed as previously described to measure DNA methylation (MethyLight) [32–34]. We used ABI 7300 (Applied Biosystems, Foster City, CA) for quantitative real-time PCR. Using five sets of primers and probes, we amplified five CIMP-specific promoters [CACNA1G (calcium channel, voltage-dependent, T type α-1G subunit), CDKN2A (p16/INK4A), CRABP1 (cellular retinoic acid binding protein 1), MLH1, and NEUROG1 (neurogenin 1)] [11]. COL2A1 (the collagen 2A1 gene) was used to normalize for the amount of input bisulfite-converted DNA [11]. Primers and probes were previously described as follows: CACNA1G, CRABP1, and NEUROG1 [11]; CDKN2A and COL2A1 [34]; and MLH1 [37]. The percentage of methylated reference (PMR) at a specific locus was calculated by dividing the GENE:COL2A1 ratio of a sample by the GENE:COL2A1 ratio of SssI-treated human genomic DNA (presumably fully methylated) and by multiplying this value by 100 [32,33]. A PMR cutoff value of 4 was based on previously validated data [11,32–34,37]. The precision and performance characteristics of bisulfite conversion and subsequent MethyLight assays have been previously evaluated, and assays have been validated [37].
CIMP-high was defined as the presence of ≥ 4 methylated promoters among five gene promoters, including CACNA1G, CDKN2A, CRABP1, MLH1, and NEUROG1 [11]. CIMP-low was defined as the presence of one to three of five methylated promoters. CIMP-0 was defined as the absence of methylation in any of the five promoters. We have demonstrated that CIMP-low tumors appear to have features different from those of CIMP-high and CIMP-0 tumors (submitted for publication). We have previously validated the use of these five markers in our CIMP-specific panel, and all of the five markers showed high sensitivity (> 90%) and/or specificity (> 90%) for the prediction of CIMP status [11].
MSI Analysis
For microsatellite analyses, whole genome amplification of genomic DNA was performed by PCR using random 15-mer primers, as previously described [40]. Methods for MSI analysis were as previously described [19]. Briefly, the status of MSI was determined by analyzing the variability in the length of microsatellite markers from tumor DNA compared to that from normal DNA. In addition to the recommended MSI panel consisting of D2S123, D5S346, D17S250, BAT25, and BAT26, we used BAT40, D18S55, D18S56, D18S67, and D18S487 (i.e., a 10-marker panel). PCR and DNA fragment analysis for all of the markers, except for D2S123, D5S346, and D17S250, was performed in duplicate. A high degree of MSI (MSI-H) was defined as having instability in 30% or more of the markers. MSI-low (MSI-L) was defined as having instability in less than 30% of the markers, and microsatellite stability (MSS) was defined as having no unstable marker. We combined MSI-L and MSS tumors into one group designated as “MSI-LVMSS.”
Tissue Microarray (TMA) Construction
TMAs were constructed at the Dana-Farber/Harvard Cancer Center TMA Core Facility, as previously described [41,42], with some modifications. H&E-stained slides were reviewed by a pathologist (S.O.) to mark highly cellular portions to guide donor tissue core selection. TMAs were constructed using Automated Arrayer (Beecher Instruments, Sun Prairie, WI). Two 0.6-mm tissue cores each from tumor and normal colonic mucosa were placed in each TMA block. Each TMA block will have a total of approximately 400 cores (100 cases). A previous validation study demonstrated that the analysis of two disks was comparable to the analysis of the whole tissue section in more than 95% of cases and that proteins retained their antigenicity for > 60 years [43]. We analyzed whole tissue sections for cases in which there was not enough tumor tissue for TMAs, or for cases in which results were equivocal or indeterminate in TMAs.
Immunohistochemistry for COX-2 and p53
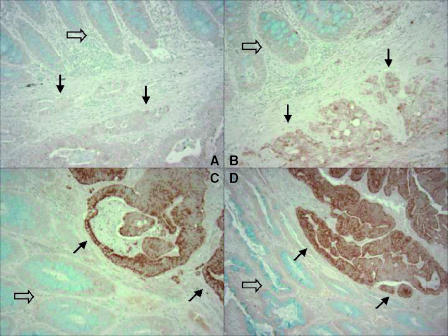
The methods for COX-2 and p53 immunohistochemistry were as previously described [19]. Using normal colonic mucosa as a reference, COX-2 expression was interpreted as negative (no overexpression compared to normal mucosa), weakly positive (1+), or strongly positive (2+) (Figure 1). Inflammatory cells served as internal positive control. Only strong and unequivocal nuclear staining for p53 in 50% or more of tumor cells was interpreted as p53+. Appropriate positive and negative controls were included in each run of COX-2 and p53 immunohistochemistry. All immunohistochemically stained slides were interpreted by a pathologist (S.O.) who was blinded from any other laboratory data.
Figure 1.
COX-2 immunohistochemistry in colorectal cancer. (A) No COX-2 overexpression (negative) in carcinoma (bottom, solid arrows) relative to normal mucosa (top, empty arrow). (B) Weak (1+) overexpression in carcinoma (bottom, solid arrows) relative to normal mucosa (top, empty arrow). (C) Strong (2+) overexpression in carcinoma (top right, solid arrows; normal mucosa at bottom left, empty arrow). (D) Strong (2+) overexpression in carcinoma (top right, solid arrows; normal mucosa at bottom left, empty arrow).
Statistical Analysis
In statistical analysis, chi-square analysis (or Fisher's exact test when any category was less than 10) was performed for categorical data, using the SAS program (version 9.1; SAS Institute, Cary, NC). All P values were two-sided, and statistical significance was set at P = .05.
Results
p53 and COX-2 Expressions and CIMP
We obtained 751 colorectal cancer specimens and quantified DNA methylation in a panel of the five CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, MLH1, and NEUROG1) [11] by MethyLight technology. Among the 751 colorectal cancer cases, 115 (15%) tumors were classified as CIMP-high (≥ 4 of 5 methylated promoters), 251 (33%) were classified as CIMP-low (1 to 3 of 5 methylated promoters), and the remaining 385 (51%) were classified as CIMP-0 (no methylated promoters). We also evaluated p53 expression by immunohistochemistry in the 751 colorectal cancer cases, with 321 (43%) showing positive staining. CIMP-high was significantly more common in p53- tumors (22%) than in p53+ tumors (6.5%; P < .0001) (Table 1).
Table 1.
Frequencies of CIMP-0, CIMP-Low, and CIMP-High in Colorectal Cancer Cases with Various COX-2 and p53 Statuses.
| Number of Methylated Promoters | CIMP-Low (1–3) [n(%)] | CIMP-High (≥ 4)[n (%)] | ||||||
| 0 (CIMP-0) [n(%)] | 1(n) | 2(n) | 3(n) | 4(n) | 5(n) | |||
| All cases (n = 751) | 385 (51) | 131 | 77 | 43 | 45 | 70 | 251 (33) | 115 (15) |
| p53+ tumors (n = 321) | 179 (56) | 76 | 29 | 16 | 9 | 12 | 121 (38) | 21 (6.5)* |
| p53- tumors (n = 430) | 206 (48) | 55 | 48 | 27 | 36 | 58 | 130 (30) | 94 (22)* |
| COX-2 strongly positive (2+) tumors (n = 398) | 228 (57) | 72 | 36 | 15 | 21 | 26 | 123 (31) | 47 (12)† |
| COX-2 weakly positive (1+) tumors)n = 114) | 59 (52) | 18 | 12 | 5 | 13 | 7 | 35 (31) | 20 (17) |
| COX-2- tumors (n = 99) | 35 (35) | 17 | 16 | 8 | 5 | 18 | 41 (41) | 23 (23)† |
p53+ vs p53- (P < .0001).
Negative COX-2 vs strongly positive (2+) COX-2 (P = .003).
Among 611 tumors in which COX-2 expression in tumors relative to normal mucosa could be evaluated, 398 (65%) tumors strongly (2+) overexpressed COX-2, 114 (19%) tumors weakly (1+) overexpressed COX-2, and 99 (16%) tumors were negative for COX-2 overexpression. CIMP-high was significantly more common in COX-2- tumors (23%) than in COX-2 strongly positive (2+) tumors (12%; P = .003) (Table 1) and COX-2+ (1+ or 2+) tumors (13%; P = .009).
Combined p53/COX-2 Status, CIMP, and MSI
Because of an important role of p53 in regulating COX-2 expression, we correlated the combined status of p53 and COX-2 expressions with CIMP in colorectal cancer cases (Table 2). Among 544 tumors for which we had data on COX-2, p53, and CIMP, there was a significant positive correlation between COX-2 and p53 expressions (P = .0003). CIMP-high was significantly less common in COX-2+/p53+ tumors (4.6%) than in COX-2+/p53- tumors (19%; P < .0001), COX-2-/p53+ tumors (17%; P = .04), and COX-2-/p53- tumors (28%; P < .0001), indicating a synergistic effect of p53 and COX-2 positivity on decreasing CIMP-high frequency. There was a trend toward higher frequencies of CIMP-high among women's tumors than among men's tumors in each COX-2/p53 subgroup of tumors, although statistical significance was not reached in any of the COX-2/p53 subgroups (Table 2).
Table 2.
Frequencies of CIMP-High in Colorectal Cancer Cases with Various Combined COX-2/p53 Statuses.
| Tumors | Frequency of CIMP-High | ||
| Men and Women | Men | Women | |
| COX-2+/p53+ | 4.6% (10/218)*†‡ | 3.1% (3/96) | 5.7% (7/122) |
| COX-2+/p53- | 19% (46/237)* | 14% (14/103) | 23% (32/137) |
| COX-2-/p53+ | 17%(4/24)† | 10% (1/10) | 21% (3/14) |
| COX-2-/p53- | 28% (18/65)‡ | 17% (5/30) | 37% (13/35) |
COX-2+/p53+ vs COX-2+/p53- (P < .0001).
COX-2+/p53+ vs COX-2-/p53+ (P = .04).
COX-2+/p53+ vs COX-2-/p53- (P < .0001).
We also correlated the combined status of p53 and COX-2 with the MSI status of colorectal cancer cases. MSI-H was less common in COX-2+/p53+ tumors [11 of 209 (5.3%)] than in COX-2+/p53- tumors [42 of 233 (18%); P < .0001 ], COX-2-/p53+ tumors [3 of 23 (13%); P = .15], and COX-2-/p53- tumors [20 of 61 (33%); P < .0001], indicating a synergistic effect of p53 and COX-2 positivity on decreasing MSI-H frequency.
Six MSI/CIMP Subtypes of Colorectal Cancer and the Combined Status of COX-2/p53
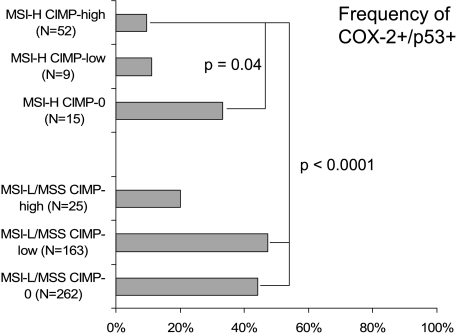
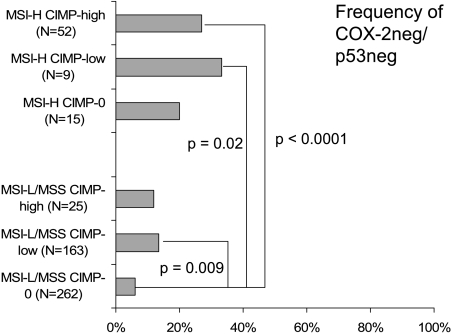
To analyze the effects of CIMP and MSI status on COX-2/p53, we subclassified colorectal cancer cases into six categories on the basis of CIMP and MSI status [i.e., MSI-H CIMP-high (n = 52), MSI-H CIMP-low (n = 9), MSI-H CIMP-0 (n = 15), MSI-L/MSS CIMP-high (n = 25), MSI-L/MSS CIMP-low (n = 163), and MSI-L/MSS CIMP-0 (n = 262)]. We assessed COX-2 and p53 expressions in each category. The frequencies of COX-2+/p53+ tumors were significantly less common in MSI-H CIMP-high tumors (9.6%) than in MSI-H CIMP-0 tumors (33%; P=.04), MSI-L/MSS CIMP-low tumors (47%; P < .0001), and MSI-L/MSS CIMP-0 tumors (44%; P < .0001) (Figure 2). The frequencies of COX-2+/p53+ tumors were also less common in MSI-H CIMP-low tumors (11 %) than in MSI-L/MSS CIMP-low tumors (47%; P= .04). In contrast, the frequencies of COX-2-/p53- tumors were significantly more common in MSI-H CIMP-high tumors (27%) than in MSI-L/MSS CIMP-low (14%; P = .02) and in MSI-L/MSS CIMP-0 tumors (6.1%; P < .0001) (Figure 3). The frequencies of COX-2-/p53- tumors were also more common in MSI-H CIMP-low tumors (33%) than in MSI-L/MSS CIMP-0 tumors (6.1%; P = .02). Therefore, MSI-H and CIMP-high were synergistically inversely associated with positivity for both COX-2 and p53.
Figure 2.
Frequencies of COX-2+/p53+ tumors in the six MSI/CIMP subtypes of colorectal cancer.
Figure 3.
Frequencies of COX-2-/p53- tumors in the six MSI/CIMP subtypes of colorectal cancer.
Discussion
We conducted this study to evaluate the interactions of COX-2 and p53 expressions and their relations with CIMP and MSI in colorectal cancer. We demonstrated that there were inverse correlations of each of the COX-2 and p53 overexpressions with CIMP-high, and that COX-2 and p53 overexpressions were synergistically inversely associated with both CIMP-high and MSI-H. Our data imply that CIMP-low/CIMP-0 tumors tend to activate the COX-2 pathway in part through mutations of p53, a COX-2 regulator, whereas CIMP-high tumors tend not to mutate the p53 gene or to overexpress COX-2. Our data also suggest that, compared to the use of COX-2 or p53 analysis alone, the use of combined immunohistochemical analysis for both COX-2 and p53 may better serve for the molecular classification of colorectal cancer. The apparent synergistic effect of COX-2 and p53 might also reflect the limitations of immunohistochemical assays in which false positives and false negatives can occur, and the combined analysis for COX-2 and p53 might select tumors that are caused by COX-2 activation, which can, in turn, be caused by mutations and functional loss of p53. It has been shown that the correlation of TP53 gene mutations and p53 positivity by immunohistochemistry is less than perfect [44]. Thus, the combination of COX-2 and p53 immunohistochemistry might be useful to decrease false positives and false negatives, or to select a more homogenous group of colorectal cancer cases.
We used quantitative real-time PCR (MethyLight) to distinguish high levels from low levels of DNA methylation. Assays to measure DNA methylation may be potentially very useful in clinical practice because many tumor-suppressor genes have been shown to be methylated and functionally silenced in a variety of human neoplasias [1] DNA methylation may be a useful marker for predicting prognosis, for monitoring the efficacy of adjuvant therapy in cancer patients [45], and for performing risk assessment in the surveillance of high-risk or low-risk individuals [46,47]. Previous studies on CIMP in colorectal cancer [3,6–8,12] have primarily used methylation-specific PCR (MSP). MSP is a qualitative assay and cannot reliably distinguish high levels from very low levels of methylation. Using quantitative MethyLight assays, we have shown that low levels of promoter methylation do not typically silence gene expression, suggesting that low levels of DNA methylation have no or little biologic significance [37]. Studies using MSP may overestimate the frequency of methylation positivity in a given methylation marker, hence the frequency of CIMP. In our study, the use of quantitative DNA methylation assays and relatively unbiased samples of colorectal cancer from two large prospective cohorts has enabled us to precisely estimate the frequency of colorectal cancer cases with a specific molecular feature (i.e., CIMP, MSI-H, or Cox-2+) at a population level.
The mechanisms of COX-2 regulation are still largely unknown. COX-2 expression has previously been shown to be repressed by wild-type p53 [30], although p53 is unlikely the only regulator of COX-2. Thus, tumors with wild-type p53 may have a lower frequency of COX-2 overexpression. Indeed, our data support this hypothesis. The frequency of COX-2- tumors was higher among p53- tumors than among p53+ tumors, and COX-2 and p53 statuses were positively correlated with each other. For therapeutic approaches targeting the COX-2 pathway, it may be important to consider the effects of wild-type or mutant p53, potentially modulating signaling through the COX-2 pathway. Further study is necessary to investigate the mechanisms of COX-2 regulation by p53.
Our data may have significant clinical implications because of the emerging importance of both COX-2 and DNA methylation as promising chemotherapeutic/chemopreventive targets. Previous studies demonstrated that MSI-H tumors, whether in sporadic or familial setting, are inversely associated with COX-2 overexpression [26,27]. In addition, our data suggest a synergistic effect of CIMP and MSI-H on lowering the frequency of COX-2 and p53 overexpression in colorectal cancer. Elucidating the molecular mechanisms of COX-2 overexpression and its action in COX-2-overexpressed tumors and the alternative mechanisms that may bypass COX-2 overfunction in COX-2- tumors is important for the purpose of developing molecularly targeted treatments against colorectal cancer.
The prognostic significance of CIMP stratified by COX-2 or p53 status has not been studied. Hawkins et al. [6] showed that patients with MSS/CIMP tumors experienced worse survival compared to patients with either MSS/non-CIMP tumors or MSI-H tumors. However, their data may have been confounded by the fact that, in their study, CIMP tumors were associated with advance stage at initial presentation [6]. In contrast, Van Rijnsoever et al. [5] demonstrated that CIMP positivity conferred improved survival among patients who received 5-fluorouracil-based adjuvant chemotherapy for stage III colorectal cancer cases. We have previously examined promoter methylation in 25 of 34 advanced colorectal cancer cases in phase I/II clinical trials of combination chemotherapy with gefitinib (an epidermal growth factor receptor tyrosine kinase inhibitor) [48], but we found no CIMP-high tumors (unpublished data). We found that combined positivity for both p21 and p53, but not COX-2 status, was a significant predictor of tumor resistance to combination chemotherapy with gefitinib [48]. Further studies are necessary to determine whether CIMP stratified by COX-2 or p53 status is significantly associated with improved or worse patient outcomes. Our prospective cohort studies, the Nurses' Health Study (n = 120,000; followed since 1976) [38] and the Health Professional Follow-up Study (n = 51,000; followed since 1986) [39], are currently ongoing. Thus, relational data on patient survival and CIMP will be available in the future.
In conclusion, COX-2 and p53 overexpression exhibits synergistic inverse correlations with CIMP-high and MSI-H. Our data suggest that a combined analysis of COX-2 and p53 may be more useful for the molecular classification of colorectal cancer than either COX-2 or p53 analysis alone.
Acknowledgements
We deeply thank the Nurses' Health Study and the Health Professional Follow-up Study cohort participants who generously agreed to provide us with biologic specimens and information through responses to questionnaires. We thank Graham Colditz, Walter Willett, and many other staff members who implemented and maintained the cohort studies. We thank deeply Peter Laird, Daniel Weisenberger, and Mihaela Campan for assisting in the development of MethyLight assays.
Abbreviations
- CACNA1G
calcium channel, voltage-dependent, T type α-1G subunit
- CDKN2A
cyclin-dependent kinase inhibitor 2A (p16/INK4A)
- CIMP
CpG island methylator phenotype
- COX-2
cyclooxygenase-2
- CRABP1
cellular retinoicacid binding protein 1
- MSI
microsatellite instability
- MSI-H
microsatellite instability—high
- MSI-L
microsatellite instability—low
- MSS
microsatellite stability
- NEUROG1
neurogenin 1
- PMR
percentage of methylated reference (degree of DNA methylation)
Footnotes
This work was supported by National Institutes of Health grants P01 CA87969-03 and P01 CA55075-13. No conflict of interest exists.
References
- 1.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. (Spec No 1) [DOI] [PubMed] [Google Scholar]
- 2.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 3.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 6.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 7.Samowitz W, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Kambara T, Simms LA, Whitehall VLJ, Spring KJ, Wynter CVA, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 10.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 11.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterized by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006 doi: 10.1136/gut.2005.082933. (in press (published online first on January 11, 2006; doi:2010.1136/gut.2005.082933)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan AO-O, Broaddus RR, Houlihan PS, Issa J-PJ, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- 15.Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JR, DuBois RN. COX-2: a molecular target for colorectal cancer prevention. J Clin Oncol. 2005;23:2840–2855. doi: 10.1200/JCO.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 17.Fux R, Schwab M, Thon KP, Gleiter CH, Fritz P. Cyclooxygenase-2 expression in human colorectal cancer is unrelated to overall patient survival. Clin Cancer Res. 2005;11:4754–4760. doi: 10.1158/1078-0432.CCR-04-2586. [DOI] [PubMed] [Google Scholar]
- 18.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10:8465–8471. doi: 10.1158/1078-0432.CCR-04-0653. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 20.Sheehan KM, O'Connell F, O'Grady A, Conroy RM, Leader MB, Byrne MF, Murray FE, Kay EW. The relationship between cyclooxygenase-2 expression and characteristics of malignant transformation in human colorectal adenomas. Eur J Gastroenterol Hepatol. 2004;16:619–625. doi: 10.1097/00042737-200406000-00017. [DOI] [PubMed] [Google Scholar]
- 21.Giovannucci E, Egan KM, Hunter DJ, Stampfer MJ, Colditz GA, Willett WC, Speizer FE. Aspirin and the risk of colorectal cancer in women [see comments] N Engl J Med. 1995;333:609–614. doi: 10.1056/NEJM199509073331001. [DOI] [PubMed] [Google Scholar]
- 22.Kazanov D, Dvory-Sobol H, Pick M, Liberman E, Strier L, Choen-Noyman E, Deutsch V, Kunik T, Arber N. Celecoxib but not rofecoxib inhibits the growth of transformed cells in vitro. Clin Cancer Res. 2004;10:267–271. doi: 10.1158/1078-0432.ccr-0412-3. [DOI] [PubMed] [Google Scholar]
- 23.Lev-Ari S, Strier L, Kazanov D, Madar-Shapiro L, Dvory-Sobol H, Pinchuk I, Marian B, Lichtenberg D, Arber N. Celecoxib and curcumin synergistically inhibit the growth of colorectal cancer cells. Clin Cancer Res. 2005;11:6738–6744. doi: 10.1158/1078-0432.CCR-05-0171. [DOI] [PubMed] [Google Scholar]
- 24.Dannenberg AJ, Lippman SM, Mann JR, Subbaramaiah K, DuBois RN. Cyclooxygenase-2 and epidermal growth factor receptor: pharmacologic targets for chemoprevention. J Clin Oncol. 2005;23:254–266. doi: 10.1200/JCO.2005.09.112. [DOI] [PubMed] [Google Scholar]
- 25.Samoha S, Arber N. Cyclooxygenase-2 inhibition prevents colorectal cancer: from the bench to the bed side. Oncology. 2005;69(1):33–37. doi: 10.1159/000086630. [DOI] [PubMed] [Google Scholar]
- 26.Sinicrope FA, Lemoine M, Xi L, Lynch PM, Cleary KR, Shen Y, Frazier ML. Reduced expression of cyclooxygenase 2 proteins in hereditary nonpolyposis colorectal cancers relative to sporadic cancers. Gastroenterology. 1999;117:350–358. doi: 10.1053/gast.1999.0029900350. [DOI] [PubMed] [Google Scholar]
- 27.Karnes WE, Jr, Shattuck-Brandt R, Burgart LJ, DuBois RN, Tester DJ, Cunningham JM, Kim CY, McDonnell SK, Schaid DJ, Thibodeau SN. Reduced COX-2 protein in colorectal cancer with defective mismatch repair. Cancer Res. 1998;58:5473–5477. [PubMed] [Google Scholar]
- 28.Toyota M, Shen L, Ohe-Toyota M, Hamilton SR, Sinicrope FA, Issa JP. Aberrant methylation of the cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 2000;60:4044–4048. [PubMed] [Google Scholar]
- 29.Goel A, Chang DK, Ricciardiello L, Gasche C, Boland CR. A novel mechanism for aspirin-mediated growth inhibition of human colon cancer cells. Clin Cancer Res. 2003;9:383–390. [PubMed] [Google Scholar]
- 30.Subbaramaiah K, Altorki N, Chung WJ, Mestre JR, Sampat A, Dannenberg AJ. Inhibition of cyclooxygenase-2 gene expression by p53. J Biol Chem. 1999;274:10911–10915. doi: 10.1074/jbc.274.16.10911. [DOI] [PubMed] [Google Scholar]
- 31.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 33.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 35.Ebert MP, Mooney SH, Tonnes-Priddy L, Lograsso J, Hoffmann J, Chen J, Rocken C, Schulz HU, Malfertheiner P, Lofton-Day C. Hypermethylation of the TPEF/HPP1 gene in primary and metastatic colorectal cancers. Neoplasia. 2005;7:771–778. doi: 10.1593/neo.05235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anacleto C, Leopoldino AM, Rossi B, Soares FA, Lopes A, Rocha JC, Caballero O, Camargo AA, Simpson AJ, Pena SD. Colorectal cancer “methylator phenotype”: fact or artifact? Neoplasia. 2005;7:331–335. doi: 10.1593/neo.04502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 39.Wei EK, Giovannucci E, Fuchs CS, Willett WC, Mantzoros CS. Low plasma adiponectin levels and risk of colorectal cancer in men: a prospective study. J Natl Cancer Inst. 2005;97:1688–1694. doi: 10.1093/jnci/dji376. [DOI] [PubMed] [Google Scholar]
- 40.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. Alphamethylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 42.Rubin MA. Use of laser capture microdissection, cDNA microarrays, and tissue microarrays in advancing our understanding of prostate cancer. J Pathol. 2001;195:80–86. doi: 10.1002/path.892. [DOI] [PubMed] [Google Scholar]
- 43.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000;80:1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 44.Curtin K, Slattery ML, Holubkov R, Edwards S, Holden JA, Samowitz WS. p53 alterations in colon tumors: a comparison of SSCP/sequencing and immunohistochemistry. Appl Immunohistochem Mol Morphol. 2004;12:380–386. doi: 10.1097/00129039-200412000-00017. [DOI] [PubMed] [Google Scholar]
- 45.Muller HM, Millinger S, Fiegl H, Goebel G, Ivarsson L, Widschwendter A, Muller-Holzner E, Marth C, Widschwendter M. Analysis of methylated genes in peritoneal fluids of ovarian cancer patients: a new prognostic tool. Clin Chem. 2004;50:2171–2173. doi: 10.1373/clinchem.2004.034090. [DOI] [PubMed] [Google Scholar]
- 46.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan S, Krouse RS, Prasad AR, Einspahr J, et al. MGMT promoter methylation and the field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 47.Giovannucci E, Ogino S. DNA methylation, field effects, and colorectal cancer. J Natl Cancer Inst. 2005;97:1317–1319. doi: 10.1093/jnci/dji305. [DOI] [PubMed] [Google Scholar]
- 48.Ogino S, Meyerhardt JA, Cantor M, Brahmandam M, Clark JW, Namgyal C, Kawasaki T, Kinsella K, Michelini AL, Enzinger PC, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]