Abstract
SUMO is a novel ubiquitin-like protein that can covalently modify a large number of nuclear proteins. SUMO modification has emerged as an important regulatory mechanism for protein function and localization. Sumoylation is a dynamic process that is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes and is readily reversed by a family of SUMO-specific proteases (SENPs). Since SUMO was discovered 10 years ago, the biologic contribution of this posttranslational modification has remained unclear. In this review, we report that SENP1, a member of the SENP family, is overexpressed in human prostate cancer specimens. The induction of SENP1 is observed with the chronic exposure of prostate cancer cells to androgen and/or interleukin (IL) 6. SENP1 upregulation modulates the transcriptional activity of androgen receptors (ARs) and c-Jun, as well as cyclin D1 expression. Initial in vivo data from transgenic mice indicate that overexpression of SENP1 in the prostate leads to the development of prostatic intraepithelial neoplasia at an early age. Collectively, these studies indicate that overexpression of SENP1 is associated with prostate cancer development.
Keywords: SUMO, Sentrin, SENP, SUMO-specific protease, prostate cancer
The Sumoylation Pathway
In late 1996 until early 1997, our laboratory and several others reported a novel ubiquitin-like protein called sentrin-1 (or SUMO1, GMP1, PIC1, SMT3, and UBL1) [1–5]. In the ensuing years, the acronym SUMO (small ubiquitin-like modifier) gained wide acceptance and, thus, will be used throughout this review. However, it should be noted that sentrin was used in our early publications.
There are three SUMO family members, which slightly vary in length. SUMO1 is a 101-amino acid protein, SUMO2 is a 103-amino acid protein, and SUMO3 is a 95-amino acid protein [6]. SUMO2 and SUMO3 are more closely related to each other (93.6% identity in 94-residue overlap) compared with SUMO1 (52.4% identity in 84-residue overlap). SUMO homologues have been reported from Arabidopsis thaliana to Homo sapiens, suggesting that SUMO is an evolutionarily conserved protein that may have unique functions in cellular metabolism.
There is a remarkable conservation of the mechanisms between ubiquitination and sumoylation pathways. However, the activating (E1), conjugating (E2), and ligating (E3) enzymes involved in sumoylation are entirely distinct from ubiquitin E1, E2, and E3.
The E1 for SUMO is composed of two subunits, Aos1 and Uba2 [7,8]. Aos1 is homologous to the N-terminal half of Uba1, the yeast-activating enzyme (E1) for ubiquitin [9], whereas Uba2 is similar to the C-terminal half Uba1 (Figure 1). Formation of the Uba2-SUMO thiol ester adduct is an ATP-dependent process that is similar to the mechanism of action of the ubiquitin-activating enzyme (E1). Our laboratory has discovered a specific SUMO-conjugating enzyme (Ubc9) in a yeast two-hybrid screen using SUMO1 as bait [10]. Ubc9 was able to form a thiol ester adduct with SUMO1, but not with ubiquitin. Thus, in contrast to the large number of E2s used in the ubiquitination pathway, Ubc9 is the only known conjugating enzyme for the sumoylation pathway [10,11].
Figure 1.
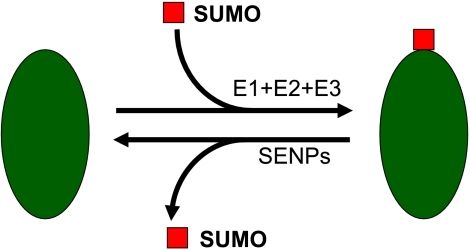
The SUMO modification pathway. On the first step of the process, the C-terminus reveals the Gly-Gly (G-G) residues for conjugation. Aos1 and Uba2 constitute the activating enzyme complex (E1). Ubc9 is the only known SUMO-conjugating enzyme (E2). E3 denotes SUMO ligases.
More recently, three types of SUMO E3s have been discovered. The first type is composed of the PIAS (protein inhibitor of the activator of STAT) family of proteins [12,13]. The second type is RanBP2, which is localized in the nuclear pore complex [14]. The third type is Pc2, which is a component of polycomb protein complexes [15]. RanGAP1 and PML are the first two sumoylated substrates to have been identified [2,3,16]. However, the list of sumoylated proteins has expanded significantly in the last few years to include p53, MDM2, topoisomerase I, topoisomerase II, androgen receptor (AR), SRC1, GRIP1, p300, and histone deacetylase (HDAC) 1 [17–25] (Figure 2). In contrast to ubiquitination, sumoylation does not target proteins for degradation. Sumoylation, in some cases, actually competes with ubiquitination on the same lysine residues, thus functioning almost like an antiubiquitin [26]. Sumoylation can also alter a protein's cellular localization. For example, sumoylated RanGAP1 is localized in the nuclear envelope, whereas unmodified RanGAP1 is localized in the cytosol [2,3]. Finally, sumoylation of many transcriptional factors serves to alter their transcriptional activity [17,18,24,27–33].
Figure 2.
SUMO-modified proteins. A partial list of SUMO-modified proteins is presented.
Reversal of Sumoylation by SUMO-Specific Proteases (SENPs)
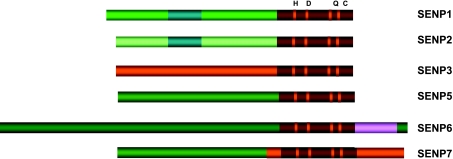
Sumoylation is a dynamic process that is readily reversed by a family of SENPs [34] (Figure 3). In the mammalian system, six SENPs have been reported [34–40] (Figure 4). SENP1 is a nuclear protease that deconjugates a large number of sumoylated proteins [40] (Figure 5). SENP2 is a nuclear envelope-associated protease that, when overexpressed, appears to have an activity similar to that of SENP1 [36,40–42] (Figure 5). There is a spliced isoform of mouse SENP2, called SuPr1, which could alter the distribution of nuclear POD-associated proteins, such as CBP and Daxx, and could convert Sp3 to a strong activator with diffuse nuclear localization [31,35]. We have recently reported SENP3 and SENP5 to constitute a family of nucleolar SENPs with preference for SUMO2/3 [43]. The third SENP family consists of SENP6 and SENP7—two nuclear proteases with unknown function. Although the ability of SENPs to reverse sumoylation is well established, the specificity of each SENP and the overall biologic relevance of the SENP family remain undefined.
Figure 3.
Sumoylation is a dynamic and reversible process. E1, SUMO-activating enzyme; E2, SUMO-conjugating enzyme; E3, SUMO ligase.
Figure 4.
The human SENP family. Six SENPs share a conserved catalytic domain with four highly conserved amino acids (H, D, Q, and C) and are grouped into three families.
Figure 5.
The subcellular localizations of SENPs. Overexpression images are presented.
Sumoylation of ARs, c-Jun, and Coregulatory Proteins
Numerous cellular targets of SUMO targets modulate transcription. Steroid receptors such as ARs, which are ligandregulated transcription factors belonging to the nuclear receptor superfamily [44], have been reported as SUMO-ylated proteins. ARs are sumoylated in vivo at lysine residues 386 and 520 [21]. Mutation of these residues increases the transactivation ability of ARs, suggesting that sumoylation is involved in the repression of AR activity [21].
c-Jun is a transcription factor that plays an important role in regulating cell growth, apoptosis, differentiation, and transformation. The transcriptional activity of c-Jun can be regulated by both phosphorylation and sumoylation. c-Jun is conjugated by SUMO at amino acids 229 and 257, with sumoylation negatively regulating c-Jun-dependent transcription [33,45].
It is well known that both ARs and c-Jun interact with various coregulatory proteins; this interaction can regulate the transcriptional activity of nuclear receptors and transcription factors.
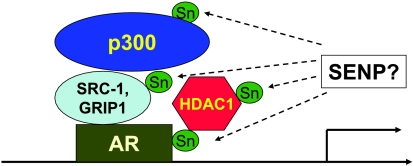
Recently, it has been found that four AR coregulators are sumoylated. These AR coregulatory proteins either transactivate or transrepress AR transcriptional activity by binding the specific functional domains of receptors (the N-terminal transactivation region, the central DNA-binding domain, and the C-terminal ligand-binding domain). The transactivator SRC1, for example, has five sumoylation sites, and two major sites are localized in a nuclear receptor box situated in the nuclear receptor-interacting region 1 [22]. It was observed that sumoylation could increase the interaction of SRC1 with progesterone receptors. For the coactivator GRIP1, two residues located in the nuclear receptor-interacting region were found to be sumoylated [46]. Substitution of these two sumoylation sites could attenuate the activity of GRIP1 on AR-dependent transcription. The transrepressors HDAC1 and HDAC4 were also found to be sumoylated [25,47,48]. Mutation of the two sumoylation sites of HDAC1 profoundly reduced HDAC1-mediated transcriptional repression [25]. An HDAC4 sumoylation mutant showed a slightly impaired ability to repress transcription, as well as reduced HDAC activity [28].
Similarly, p300 is a well-known coactivator of c-Jun [49–52] that has been shown to physically interact with c-Jun and to activate c-Jun-dependent transcription [50]. Because the transcriptional activity of p300 can be modulated by a number of signaling pathways, p300 provides an additional level of regulation for c-Jun-dependent transcription.
It has been reported that p21 regulates p300 transcriptional activity [53–55]. p21 not only inhibits p300-bound cyclin E-Cdk2 activity through repression of the histone acetyltransferase activity of p300 [56] but also stimulates p300 transactivation [55]. Within p300, a domain named CRD1 has been identified as a domain with strong transcriptional repression [55]. CRD1 functions independently of p300 histone acetyltransferase domains, but can repress the transactivational activity of p300 [55]. p21 derepresses this CRD1 activity and, thus, selectively activates p300-dependent transcription at specific promoters. Recent findings indicate that sumoylation is required for CRD1-dependent transcriptional repression [24]. The two SUMO modification sites within the CRD1 domain of p300 have been identified, and mutation at these two sites can reduce the repression of CRD1 domain and p21 inducibility [24]. Therefore, SUMO modification provides a new mechanism to control p300 function and potentially novel mechanisms for the regulation of c-Jun-dependent transcriptional activities.
As ARs, c-Jun, and coregulatory proteins are sumoylated, we believe that SENPs will play an important role in regulating AR and c-Jun activity through the deconjugation of sumoylated coregulators (see below).
SENP1 Regulates AR-Dependent Transcription
ARs and their coregulators provide us a good model with which to determine the role of the desumoylation activity of SENPs (Figure 6). We performed the initial survey by using luciferase reporter gene assay to examine whether any of the six SENPs could affect AR-dependent transcription. As shown in Figure 7, SENP1 dramatically enhanced AR transcriptional activity in LNCaP cells by 23-fold. This effect is dependent on the presence of the AR ligand, R1881. SENP1's catalytic activity is required for this effect, as the catalytically inactive mutant of SENP1 (R630L and K631M) has no effect on AR-dependent transcription. Furthermore, none of the other SENPs exerts any significant effect on AR-dependent transcription. Similarly, SUMO1, SUMO2, or Ubc9 overexpression also failed to enhance AR-dependent transcription, suggesting that these components of SUMO modification system are sufficient in cells. Thus, the effect of SENP1 on AR-dependent transcription is unique among all of the conjugation and deconjugation machinery of the sumoylation pathway.
Figure 6.
ARs and the coregulators SRC1, p300, and HDAC1 are modified by SUMO.
Figure 7.
SENP1 markedly enhances AR-dependent transcription. LNCaP cells cotransfected luciferase with plasmids, as indicated in the figure. After 12 hours of transfection, cells were treated with 10nmofR1881 for 24 hours, and luciferase activity was measured. Transfection efficiency was normalized using a β-galactosidase expression construct, and the results are presented as fold activation over an empty vector.
More interestingly, the mutation of two sumoylation sites on ARs did not abolish SENP1 action, suggesting that the SENP1 effect on AR-dependent transcription is not mediated by AR sumoylation [57]. We then tested the role of the sumoylation of SRC1, p300, and HDAC1 and showed that the main target of SENP1 is HDAC1 in the SENP1-mediated enhancement of AR-dependent transcription [57]. Sumoylation of HDAC1 is required for its transcription suppression. An HDAC1 mutant that cannot be sumoylated has lost its deacetylase activity (as demonstrated by deacetylase assay) and, subsequently, its suppressive activity [57]. Thus, in summary, when SENP1 is not present, AR-dependent transcription occurs at low levels due mainly to the suppressive effect of HDAC1. However, when SENP1 is present, HDAC1 is desumoylated and, therefore, loses its deacetylase activity and ability to suppress transcription. Thus, AR-dependent transcription would occur at high levels.
SENP1 Enhances c-Jun-Dependent Transcription
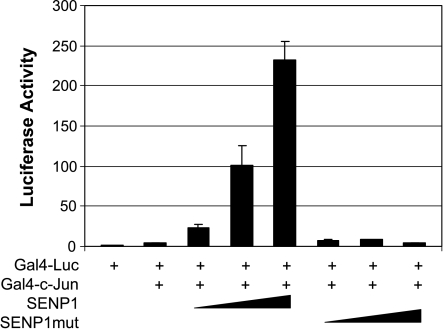
It has also been shown that c-Jun transcription can be regulated by SuPr1, an alternative spliced form of SENP2. However, SuPr1's ability to enhance c-Jun transcription is dependent on PML, but is independent of SuPr1's desumoylation activity. We showed that SENP1 also markedly enhances c-Jun's transcription activity (Figure 8). The action of SENP1 on c-Jun transcription is independent of the sumoylation and phosphorylation status of c-Jun, but is critically dependent on SENP1's desumoylation activity [58]. We further showed that p300 is essential for SENP1 to enhance c-Jun-dependent transcription because SENP1 could desumoylate the CRD1 domain of p300, thereby releasing the cis repression of CRD1 on p300 [58]. The ability of SENP1 to regulate c-Jun-dependent transcription may also play a role in its involvement in tumorigenesis.
Figure 8.
SENP1 induces c-Jun-dependent transcription. PC-3 cells were transfected with Gal4-luc and Gal4-DBD or Gal4-c-Jun plasmid in the absence or presence of increasing amounts of SENP1 wild-type or mutant plasmids. Luciferase activity was measured, and transfection efficiency was normalized by β-galactosidase expression.
SENP1 Is Overexpressed in Prostatic Intraepithelial Neoplasia (PIN) and Prostate Cancer
SENP1, acting as a strong activator for AR and c-Jun transactivation, prompted us to explore the potential role of SENP1 in prostate cancer development. Prostate cancer is the most frequently diagnosed cancer among the male population in the United States [59]. Tumor formation in prostate tissues accounts for 30% of cancer-related deaths in men. In most cases, the disease progresses from benign hyperplasia to a prostate cancer precursor state referred to as PIN (Figure 9). PIN formation is replaced with rapidly proliferating prostate cancer cells that readily metastasize to other areas of the body. This atrophy of the prostate gland is attributed to changes at the molecular level, specifically with alterations in the expression and transcriptional activity of numerous factors. The correlation between enhanced AR-dependent transcriptional activity and prostate carcinogenesis is accepted; in fact, the expression of the AR-regulated prostate-specific antigen (PSA) gene is used as a biologic marker for the diagnosis of prostate cancer [60,61]. The oncogenic property of c-Jun is well established. Recent studies suggest a correlation between c-Jun levels and prostate carcinogenesis; the expression of c-Jun is enhanced with the progression of the carcinoma [62]. Because SENP1 overexpression directly regulates these two factors, we evaluated whether expression of SENP1 is observed in human prostate cancer specimens. We first investigated the expression of SENP1 in normal prostate, PIN [63], and prostate cancer using in situ hybridization technique. Nonradioactive in situ hybridization was performed on 43 prostatectomy cases with areas of both high-grade PIN and cancer. SENP1 messenger RNA was increased in 29 of 43 cases of highgrade PIN (67%) (Figure 10A). Similarly, SENP1 expression was increased in 26 of 43 prostate cancer samples (60%) (Figure 10B). Thus, SENP1 expression is preferentially increased during the development of prostate cancer in the majority of cases.
Figure 9.
Pathogenesis of prostate cancer (reproduced with permission from the New England Journal of Medicine [59]).
Figure 10.
Overexpression of SENP1 in prostate cancer tissues. (A–C) In situ hybridization showed that SENP1 was overexpressed in high-grade PIN (black arrow; A) and cancer cells (black arrow; B and C), but not in normal epithelial cells (white arrow) in prostate cancer specimens. The insert in (C) was a sense probe control. (D) Immunohistochemical staining ofARs in normal prostate glands (white arrowhead) and prostate cancer (black arrowhead).
SENP1 Induces Cyclin D1 Expression and Cell Proliferation
Because overexpression of SENP1 modulates AR- and c-Jun-dependent transcriptional activity and both factors are known to mediate cell proliferation, we reasoned that overexpression of SENP1 could regulate the proliferation of prostate cancer cells. Indeed, when endogenous of SENP1 was silenced by SENP1 siRNA in LNCaP cells, growth was significantly decreased. Provided SENP1 can strongly increase AR transcriptional activity, it is of no doubt that the effect of SENP1 on proliferation could be attributed to its regulation for AR transcription. However, we observed similar results in PC-3, an androgen-independent prostate cancer cell line, suggesting that SENP1 could play a role in regulating cell proliferation through pathways other than the AR-dependent pathway. We also found that the number of SENP1-silenced PC-3 cells in the G1 phase was significantly increased but decreased in the S and G2/M phases, suggesting that SENP1 plays a role in G1-S phase transition in PC-3 cells. This slowing of the proliferation of SENP1-silenced PC-3 cells may thus be due to an alteration in G1-S phase progression.
There are numerous cell cycle regulators that control cell proliferation and cell cycle progression [64,65]. Among them, cyclins D1 and E are predominantly associated with cell growth regulation and G1-S phase transition [66–68]. We therefore reasoned that SENP1 might modulate cell proliferation and cell cycle progression by regulating the expression of cyclins D1 and E. Silencing SENP1 expression decreased the expression of cyclin D1, but not cyclin E, in PC-3 cells. Conversely, stably transfected SENP1 in LNCaP cells enhanced cyclin D1 expression and cell proliferation. Interestingly, cell proliferation is dependent on cyclin D1 expression induced by SENP1 (Figure 11). Thus, the regulation of cyclin D1 expression by SENP1 is another means through which prostate cancer cell growth is regulated.
Figure 11.
SENP1 regulates cyclin D1 expression in prostate cancer cells. SENP1 regulates cyclin D1 expression. (A) PC-3 cells were transfected with nonspecific siRNA or SENP1 siRNA. (B) LNCaP cells were stably transfected with an empty vector, SENP1, or SENP1 mutant. Cyclin D1 protein levels were determined in these cell clones.
We also found that the induction of cyclin D1 expression by SENP1 depended on its catalytic activity, as the mutation of SENP1 catalytic domain disrupted its activity on cyclin D1 transcription and cell proliferation. We further determined that HDAC1 is also the mediator for the SENP1 regulating cyclin D1 expression.
Androgen and Interleukin (IL) 6 Induce SENP1 Transcription in Prostate Cancer Cells
The expression of SENP1 correlated with the manifestation of prostate tumorigenesis and cell proliferation, but the mechanism for SENP1 induction remained undefined. We hypothesized that activation of ARs by their native agonist, androgen, would upregulate SENP1 levels because the biologic function of ARs is to directly modulate gene/protein expression. To test this hypothesis, the human androgen-sensitive cell line, LNCaP, was used. LNCaP cells were exposed to the synthetic androgen, R1881 (20 nM), for 24 and 48 hours, and SENP1 mRNA levels were evaluated using quantitative real-time polymerase chain reaction (PCR). SENP1 expression is enhanced five-fold and seven-fold with 24- and 48-hour R1881 treatment, respectively (Figure 12A). Hence, continuous exposure to androgen induces SENP1 expression.
Figure 12.
The combination of androgen and IL-6 synergistically enhances SENP1 and PSA expression. SENP1 expression was examined in LNCaP cells treated with R1881 (A); IL-4, IL-6, or TNF-α (B); or a combination ofR1881 and IL-6 (C) for either 24 or 48 hours by real-time PCR. PSA expression was also examined in LNCaP cells treated with R1881, IL-6, or R1881 + IL-6 by enzyme-linked immunosorbent assay (D).
Various cytokines are expressed in prostate cancer cells, including IL-1, IL-6, and tumor necrosis factor α (TNF-α) [69,70]; elevated serum levels of IL-6 and TNF-α have been reported in patients with prostate cancer [61,71]. Unlike IL-1 and TNF-α, IL-6 promotes the activation of AR-dependent transcription through the initiation of the MAPK and JAK/STAT pathways [72–74]. To determine if androgen-independent activation of ARs promotes SENP1 upregulation, LNCaP cells were treated with 25 ng/ml IL-6, IL-1, or TNF-α for either 24 or 48 hours. SENP1 levels were enhanced in cells treated with IL-6, but not with IL-1 or TNF (Figure 12B).
The relapse of prostate cancer occurs due to the adaptation of ARs to low levels of androgen and activation by alternate pathways [75,76]. To evaluate whether the combination of androgen-dependent and androgen-independent pathways on AR activation could regulate SENP1 expression, SENP1 levels were assessed following the treatment of LNCaP cells with R1881 (20 nM), IL-6 (25 ng/ml), or both for 24 hours. Treatment with either androgen or IL-6 produced approximately a five-fold increase in SENP1 levels (Figure 11). The combination of both R1881 and IL-6 profoundly enhanced SENP1 expression by more than seven-fold compared to either compound alone (Figure 12C).
To ensure that the concentrations of R1881 and IL-6 were sufficient to activate AR-dependent transcription, the expression of the AR-dependent protein PSA was evaluated. Serum PSA levels are commonly used as diagnostic tools for prostate cancer; PSA levels above 2.5 ng/ml indicate high AR activity. The addition of R1881 (20 nM) or IL-6 (25 ng/ml) for 24 hours significantly increased PSA protein levels compared to controls (a 5.6-fold and a 2.2-fold increase in PSA levels compared to controls, respectively) (Figure 12D). The combination of R1881 and IL-6 further enhanced the production of PSA (Figure 12D) with an 11-fold increase compared to controls. Therefore, the addition of R1881 and/or IL-6 enhances the transcriptional activity of AR as well as SENP1 levels. We have demonstrated that inhibition of SENP1 expression by siRNA reduces androgen-induced PSA production in LNCaP cells [57]. Thus, it is likely that SENP1 induction is essential for androgen and IL-6 to induce PSA secretion.
PIN-Like Lesion Observed in the Prostate of SENP1 Transgenic Mice as Early as 4 Months of Age
We showed that IL-6 and androgen induce SENP1 expression in prostate cancer cells. SENP1 enhances c-Jun-dependent transcription; SENP1 markedly increases AR-dependent transcription, which leads to increase in PSA secretion; SENP1 also increases cyclin D1 expression, leading to increase in cellular proliferation. Furthermore, SENP1 is highly expressed in PIN and prostate cancer, but not in normal prostate tissues. Taken together, we hypothesize that SENP1 has a great potential to initiate and promote the development of prostate cancer.
We have generated a probasin-driven murine SENP1 transgenic mouse to determine the role of SENP1 in prostate cancer development. Four transgenic mice lines with the probasin-driven SENP1 transgene were identified using specific PCR primers. At 16 weeks, histologic studies on hematoxylin/eosin-stained prostate tissue specimens of two male transgenic mice were performed. Compared to age-matched wild-type mice (Figure 13A), preliminary data show the development of PIN-like lesions in the dorsolateral lobe of male SENP1 transgenic mice (Figure 13B). Studies in the four established lines are currently underway to confirm that SENP1 expression in transgenic mice prompts the development of cancerous lesions in the prostate gland.
Figure 13.
Dysplasia in the prostate of SENP1 transgenic mice. The dorsolateral and ventral lobes of the prostate from either age-matched wild-type mice (A) or 16-week-old SENP1 transgenic mice (B) were stained with hematoxylin/eosin. Abnormal growth was observed in the dorsolateral prostate of transgenic mice (highlighted with black arrow).
Conclusion
SENP1 is overexpressed in PIN and prostate cancer tissues, but not in normal prostate tissues. The increase in SENP1 expression in PIN and prostate cancer is most likely induced by androgen and IL-6. Induction of SENP1 will enhance AR-dependent transcription, c-Jun-dependent transcription, and expression of cyclin D1, leading to increase in cellular proliferation. Transgenic mice overexpressing SENP1 in the prostate gland also showed evidence of early PIN formation. Thus, SENP1 is likely to play a significant role in the development of prostate cancer (Figure 14). This is the first demonstration of the involvement of desumoylation in cancer development.
Figure 14.
Induction of SENP1 mediates prostate cancer development. Schematic representation based on our collectively studies that indicate a relationship between prostate carcinogenesis and SENP1. Continuous exposure of prostate cancer cells to either androgen or IL-6 induces upregulation of SENP1. Elevated levels of SENP1 enhance transcriptional activity of AR and c-Jun and expression of cyclin D1. These altered pathways, in turn, involve the tumorigenesis of prostate cancer.
Footnotes
This work was supported by the National Institutes of Health (RO1 CA 80089) and the Department of Defense (CDMRP PC040121).
References
- 1.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 2.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 3.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 5.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 6.Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ET. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:11349–11353. doi: 10.1074/jbc.273.18.11349. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 9.Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 10.Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 11.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 12.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 13.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 14.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 hasSUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 15.Kagey MH, Melhuish TA, Wotton D. The polycomb protein pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 16.Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 17.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 19.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 20.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauchereau A, Amazit L, Quesne M, Guiochon-Mantel A, Milgrom E. Sumoylation of the progesterone receptor and of the coactivator SRC-1. J Biol Chem. 2003;14:14. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- 23.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. p300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 25.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 26.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 27.Hay RT, Vuillard L, Desterro JM, Rodriguez MS. Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos Trans R Soc Lond B Biol Sci. 1999;354:1601–1609. doi: 10.1098/rstb.1999.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehembre F, Muller S, Pandolfi PP, Dejean A. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene. 2001;20:1–9. doi: 10.1038/sj.onc.1204063. [DOI] [PubMed] [Google Scholar]
- 30.Kishi A, Nakamura T, Nishio Y, Maegawa H, Kashiwagi A. Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am J Physiol Endocrinol Metab. 2003;284:E830–E840. doi: 10.1152/ajpendo.00390.2002. [DOI] [PubMed] [Google Scholar]
- 31.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 32.Tojo M, Matsuzaki K, Minami T, Honda Y, Yasuda H, Chiba T, Saya H, Fujii-Kuriyama Y, Nakao M. The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J Biol Chem. 2002;277:46576–46585. doi: 10.1074/jbc.M205987200. [DOI] [PubMed] [Google Scholar]
- 33.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 34.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 35.Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI. SUMO-1 protease-1 regulates gene transcription through PML. Mol Cell. 2002;10:843–855. doi: 10.1016/s1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 36.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 37.Kim KI, Baek SH, Jeon YJ, Nishimori S, Suzuki T, Uchida S, Shimbara N, Saitoh H, Tanaka K, Chung CH. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J Biol Chem. 2000;275:14102–14106. doi: 10.1074/jbc.275.19.14102. [DOI] [PubMed] [Google Scholar]
- 38.Nishida T, Kaneko F, Kitagawa M, Yasuda H. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J Biol Chem. 2001;276:39060–39066. doi: 10.1074/jbc.M103955200. [DOI] [PubMed] [Google Scholar]
- 39.Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423–6427. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 40.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itahana Y, Yeh ETH, Zhang Y. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol Cell Biol. 2006;26:4675–4689. doi: 10.1128/MCB.01830-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 44.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 45.Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem. 2002;277:30283–30288. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- 47.Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tussie-Luna MI, Bayarsaihan D, Seto E, Ruddle FH, Roy AL. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc Natl Acad Sci USA. 2002;99:12807–12812. doi: 10.1073/pnas.192464499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn C, Wiltshire C, MacLaren A, Gillespie DA. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 2002;14:585–593. doi: 10.1016/s0898-6568(01)00275-3. [DOI] [PubMed] [Google Scholar]
- 50.Lee JS, See RH, Deng T, Shi Y. Adenovirus E1A downregulates cJun- and JunB-mediated transcription by targeting their coactivator p300. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vries RG, Prudenziati M, Zwartjes C, Verlaan M, Kalkhoven E, Zantema A. A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 2001;20:6095–6103. doi: 10.1093/emboj/20.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albanese C, D'Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, Kitsis RN, Henglein B, Avantaggiati M, Somasundaram K, et al. Activation ofthecyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 53.Gregory DJ, Garcia-Wilson E, Poole JC, Snowden AW, Roninson IB, Perkins ND. Induction of transcription through the p300 CRD1 motif by p21WAF1/CIP1 is core promoter specific and cyclin dependent kinase independent. Cell Cycle. 2002;1:343–350. [PubMed] [Google Scholar]
- 54.Ait-Si-Ali S, Ramirez S, Barre FX, Dkhissi F, Magnaghi-Jaulin L, Girault JA, Robin P, Knibiehler M, Pritchard LL, Ducommun B, et al. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 55.Snowden AW, Anderson LA, Webster GA, Perkins ND. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol Cell Biol. 2000;20:2676–2686. doi: 10.1128/mcb.20.8.2676-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 57.Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase I. Mol Cell Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng J, Perkins ND, Yeh ET. Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases. J Biol Chem. 2005;280:14492–14498. doi: 10.1074/jbc.M412185200. [DOI] [PubMed] [Google Scholar]
- 59.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 60.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 61.Culig Z, Klocker H, Bartsch G, Steiner H, Hobisch A. Androgen receptors in prostate cancer. J Urol. 2003;170:1363–1369. doi: 10.1097/01.ju.0000075099.20662.7f. [DOI] [PubMed] [Google Scholar]
- 62.Tiniakos DG, Mitropoulos D, Kyroudi-Voulgari A, Soura K, Kittas C. Expression of c-jun oncogene in hyperplastic and carcinomatous human prostate. Urology. 2006;67:204–208. doi: 10.1016/j.urology.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 63.Meng MV, Shinohara K, Grossfeld GD. Significance of highgrade prostatic intraepithelial neoplasia on prostate biopsy. Urol Oncol. 2003;21:145–151. doi: 10.1016/s1078-1439(03)00009-7. [DOI] [PubMed] [Google Scholar]
- 64.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 65.Harper JW, Adams PD. Cyclin-dependent kinases. Chem Rev. 2001;101:2511–2526. doi: 10.1021/cr0001030. [DOI] [PubMed] [Google Scholar]
- 66.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Cyclin D1: normal and abnormal functions [minireview] Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 67.Wang C, Li Z, Fu M, Bouras T, Pestell RG. Signal transduction mediated by cyclin D1: from mitogens to cell proliferation: a molecular target with therapeutic potential. Cancer Treat Res. 2004;119:217–237. doi: 10.1007/1-4020-7847-1_11. [DOI] [PubMed] [Google Scholar]
- 68.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 69.Culig Z, Hobisch A, Herold M, Hittmair A, Thurnher M, Eder IE, Cronauer MV, Rieser C, Ramoner R, Bartsch G, et al. Interleukin 1beta mediates the modulatory effects of monocytes on LNCaP human prostate cancer cells. Br J Cancer. 1998;78:1004–1011. doi: 10.1038/bjc.1998.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizokami A, Gotoh A, Yamada H, Keller ET, Matsumoto T. Tumor necrosis factor-alpha represses androgen sensitivity in the LNCaP prostate cancer cell line. J Urol. 2000;164:800–805. doi: 10.1097/00005392-200009010-00053. [DOI] [PubMed] [Google Scholar]
- 71.Nakashima J, Tachibana M, Ueno M, Miyajima A, Baba S, Murai M. Association between tumor necrosis factor in serum and cachexia in patients with prostate cancer. Clin Cancer Res. 1998;4:1743–1748. [PubMed] [Google Scholar]
- 72.Lee HJ, Chang C. Recent advances in androgen receptor action. Cell Mol Life Sci. 2003;60:1613–1622. doi: 10.1007/s00018-003-2309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L, Wang L, Lin HK, Kan PY, Xie S, Tsai MY, Wang PH, Chen YT, Chang C. Interleukin-6 differentially regulates androgen receptor transactivation via PI3K-Akt, STAT3, and MAPK, three distinct signal pathways in prostate cancer cells. Biochem Biophys Res Commun. 2003;305:462–469. doi: 10.1016/s0006-291x(03)00792-7. [DOI] [PubMed] [Google Scholar]
- 74.Ueda T, Bruchovsky N, Sadar MD. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. J Biol Chem. 2002;277:7076–7085. doi: 10.1074/jbc.M108255200. [DOI] [PubMed] [Google Scholar]
- 75.Mohler JL, Gregory CW, Ford OH, III, Kim D, Weaver CM, Petrusz P, Wilson EM, French FS. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10:440–448. doi: 10.1158/1078-0432.ccr-1146-03. [DOI] [PubMed] [Google Scholar]
- 76.Chlenski A, Nakashiro K, Ketels KV, Korovaitseva GI, Oyasu R. Androgen receptor expression in androgen-independent prostate cancer cell lines. Prostate. 2001;47:66–75. doi: 10.1002/pros.1048. [DOI] [PubMed] [Google Scholar]