Abstract
We recently showed that adenoviral transfer and expression of the Lpsd/Ran gene isolated from endotoxin-resistant C3H/HeJ mice could protect endotoxin-sensitive mice from endotoxic shock. Elevation of proinflammatory cytokines, such as tumor necrosis factor α (TNF-α), is thought to be essential for the development of septic shock. To investigate the extent to which Lpsd/Ran affects TNF-α production, we transduced primary macrophages from endotoxin-sensitive and -resistant mice with adenoviral vectors expressing the wild-type and the mutant Lps/Ran cDNAs and other control genes, and compared the amount of TNF-α produced by these various transduced macrophages. Successful transfer and expression of Lpsd/Ran cDNA in endotoxin-sensitive C3H/HeOuJ macrophages reduced TNF-α production upon lipopolysaccharide (LPS) stimulation, as compared with macrophages transduced with vectors expressing the wild-type Lpsn/Ran cDNA, the green fluorescent protein gene, or the lacZ gene. On the other hand, successful transfer and expression of the wild-type Lpsn/Ran cDNA in primary macrophages from endotoxin-resistant C3H/HeJ mice failed to induce TNF-α production to any significant extent unless a very high LPS concentration was used. Given our previous demonstration that Lpsn/Ran functions effectively in restoring LPS responsiveness in B cells from C3H/HeJ mice, we conclude that Lps/Ran is involved in a CD14-independent signal transduction pathway. This dominant negative down-regulation by Lpsd/Ran on TNF-α production by macrophages and probably other innate immune responses may be key to the development of an effective gene therapy for endotoxic or septic shock.
Keywords: gene therapy, septic shock, Ran GTPase, adenovirus
Endotoxin lipopolysaccharide (LPS), which is a major component of Gram-negative bacteria, can cause endotoxic shock and death in humans. At the cellular level, activation of macrophages by endotoxin is one of the key events that results in the production of a number of proinflammatory cytokines. Among the major proinflammatory cytokines involved in endotoxic or septic shock is tumor necrosis factor α (TNF-α) (1–4). Mice deficient in the 55-kDa TNF-α receptor are resistant to endotoxic shock (5). Passive immunization with anti-TNF-α antiserum or soluble recombinant human TNF receptor-Ig chimeric proteins protects mice against lethal endotoxin challenges (1). Despite these findings, a number of clinical trials based on these and other related findings were not successful (6). Thus, although TNF-α is an important hallmark in endotoxemia or sepsis, several factors in probably more than one signal-transduction pathway are likely to be involved in endotoxic or septic shock (7).
C3H/HeJ inbred mice have a natural resistance to endotoxin challenge at a dose that is lethal to other inbred strains of mice (8). The original experiments used to define the type of genetic inheritance governing the LPS response in mice were conducted by using the mitogenic assay for B lymphocytes, whose response to endotoxin stimulation is mediated through a CD14-independent pathway (7, 9–11). By means of this B cell mitogenic assay, the consensus from several studies is that a pair of autosomal codominant alleles controlled the mitogenic response of B cells to LPS in mice (9–11). The results of more investigations suggest that a Lps locus was on chromosome 4 and was linked to the major urinary protein locus (Mup-1) but downstream from Mup-1 and the Lyb2:4:6 genes (12–14). Recent genetic mapping analyses show that the Tlr4 gene is located within this Lps locus, and a missense point mutation is found in the coding region at position 712 of the Tlr4 gene from C3H/HeJ mice (15, 16). However, no essential functional reconstitution data by which this defect exerts its profound biological effects have been published to date. The importance of Tlr4 in innate immunity is beyond doubt. Whether or not the Tlr4d gene is the HeJ Lpsd gene remains unclear. More recent investigations complicate even further the implication that the Tlr4d gene is the HeJ Lpsd gene. Introduction of Tlr4 into cell lines did not confer the ability to respond to LPS but constitutively activated NF-κB, and additional molecules are required for the induction of LPS response (17, 18). Furthermore, Vogel et al. (19) studied endotoxin response by measuring TNF-α production after LPS stimulation and showed that F1 progeny (Lps0 × Lpsd) from crosses between Tlr4null (Lps0 × Lps0) and C3H/HeJ (Lpsd × Lpsd) mice had a serum TNF-α with an average value of 1,000–1,100 units/ml. The same amount of serum TNF-α was obtained from F1 progeny (Lpsn × Lpsd) from crosses between a responder strain (Lpsn × Lpsn) and C3H/HeJ. Vogel et al. concluded that the HeJ defect exerts a dominant negative effect on LPS sensitivity and that Tlr4 apparently is not required.
Using a functional cDNA cloning strategy, we previously isolated a cDNA whose expression in B cells from C3H/HeJ mice enables them to proliferate and differentiate in the presence of a high dose of LPS, giving rise to plaque formation (20). This cDNA is identical to a gene encoding for Ran GTPase. We subsequently sequenced the cDNA of its counterpart from the C3H/HeJ genome and found that the two cDNA sequences are identical except at position 870 of the 3′ untranslated region, where a thymidine of the wild-type cDNA has been replaced by a cytidine in the C3H/HeJ cDNA (21). By studying them in parallel, we showed that the Lpsn/Ran, but not Lpsd/Ran, could restore LPS-mediated proliferation of B cells from C3H/HeJ mice, and that Lpsd/Ran could protect sensitive mice against endotoxin challenge (21).
Materials and Methods
Construction of Adenoviral Vectors.
For the construction of Ad5-green fluorescent protein (GFP) vector, pEGFP-1 (CLONTECH) was digested with BamHI and NotI to release the 800-bp GFP fragment, which then inserted into corresponding sites of the expression vector pcDNA3 (Invitrogen). The expression cassette containing the cytomegalovirus (CMV), the GFP, and the bovine growth hormone polyadenylation signal was released by Bgl2 and PvuII digestion and then inserted into the E1 region of Ad5. For the construction of Ad5-lacZ vector, pCMVβ (CLONTECH) was digested with EcoRI and SalI, and the expression cassette (4.5 kb) containing CMV promoter, lacZ, and simian virus 40 polyadenylation signal then was inserted into E1 region of Ad5 vector.
Viral Titer Determination.
For each virus, we prepared a large-scale virus stock as described (22). Briefly, subconfluent 293 human embryonic kidney cells (American Type Culture Collection), cultured in D10 medium (DMEM supplemented with 10% FCS), were infected with a diluted adenovirus stock at a multiplicity of infection (MOI) of about 100:1. Typically, cytopathic effect would be evident in 4 days, defined by the appearance of many nonadherent cells from a monolayer of adherent cells. The infected cells were collected and lysed in PBS with 1/10 vol of 5% sodium deoxycholate. The lysate then was sheared by means of a probe-type homogenizer. Next, the cell extracts were centrifuged at 12,000 g for 10 min to pellet the cell debris. CsCl banding twice purified virus particles in the clear suspension. The twice-banded virus solution then was dialyzed for 6 hr, at 4°C, against two changes of 100 vol of 10 mM Tris⋅HCl, pH 8.0, and PBS, pH 7.4. After dialysis, the virus solution then was filtered through a 0.4-μm filter, aliquoted, and stored at −80°C until use. For determining virus titer by cytopathic assay, we followed the method of Nyberg-Hoffman et al. (23). Briefly, 293 cells were seeded in 96-well plates at 1 × 104 cells per well. The next day, cells were infected with various dilutions of a particular virus stock, which ranged from 1 × 10−9 to 1 × 10−12. The plates were incubated for 7–10 days. Typically, while the control wells did not exhibit cytopathic effect in terms of visible clear plaques, the test wells exhibited plaques with increase frequency at lower dilutions. The titer was calculated by multiplying the number of wells with obvious cytopathic effect at the highest dilution and dividing it by the total volume of viral supernatant used for testing. Each titration was repeated thrice.
Virus Infection of Primary Macrophages.
To infect primary macrophages, we prepared various adenovirus stocks carrying one of the following genes: Lpsn/Ran-HA, Lpsd/Ran-HA, GFP, and lacZ. Virus titers were normalized and adjusted as described in Fig. 2. For infection, 2.5 × 105 peritoneal macrophages from C3H/HeOuJ mice, primed with 3 ml of sodium thioglycollate 3 days previously, were seeded in each well of a 24-well plate. The infection MOI was 100,000:1 adjusted with R10 (RPMI 1640 with 10% FBS) to final volume of 250 ml per well, and the infection time was 6 hr. At the end of 6 hr, the spent viral supernatant was replaced with fresh R10 medium, and the plate was left incubated for an additional 14 hr at 37°C. At this time, 250 μl of either R10 medium or LPS from Salmonella typhimurium (final concentration = 1 μg/ml) was added to the cultures. Another 2 hr later, the supernatant from each well was harvested and stored at −70°C until needed for TNF-α determination as measured by ELISA or bioactivity assay. Each sample was a pool of three different wells. Details of the ELISA assay or the bioactivity assay have been described (24–26).
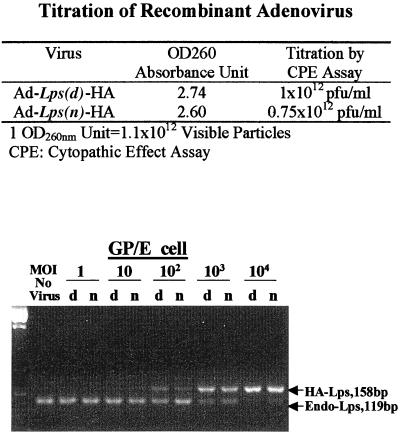
Figure 2.
Adenoviral titer determination. Cytopathic effect assay was performed as described in Materials and Methods. pfu, plaque-forming unit. (Lower) PCR results on GP/E fibroblasts with or without infection with various dilutions of either Ad5-Lpsn/Ran (n) or Ad5-Lpsd/Ran (d) virus. MOI is a ratio of 1 cell/number of infectious virus particles. The 127-bp band shows the molecular weight visible upon brighter exposure.
PCR on DNA from Adenovirus-Infected Cells.
For DNA extraction, 250,000 cells were seeded into each well of a 24-well plate and were infected with Ad5-Lpsd/Ran-hemagglutinin (HA) or with Ad5-Lpsn/Ran-HA virus at a different MOI. Infection time was 1 hr, and infection volume was 0.2 ml. After infection, we removed the viral supernatant, added 1 ml of R10 into each well, and incubated the plate for an additional 20 hr, at 37°C. Before harvest, the wells were washed thrice with PBS, pH 7.4, and once by PBS, pH 4.5. We next added 100 μl per well of DNAzol reagent and followed the procedure described by the manufacturer (GIBCO/BRL). About 0.1 μg of genomic DNA was used in each PCR. Both primers were specific for Lps/Ran sequence. Sense primer sequence was 5′-TTGTTGCCAT,GCCTGCTCTT,G-3′, and antisense primer sequence was 5′-GGTCATCATC,CTCATCTGGG,A-3′. For 30-cycle PCR, denaturation was 95°C for 5 min; annealing was 60°C for 30 sec; and extension was 72°C for 30 sec. The extension time in the last cycle was 72°C for 10 min. One-tenth of the PCR products were analyzed on a 3% agarose gel.
Reverse Transcription–PCR (RT-PCR) on RNA of Adenoviral-Infected Culture.
For RT-PCR, the cells used and infection conditions were the same as for PCR. Total RNA extraction was obtained by using Trizol reagents (GIBCO/BRL). RNA amount was normalized by OD260/280 and gel analysis. About 1 μg of RNA each was reverse-transcripted, and one-twentieth of DNA products were used in subsequent PCRs. The same sense and antisense primer sequences, as well as reaction conditions as described above, were used.
Western Blot Analysis.
The harvested macrophages from primed mice were infected with various adenovirus stocks as described above except at an MOI of 10,000:1 and stimulated with LPS for 24 hr. After 24 hr, the cells from various cultures were trypsinized, washed once, and lysed in lysis buffer (20 mM Tris⋅HCl, pH 7.5/5 mM EDTA/130 mM NaCl/1% Triton X-100/complete protease inhibitor mixture) at a concentration of 100 μl buffer per 1 million cells. About 5 μg of total lysate was loaded per lane in 12% SDS/PAGE. The gel then was electrophoresed and transferred onto a poly(vinylidene difluoride) membrane. The membrane then was rinsed briefly with TBST buffer (25 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.05% Tween 20), then incubated in 1% blocking buffer (Roche) for 1 hr with gentle shaking. The membrane was further incubated with the first antibody, which was either anti-HA antibody (Roche), anti-GFP antibody (CLONTECH), or antitubulin (Calbiochem). It then was incubated with biotinylated second antibodies. Finally, it was incubated with 25 milliunits/ml streptavidin-peroxidase conjugate. After the last incubation, chemiluminescent substrate luminal/iodophenol (Roche) was added per the manufacturer's instruction. The membrane was exposed to x-ray films for 3 sec to 10 min. The filters usually were stripped and rehybridized with other antibodies.
TNF-α ELISA Assay.
The procedure was done as described (24–26). To prepare the ELISA plate, we coated a Nunc microwell plate with 50 μl per well of the capture antibody, rat monoclonal anti-mouse TNF-α antibody (PharMingen) at a concentration of 4 μg/ml. We sealed and kept the plate at 4°C overnight. The next morning, we washed the wells with washing buffer (PBS, pH 7.4, containing 0.05% Tween-20) twice by flipping. Next we blocked the plate, which included the addition of 200 μl of PBS containing 1% BSA to each well for 1 hr at 37°C. We then aspirated the blocking solution. After two washes with washing buffer, we added 50 μl of test supernatant to each well in triplicate. For each assay, a TNF-α standard curve was established, by using recombinant mouse TNF-α purchased from PharMingen. Various concentrations of TNF-α standards ranging from 20 pg/ml to 5,000 pg/ml diluted in R10 medium were included in the standard curve. The detection antibody was biotinylated rat monoclonal anti-mouse TNF-α antibody (PharMingen). Fifty microliters of 1 μg/ml antibody was added to each well, and the plate was incubated at room temperature for 3–4 hr and washed thrice. Next, we added 50 μl of 1:1,000 diluted streptavidin-conjugated horseradish peroxidase (PharMingen) to each well. After another incubation of 1 hr at room temperature, the plate was washed thrice. We then added 50 μl of freshly prepared developing solution, prepared by dissolving one ABTS tablet (Roche) into 50 ml of ABTS buffer (Roche). After 30 min of color development, we read the OD405 of each sample by using the microplate reader. Using the OD405 values of the standards, we extrapolated the actual amount of TNF-α in each well.
TNF-α Bioactivity Assay.
The procedure was similar to those published previously (24–26). Briefly, we first inactivated the transcriptional activity of L929 cells with actinomycin D treatment. This was done by mixing 5 × 105/ml L929 cells (American Type Culture Collection) with 2 μg/ml actinomycin D (Sigma). We then aliquoted 100 μl of the mixture into each well of a 96-well plate (final concentration = 5 × 104 cells/well) and incubated it at 37°C for 1 hr. Next we added 100 μl of test supernatant or R10 (as control) to each well, in triplicate, and incubated the plate at 37°C for 20 hr. We then added 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Sigma) to each well, and incubated the plate at 37°C for another 4 hr. Next, we aspirated the spent solution in the wells and added 100 μl of isopropanol in 0.04 M HCl. After dissolving the dark blue formazan crystals, we determined the OD570/690 values by the microplate reader. Using these values, we then determined the percentage of dead cells, which is equal to 1 − [OD570/690 of test supernatant/OD570/690 of controls] × 100%.
Results
As TNF-α is a major proinflammatory cytokine in endotoxic shock, one possible way Lpsd/Ran exerts its protective effect against endotoxin challenge in sensitive mice could be via down-modulating TNF-α production. To test this hypothesis, we generated a number of adenovirus vectors, each of which carries either the wild-type or the mutant Lps/Ran cDNA, a GFP gene, or a lacZ gene as controls (Fig. 1). We also found the same amount of Ran protein in endotoxin-sensitive cells as in C3H/HeJ cells, which probably is the result of the expression of more than one Ran isoform gene, and each of these isoform genes may have distinct biological function (27, 28). To circumvent this potential complication, we inserted a 40-base HA-epitope sequence in position 527 of the Lpsn/Ran cDNA and Lpsd/Ran cDNA. This design enabled us to directly measure the presence of the foreign Lps/Ran cDNA in the transduced cells and its expression at both the RNA and protein levels over a background of Ran-isoform genes and their products. Also, its copy number and expression can be compared quantitatively with those of endogenous Ran. This is achieved through PCR or RT-PCR by a design where the reactions include the use of Lps/Ran sequence primers specific for and therefore competed by both the HA-tagged Lps/Ran and the endogenous Ran sequences.
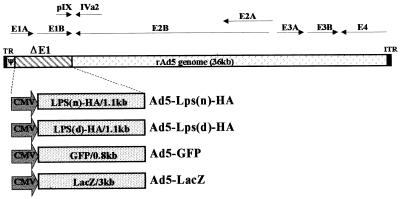
Figure 1.
Generation of recombinant adenoviral vectors. Depicted are the recombinant adenovirus type 5 (Ad5) genome. Ad5-Lps(n)-HA and Ad5-Lps(d)-HA are the vectors containing the Lps(d)/Ran cDNA and Lps(d)/Ran cDNAs tagged with an HA-epitoped in-frame with the codon. Ad5-GFP and Ad5-LacZ are vectors containing the GFP and lacZ genes. Expression cassettes replacing the E1 region include the cytomegalovirus promoter, the inserted cDNAs with their own polyadenylation signals, flanked by 5′ inverted terminal repeat (ITR), and encapsidation signal (ψ).
To increase the validity of our results, we worked on primary macrophages from LPS responder C3H/HeOuJ mice and hyporesponder C3H/HeJ mice. The use of adenovirus vectors provides high efficiency of gene transfer but, as evident from a recent report (29), standardization of the technology in infectivity and virus titer is still required. To normalize the efficiency of gene transfer using various virus stocks, we compared the OD values and the virus titers as determined by the cytopathic effect assay (23) with PCR results on DNA extracted from GP/E fibroblasts infected with varying dilutions of the adenovirus stocks. As shown in Fig. 2, an MOI of 100 to 1,000 for both Ad5-Lpsn/Ran or Ad5-Lpsd/Ran could achieve a near 100% efficiency on GP/E fibroblasts. This result is consistent with Ad5-GFP infection, where similar dilutions based on the titer determined by all three methods provided a near 100% of the cells infected when an MOI of 100 was used. As shown in Fig. 2, all three methods showed that Ad5-Lpsn/Ran has a lower titer compared with Ad5-Lpsd/Ran. Based on the competitive PCR results, where the intensity of the 158-bp HA-tagged Lps(d) band was about 1.5 times that of HA-tagged Lps(n) band, we diluted the Ad5-Lpsd/Ran accordingly so that the titer for both viruses was the same for all subsequent experiments.
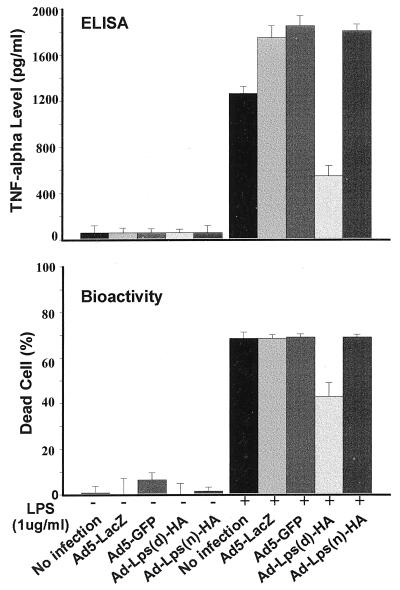
To assess the biological effects of the Lps/Ran genes, we infected C3H/HeOuJ peritoneal macrophages with Ad5-Lps/Ran viruses and other control viruses. After a typical 6-hr infection and another 14-hr incubation to permit a predetermined optimal time for the expression of the transgenes, the culture was replaced with fresh medium, with or without 1 μg/ml LPS. Typically, after another 2 hr of incubation, the supernatants were collected and analyzed either by ELISA or cytotoxicity assay. The latter assay was to measure the bioactivity of TNF-α in the supernatant (24). In all 10 experiments conducted, there was a consistent reduction of TNF-α level or activity in macrophages transduced with Ad5-Lps(d)-HA upon LPS stimulation, as compared with those transduced with either Ad5-Lps(n)-HA, Ad5-GFP, or Ad5-lacZ. By ELISA, which measures the overall TNF-α protein, there was a 50–70% reduction, and by bioactivity, which measures TNF-α biological activity, there was a 30% reduction (Fig. 3). To eliminate possible artifacts that this reduction could be the result of epitope sequence insertion into the Lpsd/Ran cDNA, we also infected macrophages with the original wild-type Ad5-Lpsn/Ran and the original mutant Ad5-Lpsd/Ran virus, as we reported recently (21). Similar results were obtained in three independent experiments (not shown).
Figure 3.
Down-modulation of TNF-α production by C3H/HeOuJ macrophages after Lpsd/Ran adenoviral transfer. Primary macrophages from sodium thioglycollate-primed C3H/HeOuJ mice were seeded (250,000/well) and infected with various adenoviral stocks at MOIs of 1:100,000 for 6 hr. TNF-α from the supernatants was measured by ELISA (Upper) or its bioactivity (Lower) was assayed as described in Materials and Methods. Each point was from three replicates, with error bars indicating SD.
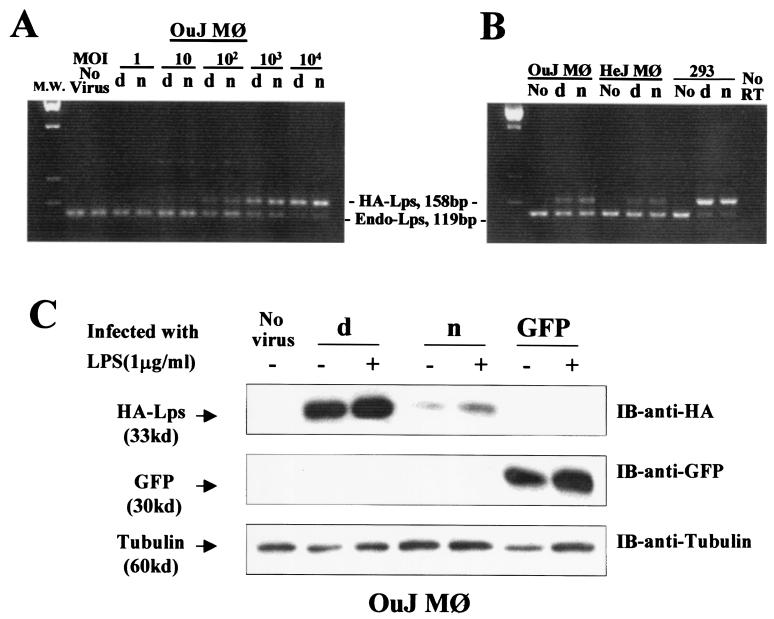
To ascertain that the transduced macrophages contained similar copies of Ad5-Lpsn/Ran DNA or Lpsd/Ran DNA, we performed PCR on DNA extracted from cells infected with either virus at MOIs ranging from 1:1 to 10,000:1. As shown in Fig. 4A, starting at MOI of 100:1, the 158-bp viral Lps/Ran-HA DNA fragment appeared; its intensity peaked at a MOI of 10,000:1 and was inversely proportional to that of the 127-bp endogenous Lps/Ran genomic DNA fragment (Fig. 4A). Furthermore, the intensity between the viral Lpsn/Ran DNA and the viral Lpsd/Ran DNA was comparable at all MOIs examined (Fig. 4A). The observed PCR signals remained after acid washing of the infected macrophages, and were absent from cells exposed to heat-inactivated virus (by boiling), suggesting that the signals were not caused by passive virus adsorption. To rule out the possibility that the macrophages might have phagocytized the adenoviral particles instead of being infected by the virus and expressing the transgene, we proceeded to measure the degree of gene expression at the RNA or protein level. After adenoviral infection, RNA from the virus-producing cells, 293 human embryonic kidney cells, and C3H/HeOuJ and C3H/HeJ macrophages were subjected to RT-PCR using primers specific for both the epitope-tagged and the endogenous Lps/Ran. A comparable level of Lpsn/Ran mRNA and Lpsd/Ran mRNA was observed in all of these cell lines (Fig. 4B).
Figure 4.
Evidence of gene transfer and expression in transduced C3H/HeOuJ macrophages. (A) The result of PCR on DNA extracted from peritoneal macrophages of C3H/HeOuJ mice primed i.p. with 3 ml of 3% thioglycollate solution (Sigma) 3 days before cell collection. The amount of genomic DNA used in each lane was estimated to be equivalent to 4,000 cells. OuJ Mφ represents macrophages from C3H/HeOuJ mice. (B) The result of RT-PCR on C3H/HeOuJ macrophages after adenoviral gene transfer. HeJ Mφ represents macrophages from C3H/HeJ mice; 293 represents human embryonic kidney 293 cells. (C) Western blot analysis on lysates from C3H/HeOuJ cells after adenoviral gene transfer. d, Ad5-Lpsd/Ran virus; n, Ad5-Lpsn/Ran virus; no, no virus; IB, immunoblot.
Despite comparable expression of the transgenes at the RNA mRNA level, measurement of the amount of proteins in the transduced cells, however, reveals interesting differences between Lpsn/Ran and Lpsd/Ran. In Fig. 4C, Western blot analysis on lysates of various transduced cells shows that the level of Lpsd/Ran-HA was higher than that of Lpsn/Ran-HA. Both Lpsd/Ran and Lpsn/Ran appeared to increase as a result of LPS stimulation, whereas the level of GFP remained unchanged in the transduced cells with or without LPS stimulation (Fig. 4C). This difference could not be related to adenoviral infection, as we obtained similar results in cells infected with retrovirus vectors carrying the Lps/Ran cDNAs (H. Chen and P.M.C.W., unpublished work).
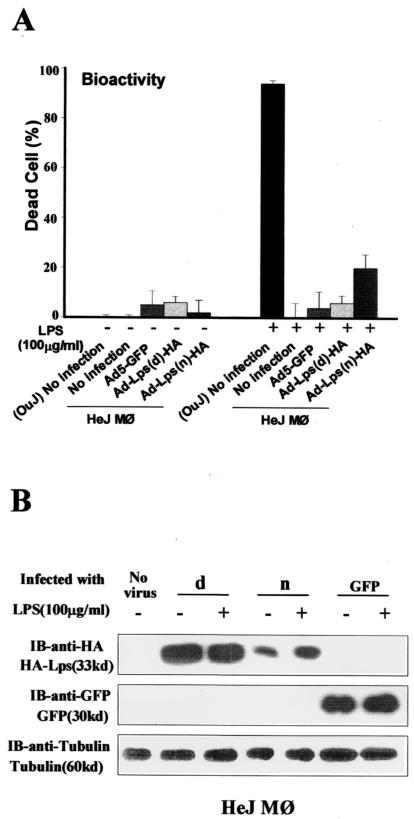
With identical experimental design as in Fig. 3, transduction of Lpsn/Ran cDNA into macrophages from C3H/HeJ mice did not restore their LPS response in terms of TNF-α production, despite evidence of successful gene transfer and expression in these cells (Fig. 4B). Considering the possibility that Lpsd/Ran might exert a dominant negative function on the wild-type protein, we stimulated the transduced cells with a high dose of LPS (100 μg/ml). At this concentration, C3H/HeJ macrophages transduced with Lpsn/Ran responded to the stimulation, albeit its response was only 20% of uninfected C3H/HeOuJ macrophages, whereas uninfected, Ad5-GFP-infected, or Ad5-Lpsd/Ran-infected C3H/HeJ macrophages did not produce a significant amount of TNF-α even at this LPS dose. Western blot analysis showed that, like in C3H/HeOuJ responder macrophages, Lpsd/Ran protein was higher than Lpsn/Ran (Fig. 5B). This increased Lpsd/Ran protein level compared with that of the wild-type Lpsn/Ran could explain the apparent dominant negative function of Lpsd/Ran on TNF-α production, as predicted in reports by other investigators (19, 30, 31). Mechanistically, this increase could be the result of different translational efficiency, different intracellular protein localization, or different protein degradation rates. Thus overproduction of Lps/Ran in Ad5-Lpsd/Ran-transduced macrophages produces a negative biological feedback signal, down-modulating TNF-α production, which may be akin to an autocrine negative feedback regulation for growth factor receptors where their corresponding ligands are overproduced within the same cells (32, 33).
Figure 5.
Restoration of TNF-α production by C3H/HeJ macrophages after Lpsn/Ran adenoviral transfer. The infection conditions and the Western blot analysis were identical to those in the legend to Fig. 3. (OuJ)No infection, uninfected macrophages from C3H/HeOuJ mice.; d, Ad5-Lpsd/Ran virus; n, Ad5-Lpsn/Ran virus; IB, immunoblot.
Discussion
Because expression of the Lpsd/Ran in responding macrophages reduces but does not abrogate TNF-α production upon LPS stimulation, the data suggest the presence of more than one LPS signaling pathway. These data are also consistent with the results in Fig. 5, showing that successful transfer and expression of Lpsn/Ran in C3H/HeJ macrophages could not restore TNF-α production upon LPS stimulation unless a very high LPS dose is given. Yet Lpsn/Ran is effective in restoring LPS responsiveness of splenic B cells from C3H/HeJ mice (20, 21). B lymphocytes do not express CD14 and therefore their stimulation by endotoxin is likely achieved via a CD14-independent signaling pathway.
Upon LPS stimulation, no elevation of TNF-α is detected in the serum of C3H/HeJ mice (19). In this study, expression of Lpsd/Ran clearly reduced TNF-α production but the reduction was incomplete. This finding is consistent with the idea that more than one genetic mutation has occurred in the genome of C3H/HeJ mice, accounting for its resistance to endotoxic challenge (9–11, 30, 31). In addition to Lps/Ran that we described previously (20, 21), Tlr4 has been mapped to the Lps locus and is found to have a point mutation in the C3H/HeJ genome (15, 16). Its involvement in innate immunity via a CD14-dependent signal transduction pathway has been extensively documented (34–36). Although Lps/Ran is likely to be involved in a CD14-independent pathway, it also may have an indirect effect on a CD14-dependent pathway, as it is a nuclear protein that can function downstream in more than one signal transduction pathway.
We recently showed that Lpsd/Ran could protect endotoxin-sensitive mice against LPS challenge (21). Our study here also suggests that the protective effect can be caused by the ability of Lpsd/Ran to down-modulate TNF-α production. Importantly, this down-modulation may confer a dampened response signal on the responding cells so that they elicit a competent but not overly reactive immune response against the endotoxin challenge. The finding in this report provides one clear scientific basis on which gene therapy for endotoxic or septic shock can be applied.
Acknowledgments
We thank Hong Chen for the Lps-HA cDNA plasmids and Rachel Sheppard for helpful discussions. This work was supported in part by National Institutes of Health Grants RO1CA70854 and RO1A139159 to P.M.C.W. and by StemCell Therapeutics. Y.W.K. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- TNF-α
tumor necrosis factor α
- LPS
lipopolysaccharide
- GFP
green fluorescent protein
- MOI
multiplicity of infection
- HA
hemagglutinin
- RT-PCR
reverse transcription–PCR
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040567797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040567797
References
- 1.Beutler B, Milsark I W, Cerami A C. Science. 1985;229:869–871. [PubMed] [Google Scholar]
- 2.Tracy K J, Beutler B, Lowry S F, Merryweather J, Wolpe S, Milsark I W, Hariri R J, Fahey T J, 3rd, Zentella A, Albert J D, et al. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 3.Morrison D C, Ryan J L. Annu Rev Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 4.Miethke T, Wahl C, Heeg K, Echtenacher B, Krammer P H, Wagner H. J Exp Med. 1992;175:91–98. doi: 10.1084/jem.175.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeffer K, Matsuyama T, Kundig T M, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi P S, Kronke M, Mak T W. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 6.Grau G E, Maennel D N. Nat Med. 1997;3:1193–1195. doi: 10.1038/nm1197-1193. [DOI] [PubMed] [Google Scholar]
- 7.Wong, P. M. C., Chung, S. W. & Sultzer, B. M. (2000) Scand. J. Immunol., in press.
- 8.Sultzer B M. Nature (London) 1968;219:1253–1254. doi: 10.1038/2191253a0. [DOI] [PubMed] [Google Scholar]
- 9.Glode L M, Rosenstreich D L J. Immunology. 1976;117:2061–2065. [PubMed] [Google Scholar]
- 10.Sultzer B M. Infect Immun. 1976;13:1579–1584. doi: 10.1128/iai.13.6.1579-1584.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly K, Watson J. Immunogenetics. 1977;5:75–84. [Google Scholar]
- 12.Watson J, Riblet R. J Exp Med. 1974;140:1147–1161. doi: 10.1084/jem.140.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson J, Kelly K, Largen M, Taylor B A. J Immunol. 1978;120:422–424. [PubMed] [Google Scholar]
- 14.Rosenstreich D L, Vogel S N, Jacques A R, Wahl L M, Oppenheim J J. J Immunol. 1978;121:1664–1670. [PubMed] [Google Scholar]
- 15.Poltorak A, He X, Smirnova I, Liu M Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1999;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschning C J, Weshe H, Ayres T M, Roche M. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel S N, Johnson D, Perera P Y, Medvedev A, Lariviere L, Qureshi S T, Malo D J. Immunology. 1999;162:5666–5670. [PubMed] [Google Scholar]
- 20.Kang A, Wong P M C, Chen H, Castagna R, Sultzer B. Infect Immun. 1996;64:4612–4617. doi: 10.1128/iai.64.11.4612-4617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong P M C, Kang A, Chen H, Fan P, Kan Y W, Sultzer B M, Zhuo Y, Yuan Q, Chung S W. Proc Natl Acad Sci USA. 1999;96:11543–11548. doi: 10.1073/pnas.96.20.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray E J, editor. Methods in Molecular Biology: Gene Transfer and Expression Protocols. Clifton, NJ: Humana; 1991. [Google Scholar]
- 23.Nyberg-Hoffman C, Sharbram P, Li W, Giroux D, Aguilar-Cordova E. Nat Med. 1997;3:808–811. doi: 10.1038/nm0797-808. [DOI] [PubMed] [Google Scholar]
- 24.Espevik T, Nissen-Meyer J. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 25.Lasfargues A, Chaby R. Cell Immunol. 1988;115:65–178. doi: 10.1016/0008-8749(88)90171-2. [DOI] [PubMed] [Google Scholar]
- 26.Sheehan K, Ruddle N H, Schreiber R D. J Immunol. 1989;142:3884–3893. [PubMed] [Google Scholar]
- 27.Coutavas E E, Hsieh C M, Ren M, Drivas G T, Rush M G, D'Eustachio P. Mamm Genome. 1994;5:623–628. doi: 10.1007/BF00411457. [DOI] [PubMed] [Google Scholar]
- 28.Drivas G T, Shih A, Coutavas E, Rush M, D'Eustachio P. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyberg-Hoffman C, Aguilar-Cordova E. Nat Med. 1999;5:955–957. doi: 10.1038/11400. [DOI] [PubMed] [Google Scholar]
- 30.Perera P Y, Vogel S N, Detore G R, Haziot A, Goyert S M. J Immunol. 1997;158:4422–4429. [PubMed] [Google Scholar]
- 31.Haziot A, Lin X Y, Zhang F, Goyert S M. J Immunol. 1998;160:2570–2572. [PubMed] [Google Scholar]
- 32.Sporn M B, Roberts A B. Nature (London) 1985;313:745–747. doi: 10.1038/313745a0. [DOI] [PubMed] [Google Scholar]
- 33.Stiles C D. Nature (London) 1984;311:604–605. doi: 10.1038/311604a0. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov R, Janeway C A. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 35.Wright S D. J Exp Med. 1999;189:605–609. doi: 10.1084/jem.189.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman J A, Kafatos F C, Janeway C A, Ezekowitz R A B. Science. 1999;284:1313–1317. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]