Abstract
Regulation of p53 by the ubiquitin-proteasomal pathway has been studied considerably. Studies have also demonstrated that the ubiquitin-like proteins SUMO-1 and NEDD8 modify p53. Similarly, p63 and p73 are subject to regulation by ubiquitin and ubiquitin-like modifications, and perturbations of these pathways in the regulation of the p53 family have been implicated in tumorigenesis and developmental abnormalities. Here, we provide an overview of the current understanding of the regulation of the p53 family by covalent modification by ubiquitin, SUMO-1, and NEDD8.
Keywords: p63, p73, ubiquitin, SUMO, NEDD8
Introduction to the p53 Family
The p53 family consists of three members: p53, p63, and p73. Since the discovery of p63 and p73 almost 10 years ago, it has become apparent that they are not merely redundant “p53-like” genes. Although there are several similarities between the three genes and their protein products, there are also interesting differences, suggesting that each protein may have a unique role in diverse processes ranging from development to tumorigenesis. Thus, knowledge of the distinct pathways that regulate the levels and activity of each p53 family protein will likely shed light on the functions of these proteins. The regulation of the activity and stability of p53 by ubiquitination has been studied extensively. Ubiquitin is best known as a posttranslational modification that targets proteins for degradation through the 26S proteasome; however, the role of ubiquitin has expanded to involve additional functions. Along those lines, ubiquitin-like (UBL) proteins, which consist of a family of at least 10 members, have diverse functions that are not necessarily associated with proteasomal degradation. p53 function is regulated by at least two UBL proteins SUMO-1 and NEDD8, and early data suggest that ubiquitin, SUMO-1, and NEDD8 modifications modulate both p63 and p73 functions. This review will concentrate on ubiquitination, sumoylation, and neddylation of the p53 family, with particular focus on p63 and p73.
Structure and Function of the p53 Family
p53 is a sequence-specific DNA-binding transcription factor that plays a central role in the cellular response to oncogenic stimuli and cytotoxic stress, such as DNA damage, by initiating cell cycle arrest and apoptosis, predominantly through its ability to enhance the transcription of genes that regulate these processes (e.g., p21, PUMA, and BAX). p63 and p73 share significant homology in three functional protein domains. p63 and p73 proteins share approximately 25%, 60%, and 35% amino acid identity with p53 in N-terminal transactivation (TA), central DNA binding, and C-terminal oligomerization domains, respectively. Certain isoforms of p63 and p73 have additional domains not found in p53. For example, both p63α and p73α C-terminal isoforms have a sterile α motif (SAM), which usually functions as a protein-protein interaction motif. p63 and p73 are also able to bind canonical p53 DNA-binding sites [1,2], transactivate 53 target genes [2,3], and induce cell cycle arrest and apoptosis [2,4,5]. Although p63 and p73 can bind to known p53-responsive elements in the promoter of p53 target genes, there are clear differences in the preferred binding site sequence for p63 [6] and likely differences in target p73-responsive elements as well. As a result, p53, p63, and p73 differentially induce target genes. Several unique p63 and p73 target genes have been identified, including PERP for p63 [7], Aquaporin 3 for p73 [8], and JAG1/2 for p63/p73 [9]. Further identification of genes that are specific targets of each p53 family protein will likely provide insight into their unique functions.
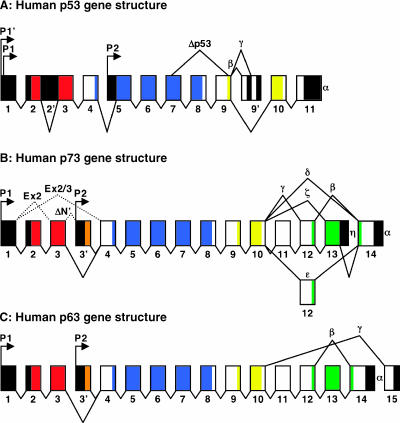
p63 and p73 genes give rise to multiple mRNA that, when translated, produce several different protein isoforms (Figure 1). For more than two decades, it was believed that, in contrast to p63 and p73, the p53 gene encoded one predominant mRNA, resulting in a single protein. However, recent studies clearly demonstrate the existence of multiple p53 protein isoforms as well (reviewed in Murray-Zmijewski et al. [10]). Multiple p53, p63, and p73 protein isoforms contain different protein domains as a result of alternative splicing, alternative promoter usage, and alternative initiation of translation. The p53 gene encodes at least two N-terminally truncated isoforms Δ40p53 and Δ133p53, which lack the TA domain. Δ40p53 is generated by alternative initiation of translation at a second ATG-40 located within exon 4 [11–13] and by alternative splicing of intron 2 (Figure 1A), which also results in translation from the second ATG [12]. Furthermore, there are at least four alternatively spliced C-terminal p53 isoforms, which include full-length p53 (α), p53β (formerly known as p53i9) [14,15], p53γ [14], and Δp53 [16]. Interestingly, p53β and p53γ isoforms lack the oligomerization domain, and Δp53 lacks the extreme C-terminus of the DNA-binding domain. For p73, there are at least seven C-terminal isoforms generated either by alternative splicing (α, β, γ, δ, ɛ, and ζ) [3,17,18] or by alternative termination of translation (η) [19] (Figure 1B). In addition, the p73 gene encodes four N-terminal isoforms that include the full-length TAp73 and the N-terminally truncated isoforms that are collectively termed ΔTAp73 or ΔNp73 due to the fact these isoforms lack the TA domain. The N-terminally truncated isoforms are generated as a result of transcription from an alternative promoter within intron 3 (ΔNp73) [20], translation from an alternative initiation site (ΔN′p73) [19], and alternative N-terminal splicing (ΔEx2p73 and ΔEx2/3p73) [3]. ΔN′p73 isoforms are transcribed from the same promoter used to generate the TA isoforms of p73; however, alternative N-terminal splicing of intron 3 (exon 3′) allows for initiation of translation within exon 3′, producing a protein indistinguishable from ΔNp73 [19]. Theoretically, p73 can be expressed as more than 30 mRNA variants encoding for multiple proteins; however, only 14 have been described. Lastly, p63 exists as three alternatively spliced C-terminal isoforms (α, β, and γ) (Figure 1C) [2]. Similar to p73, p63 encodes two N-terminal isoforms that include the full-length TAp63 and the N-terminally truncated ΔNp63 isoforms generated by transcription from an alternative promoter within intron 3. Thus, p63 can be expressed as six mRNA variants that encode six different p63 proteins.
Figure 1.
Schematic representation of the gene structure of the p53 family. The approximate exon regions encoding the unique amino acids for ΔN isoforms (orange), TA domain (TAD; red), DNA binding (DBD; blue), oligomerization (OD; yellow), and SAM domains (green) are indicated by color. Untranslated regions are shaded black. Arrows indicate transcriptional start sites. (A) The C-terminal splicing patterns generating full-length p53α, p53β, p53γ, and Δp53 are shown. The p53 isoforms that include the entire TA domain are transcribed from P1 and the recently described P1′ transcription initiations sites, and Δ 133p53 is transcribed from the P2 promoter located within intron 4. The alternative N-terminal splicing of intron 2 is indicated. (B) The C-terminal splicing patterns generating p73α, p73β, p73γ, p73δ, and p73ɛ are shown. The ΔNp73 isoforms are transcribed from the P2 promoter located within intron 3 (designated exon 3′). The alternative N-terminal splicing generating ΔEx2p73 and ΔEx2/3p73 is indicated. (C) The C-terminal splicing patterns generating p63α, p63β, and p63γ are shown. The ΔNp63 isoforms are transcribed from the P2 promoter located within intron 3 (designated as exon 3′). Exon size and approximate contribution of exons to the indicated functional domains are not drawn to scale.
The N-terminally truncated ΔN isoforms of p63 and p73 lack the TA domain and, in general, have antiapoptotic properties. Thus, although both the TA and ΔN isoforms of p63 and p73 can bind p53 DNA-binding sites [1,2,20], in general, only TA isoforms can transactivate promoters of p53 target genes and induce apoptosis [2,4,5,21,22]. ΔN isoforms act as dominant-negative inhibitors for TA isoforms of all three p53 family members by forming heterooligomers that generate an abortive transcriptional complex [2,19,22–24] and by competing directly for p53 DNA-binding sites [2,20,21]. Studies have also shown that ΔNp73 can enhance transformation by oncogenes such as Ras [25]. The complexity of this scenario has recently increased, given recent data suggesting that some ΔN isoforms of p63 and p73 can transactivate distinct target genes and, in certain cases, suppress growth [26]. Furthermore, different p63 and p73 C-terminal isoforms include significant variations in coding sequences, resulting in functional differences in terms of their ability to transactivate target genes and induce apoptosis. The different isoforms are likely subject to specific posttranslational modifications. Importantly, the variation in p53 family protein splice forms is conserved in lower organisms, including Drosophila and zebrafish [10], suggesting that isoform-specific functions and posttranslational regulation are likely to be important.
Roles in Cancer and Development
Perhaps the most surprising discoveries regarding p63 and p73 functions emerged as a result of data from genetically engineered knockout mice. p63-/- and p73-/- mice had significant neuronal and ectodermal developmental abnormalities, respectively [20,27,28]. Of note, the original knock-out mice generated for both p63 and p73 had deletions of all TA and ΔN isoforms. p73-/- mice have significant neurologic abnormalities due to either the absence and/or the loss of specific populations of neurons [20]. ΔNp73 is the predominant isoform in the murine fetal nervous system, and loss of this antiapoptotic p73 isoform leads to enhanced apoptosis in cortical and sympathetic ganglia neurons. The mechanism whereby ΔNp73 promotes survival is likely a combination of inactivation of full-length proapoptotic p53 family proteins (p53, TAp63, and TAp73) and activation of mitochondrial pathways (reviewed in Irwin and Miller [29]). p63-/- mice have significant limb and craniofacial malformations, as well as failure of development of the skin and other epithelial tissues [28]. Interestingly, germline mutations in p63 have been reported in patients with ectodermal dysplasia syndromes, including ectrodactyly-ectodermal dysplasia and facial cleft (EEC), ankyloblpharon-ectodermal dysplasia-clefting (AEC), limb-mammary syndrome (LMS), and non-syndromic split-hand/foot malformation (SHFM) [30]. Like p63-/- mice, patients with these clinical syndromes have varying degrees of craniofacial (cleft lip and palate), limb, skin, and hair abnormalities, and p63 genotype-phenotype correlations are apparent. Certain mutations solely affect specific isoforms (e.g., AEC patients have p63 mutations in exon 13, which includes the SAM domain that is only present in α isoforms). Furthermore, some of these mutations affect amino acid residues that undergo posttranslational modifications, such as sumoylation and ubiquitination (see below). Additional evidence for the importance of isoform-specific expression during development includes a recent report by Jacobs et al. [31] demonstrating that, in the developing nervous system, only TAp63α and TAp63γ isoforms are expressed. TAp63 was shown to be an essential proapoptotic protein in neurons, both alone and in combination with p53 [31]. Taken together, these findings again support that each isoform is likely to have specific biologic and biochemical activities. To date, there have been no reported isoform-specific knockout mice, but their generation is likely to reveal important information.
The p53 family proteins also appear to have distinct roles in tumorigenesis. Unlike p53, which is mutated in over 50% of all human cancers and is inactivated in a further 20%, p63 and p73 mutations are rarely observed in human cancer [32]. In addition, p53-/- mice, as well as mice engineered to express tumor-derived p53 mutant proteins, develop cancers [33,34]; however, initial reports suggested that p63-/- and p73-/- mice were not tumor-prone. Although not mutated, accumulating evidence suggests that the relative expression and stability of the different N-terminal isoforms of p63 and p73 may contribute to a role in tumorigenesis. The full-length TA isoforms of p63 and p73 have proapoptotic “tumor-suppressor-like” properties, whereas the ΔN isoforms of p63 and p73 generally have antiapoptotic “oncogene-like” properties. TAp73 is induced by a wide variety of chemotherapeutic agents [35–37], and blocking TAp73 function promotes survival and leads to enhanced chemoresistance [38–40]. Further support for a role of p63 and p73 in tumorigenesis was provided by a recent study of heterozygous p63+/- and p73+/- mice and compound p53/p63/p73 knock-out mice [41]. Aged p63+/- and p73+/- mice develop spontaneous tumors and premalignant lesions, and loss of the second allele of p63 and p73 was demonstrated in several of these tumors, suggesting that at least certain isoforms of p63 and p73 are tumor-suppressor proteins. In addition, loss of p63 or p73 cooperates with loss of p53 in tumor development because compound p53+/-;p63+/- and p53+/-;p73+/- mice develop a spectrum of tumors different from that of p53+/- mice. Finally, in comparison to p53+/- mice, mice heterozygous for both p53 and p63 or p73 have both larger tumor burdens and a higher incidence of metastatic lesions. Of note, another p63+/- mouse generated on a different genetic background, using an alternative gene targeting strategy, did not develop tumors but demonstrated features of premature aging [42,43].
Recent evidence from human tumors also supports the idea that, for many types of cancer, the relative balance between the TA and ΔN isoforms may be important in tumor development and/or progression. The “oncogenic” ΔN isoforms of p53 family proteins are overexpressed in a number of human cancers. Specifically, ΔNp73 expression has been shown to be elevated in breast, ovarian, hepatocellular, prostate, colon, and neuroblastoma tumors [22,44–48]. Increased ΔNp73 expression in several of the abovementioned tumors has been associated with poor prognosis in patients, and this has been attributed to the ability of ΔNp73 to inhibit p53 and TAp73, resulting in decreased apoptotic response and chemoresistance [22,45,49,50]. Furthermore, ΔNp63 expression is elevated in primary head and neck squamous cell carcinoma (HNSCC) and other squamous epithelial malignancies such as cervical, lung, and esophageal cancers [51–53]. Recently, ΔNp63α overexpression in HNSCC cells was shown to promote the survival of these tumor cells through inhibition of TAp73-dependent apoptosis both by competition for promoter binding and by physical interaction with TAp73 [40]. Lastly, Bourdon et al. [14] reported that Δ133p53 mRNA, a p53 mRNA that (like the ΔN isoforms of p63/p73) lacks the TAD, was detected in human breast tumor samples but not in normal breast sample controls. Whether Δ133p53 protein levels are similarly increased in tumors has not been addressed. Conversely, loss of expression of the full-length TAp63 and TAp73 isoforms has been reported in many tumors, including leukemias, bladder cancers, mammary tumors, and squamous cell carcinomas (reviewed in Moll and Slade [32]). Taken together, the data from mouse models and human tumors suggest that the balance between the expression of p53, p63, and p73 and the balance between various TA and ΔN isoforms likely affect the final signaling pathway leading to apoptosis or survival. Therefore, understanding the regulatory mechanisms, such as posttranslational modifications, that differentially modulate TA and ΔN isoform activity and stability are of particular interest because therapeutic modulation of the proapoptotic and antiapoptotic isoforms of the p53 family has potential therapeutic benefits in treating human cancers.
The activity of p53 is highly regulated by posttranslational modifications, protein-protein interactions, and protein stabilization [54,55]. p53 stability is regulated through the ubiquitin-proteasomal pathway by a number of E3 ubiquitin ligases, many of which have been shown to be involved in negative autoregulatory feedback loops with p53 [56]. In contrast, only a few E3 ubiquitin ligases with specificity toward p63 and p73 have been identified. None of the known p63/p73-specific E3 ubiquitin ligases is involved in negative autoregulatory feedback loop mechanisms, although there is evidence for such regulation [57–59]. There has been almost no overlap of specificity of the known E3 ubiquitin ligases for the different members of the p53 family; however, many of the same posttranslational modifications, such as phosphorylation, acetylation, and sumoylation, and their respective regulators modulate the activity of multiple p53 family members. It is conceivable that differential regulation of the p53/p63/p73 protein isoforms, perhaps by UBL molecules, may account for some of the different functional activities and stabilities for the various isoforms.
Ubiquitination of the p53 Family
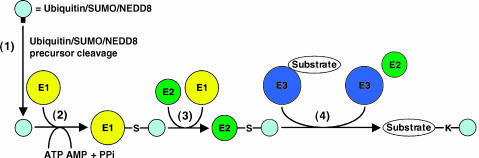
Ubiquitin is an evolutionarily conserved 76-amino-acid polypeptide. Ubiquitin was the first example of a protein that can modify another protein and act as a posttranslational modification. Although ubiquitin modifications have been shown to have multiple cellular functions, its most commonly reported function is targeting substrate proteins for degradation through the 26S proteasome [60,61]. The process of covalent attachment of ubiquitin, known as ubiquitination or ubiquitylation, occurs through sequential steps catalyzed by ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes (as summarized in Figure 2). A ubiquitin conjugation cascade is hierarchical, as eukaryotic genomes encode a single or (at most) a few E1s, a moderate number of E2s (approximately 60 in mammals), and a larger number of E3s [62]. Substrates can be modified by monoubiquitins, multiple monoubiquitins, and polyubiquitin chains that may be linked at any of the seven lysine residues within ubiquitin (K6-, K11-, K27-, K29-, K33-, K48-, and K63-linked chains), and both linkage type and chain length can act as functionally distinct signals [63,64]. For example, it is well established that K48-linked chains promote proteasomal degradation, whereas both K63-linked chains and monoubiquitination have nonproteolytic functions, which include kinase activation, DNA repair, ribosomal regulation, transcriptional modulation, and protein localization and trafficking [61,63,65].
Figure 2.
General overview of the ubiquitin and UBL protein conjugation pathways. (1) Ubiquitin, SUMO, and NEDD8 are synthesized as precursors that are processed at a conserved C-terminal glycine residue by the hydrolase activity of deubiquitinating, desumoylating, and deneddylating enzymes, generating an exposed Gly-Gly motif that serves as the attachment site to target substrates. (2) The exposed C-terminal glycine of ubiquitin/SUMO/NEDD8 is adenylated by an activating (E1) enzyme in an ATP-dependent manner and is transferred to an active E1 cysteinyl side chain through a thiol ester linkage. (3) Activated ubiquitin/SUMO/NEDD8 is subsequently transferred to a conjugating (E2) enzyme, forming another thiol ester linkage. (4) A ligase (E3) transfers ubiquitin/SUMO/NEDD8 to the ɛ amino group of a substrate lysyl residue of target substrates, resulting in the formation of an isopeptide bond.
E3 ubiquitin ligases are generally categorized into two broad classes: HECT (homologous to E6-AP carboxyl terminus) domain E3s, RING (really interesting novel gene) finger E3s [61,62,66], and its relatives, including PHD (plant homeodomain) and U-box domain-containing proteins [67]. The HECT domain is an approximately 350-amino-acid C-terminal region that was originally identified in the cellular protein, E6-AP (E6-associated protein) [68]. Approximately 35 amino acids upstream of the C-terminus of the HECT domain lies an active cysteine that accepts ubiquitin from a bound E2 forming a thiol ester intermediate, which subsequently transfers ubiquitin to substrates [61,65]. In contrast, RING finger E3s are adaptor proteins, where the RING domain serves to both recruit E2-conjugating enzymes to the substrate and act as cofactors that enhance substrate modification by E2 [62]. RING finger domains possess the consensus sequence CX2CX(9–39)CX(1–3)HX(2–3)C/HX2CX(4–48)CX2C, where the cysteines and histidines function to coordinate zinc binding [66]. RING finger E3s can function as single proteins or in multiprotein complexes. The stability of all three p53 family members is regulated by various RING and HECT E3 ligases. These E3 ligases play important roles in regulating protein stability under normal conditions and following a stress response.
p53 is ubiquitinated by a number of cellular E3 ubiquitin ligases (Figure 3A). Initially, it was thought that proteasomal-dependent regulation of p53 stability was solely determined by the RING finger E3 ubiquitin ligase, Mdm2. Initial studies determined that Mdm2 interacted with p53 [69]; however, only later did studies demonstrate that Mdm2 promotes p53 ubiquitination and degradation [70–73]. Recently, Li et al. [74] determined that, in contrast to polyubiquitination, which promotes p53 degradation, Mdm2-mediated monoubiquitination of p53 signals its nuclear export [56]. Furthermore, additional cofactors, including p300 and YY1, are involved in promoting Mdm2-mediated polyubiquitination [75–77]. Additional p53-specific E3 ubiquitin ligases that target p53 for degradation have also been described. These include two RING finger E3 ubiquitin ligases, Pirh2 and COP1 [78,79]. Both Pirh2 and COP1 are also p53 target genes and, as a result, participate in a negative autoregulatory feedback loop analogous to Mdm2. More recently, Chen et al. [80] reported the discovery of ARF binding protein 1 (ARF-BP1), a HECT domain-containing E3 ubiquitin ligase capable of ubiquitinating p53. ARF-BP1-mediated ubiquitination of p53 is inhibited by ARF binding, providing an additional mechanism by which ARF mediates tumor suppression. The discovery of the abovementioned p53-specific E3 ubiquitin ligases has greatly increased the understanding of the regulation of both p53 stability and subcellular localization. Finally, alterations in p53-specific E3 ligases have been described in cancer. Mdm2 amplification is observed in approximately 7% of human tumors [81], most commonly in sarcomas. To date, studies have detected COP1 overexpression in breast and ovarian adenocarcinoma tissues [82], and Pirh2 overexpression in lung tumor samples [83]. Thus, the modulation of p53-specific E3 ubiquitin ligase expression likely plays an important role in tumorigenesis and in the cellular response to chemotherapy. In contrast to p53, the regulation of p63 and p73 stability through the ubiquitin-proteasomal pathway is less well characterized.
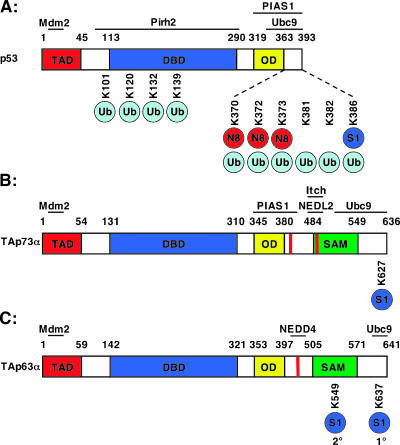
Figure 3.
Lysyl residues modified by ubiquitin and UBL proteins. The known lysines modified by ubiquitin (Ub), SUMO-1 (S1), and NEDD8 (N8) are indicated for the p53 family. Approximate binding regions of the E2-conjugating enzymes and E3 ligases are shown. (A) Mdm2 ubiquitinates multiple p53 C-terminal lysines (K370, K372, K373, K381, K382, and K386), as well as additional lysines located in the DNA-binding domain (K101, K120, K132, and K139) [139]. The specific lysines ubiquitinated by Pirh2, COP1, and ARF-BP1, and the binding regions of COP1 and ARF-BP1 have not been reported. Mdm2 also promotes NEDD8 (N8) modification of at least three C-terminal lysines (K370, K372, and K373). p53 is sumoylated at K386 by PIAS1 and PIASxβ. (B) PIAS1 binds all p73 isoforms, but only TAp73α and ΔNp73α contain the lysine residue (627) that is sumoylated. NEDL2 and Itch bind the second C-terminal proline-rich (PY) motif; however, the specific lysines ubiquitinated by these HECT E3 ubiquitin ligases are not known. (C) p63α is sumoylated at lysine 637. A secondary sumoylation site has been reported at lysine 549. NEDD4 binds to the C-terminal PY motif of p63. Studies employed yeast two-hybrid screen, in vitro binding, and coimmunoprecipitation assays, or a combination of the abovementioned techniques, to determine the binding domains shown in the figure, and specific studies are referenced in the manuscript.
Preliminary studies suggested that the ubiquitin-proteasomal pathway regulates p73 stability. First, the proteasome inhibitor lactascystin (LLnL) stabilized p73 protein levels [84]. Second, cotransfection experiments with exogenous ubiquitin resulted in the accumulation of ubiquitinated p73 proteins [85]. Third, in ts20 cells possessing a thermolabile E1 enzyme, p73 was stabilized only when the ubiquitination pathway was inactivated [85]. Interestingly, the stability of the proapoptotic TA isoforms and the antiapoptotic ΔN isoforms of p63 and p73 appears to be differentially regulated by ubiquitination in response to DNA-damaging agents, such as chemotherapeutic agents. Maisse et al. [86] demonstrated that ΔNp73, but not p53 and TAp73, is rapidly degraded in response to DNA-damaging agents in a proteasomal-dependent manner. Westfall et al. [87] also observed increased ubiquitination and decreased total ΔNp63α protein levels in a proteasome-dependent manner in response to ultraviolet radiation and paclitaxel treatment. Thus, downregulation of the ΔN isoforms of p63 and p73 may result in an enhanced cellular apoptotic response to chemotherapy treatment. However, these studies have not identified a ΔNp63-specific or a ΔNp73-specific E3 ubiquitin ligase.
Initial studies examining p73 stability naturally investigated the E3 ligase, Mdm2. The three residues (F19, W23, and L26) in the p53 N-terminus that directly contact Mdm2 are conserved in p63 and p73 [2,3,88,89]. As expected, Mdm2 binds to TAp73; however, in stark contrast to the known relationship with p53, Mdm2 does not degrade p73. Instead, Mdm2 overexpression results in p73 stabilization [90,91]. Subsequent studies aimed at understanding this differential regulation of p53 and p73 by Mdm2 used p53–p73 chimeric proteins and determined that amino acids 92 to 112 of p53, which are absent in p73, contain the region responsible for Mdm2-mediated degradation [92]. These results suggest that this unique p53 sequence element functions as a degradation signal. There are conflicting data as to whether TAp63 isoforms bind Mdm2 and whether this interaction affects p63 stability and transcriptional activity [93–95].
Following these initial results suggesting that Mdm2 was not an E3 ubiquitin ligase for p73, additional E3 ligases were identified in screens for p73-interacting proteins (Figure 3B). The first E3 ligase that was found to promote the ubiquitination of p73 was NEDL2, a NEDD4-related HECT-domain-containing E3 ubiquitin ligase [96]. The family of NEDD4 proteins contains WW domains, which are protein-protein interaction domains similar to SH3 domains that mediate binding to proline-rich (PY) motifs. The WW domains of NEDL2 interact with the C-terminal PY motifs of p73α and p73β; however, NEDL2 does not bind p53, which lacks these PY motifs. Although NEDL2 promotes p73 ubiquitination, unexpectedly, this interaction results in stabilization and increased TAp73 transcriptional activity. The exact mechanism of NEDL2-mediated stabilization has not been determined, and whether NEDL2 promotes specific ubiquitin-linked chains that do not result in degradation, but rather modulate transcription through alternative mechanisms, is unknown. Similarly, in a search for binding partners of the PY motifs found in the p73 C-terminus, Rossi et al. [97] identified Itch, another NEDD4-related E3 ligase that interacts with p73 through its WW domains. Itch was shown to ubiquitinate p73α, but not p73δ or p53, that lacks PY motifs. Itch-mediated ubiqutination resulted in proteasomal-dependent degradation of p73 [97]. Importantly, Itch was found to degrade both TAp73α and ΔNp73α isoforms. In response to DNA damage, Itch was downregulated, potentially explaining one mechanism by which TAp73 is stabilized following treatment with DNA-damaging agents, such as chemotherapeutic agents. However, the discovery of Itch does not explain the preferential ubiquitination and degradation of the ΔNp73 isoforms in response to DNA damage. Instead, the authors suggest that Itch plays a role in maintaining both TA and ΔN isoforms at low levels under normal unstressed conditions.
Only a few published reports have revealed potential p63-specific E3 ubiquitin ligases (Figure 3C). One mechanism of the preferential degradation of ΔNp63 isoforms in response to genotoxic stress has been proposed to involve stratifin-mediated nuclear export of ΔNp63α followed by RACK1 (receptor for protein kinase C)-mediated proteasomal degradation [98]. Stratifin (14-3-3σ) expression is regulated by several p53 family proteins, and RACK1 has been shown to be a scaffolding protein in pathways involved in limb development. The authors demonstrate that RACK1 promotes the ubiquitination of ΔNp63 and suggest that RACK1, or a RACK1-containing complex, functions as one of the E3 ligases that may regulate the level of ΔNp63α in HNSCC. RACK1 itself does not possess any consensus HECT or RING-type domains characteristic of typical E3 ubiquitin ligases. Because HNSCCs often overexpress ΔNp63α, cisplatin-mediated downregulation of ΔNp63 through RACK1 may contribute to chemosensitivity by decreasing the levels of ΔNp63α available to inactivate proapoptotic p53, TAp63, and TAp73 isoforms. Interestingly, RACK1 interacts with p73α, inhibiting its transcriptional activity and ability to induce apoptosis; however, RACK1 does not appear to negatively regulate p73α stability [99]. Recent data suggest that two additional E3 ubiquitin ligases may be involved in p63 ubiquitination. The HECT domain-containing E3 ligase NEDD4 has been shown to promote the ubiquitination and degradation of ΔNp63α, but not ΔNp63β, and this modification affects dorsoventral patterning in zebrafish [100]. In addition, the HECT E3 ubiquitin ligase Itch can associate with and promote ubiquitin-mediated degradation of p63 [101]. Furthermore, two critical lysyl residues of p63 that are mutated in the limb malformation syndrome, SHFM, are involved in Itch-mediated degradation of p63.
To date, there have been no reports of E3 ubiquitin ligases that act in a negative autoregulatory feedback loop with p63 and p73, although there is evidence for such regulation for both of these p53 family members. Studies have shown that p63 and p73 mutants with compromised transactivation potential are more stable than their full-length counterparts, and that transactivation-competent TA isoforms can act in trans to promote the turnover of either ΔN isoforms or transactivation-incompetent mutants [57–59]. However, to date, there has been no reported E3 ubiquitin ligase involved in an autoregulatory negative feedback loop with p63 and p73. Interestingly, cyclin G, a transcriptional target of both p53 and p73, has been implicated in the negative regulation of p53 and p73 stability, which is mediated by an unknown mechanism that is both ubiquitin-independent and proteasomal-independent [102].
A number of ubiquitin-independent mechanisms affecting the protein stability of the p53 family through proteasomes have been reported. Studies from Asher and Shaul [103] have described a ubiquitin-independent proteasomal-dependent mechanism of regulation for both p53 and p73 through NADH quinone reductase (NQO1). Studies have demonstrated that dicumarol and other inhibitors of NQO1 induce the degradation of p53, and the majority of NQO1 associates with 20S proteasomes [104–106]. NQO1 binds both p53 and p73 in an NADH-dependent manner, and it has been proposed that NQO1 acts as a gatekeeper of 20S proteasomes, protecting both proteins from proteasomal degradation. Furthermore, a U-box domain-containing E3/E4 ligase, UFD2a, was also recently shown to promote the proteasomal degradation of p73α in a ubiquitin-independent manner and, interestingly, this effect was inhibited by cisplatin treatment [107]. In summary, there is evidence that p73 is regulated by the proteasome through both ubiquitin-dependent and ubiquitin-independent pathways. Several E3 ubiquitin ligases interact with p73, but only a subset has been shown to induce ubiquitination and degradation in vivo; to date, these ligases do not clearly discriminate between the TA and ΔN isoforms. Nevertheless, ubiquitin-mediated regulation of the stability and activity of the various “tumor-suppressor-like” TA and “oncogenic” ΔN isoforms of p63 and p73 may play a role in cancer development and in response to chemotherapy.
Sumoylation of the p53 Family
The SUMO (small UBL modifier) family consists of the three paralogues: SUMO-1 (also known as Smt3c, PIC1, GMP1, Sentrin, and UBL1), SUMO-2 (also known as Smt3a and Sentrin3), and SUMO-3 (also known as Smt3b and Sentrin2) [108]. SUMO-1 is a 101-amino-acid protein that is 18% identical and 48% homologous to human ubiquitin [109]. Processed SUMO-2 and SUMO-3 differ only by three N-terminal amino acids and are approximately 50% identical to SUMO-1 [110]. The SUMO conjugation pathway involves the concerted actions of SUMO E1-activating enzymes (SAE1/SAE2, also known as Aos1 and Uba2 in yeast), E2-transferring enzyme (Ubc9), and E3 ligases (as summarized in Figure 2), which include the PIAS (protein inhibitor of activated STAT) family of RING finger proteins (siz family in Saccharomyces cerevisiae) (reviewed in Hay [108]). One of the interesting features of the SUMO-specific Ubc9-conjugating (E2) enzyme is that it can directly modify substrate proteins in the absence of E3 [111]. In most cases, SUMO modification occurs within the SUMO modification consensus motif, ψKxE (where ψ is a hydrophobic acid and x is any residue) [112]. Sumoylation has been reported to have diverse functional effects involved in the regulation of subcellular transport, transcriptional activity, chromosome segregation, and cell cycle control [108]. All three p53 family members are regulated by SUMO-1 modification, affecting their stability, transcriptional activity, and ability to induce cell cycle arrest and apoptosis.
The first indication that p53 was a target of SUMO-1 conjugation came from a report that human Ubc9 associates with p53 in yeast [113]. Subsequently, Gostissa et al. [114] discovered SUMO-1 as a p53-interacting protein in a yeast two-hybrid screen, and Rodriguez et al. [115] investigated SUMO-1 modification as a mechanism of p53 stabilization in response to genotoxic stress. Both studies demonstrated that p53 is covalently modified by SUMO-1 in the C-terminus (K386) and that sumoylation results in increased p53 transcriptional activity (Figure 3A). In support, Muller et al. [116] reported that the p53 mutant (K386R) that is defective for SUMO-1 conjugation had slightly impaired apoptotic activity. However, since the initial publications describing p53 sumoylation, there have been conflicting reports as to the functional effects of this p53 modification (reviewed in Melchior and Hengst [117]). Three members of the PIAS family of E3 SUMO ligases (PIAS1, PIASxβ, and PIASy) were later found to interact with p53, and both PIAS1 and PIASxβ were reported to promote sumoylation of p53 [118–121]. The role of the different PIAS proteins has also been controversial. Schmidt and Muller [121] reported that both PIAS1 and PIASxβ strongly repressed the transcriptional activity of p53, and Megidish et al. [119] reported that PIAS1 is an activator of p53 transcription that stimulates p53-dependent G1 arrest of the cell cycle. Interestingly, both studies reveal that PIAS-mediated effects were independent of its sumoylation function. In addition, PIASy was reported to inhibit p53 transcriptional activity, but not its ability to induce apoptosis [120]. The difficulty in the elucidation of the function of sumoylation may be explained by the limitations of the techniques employed and, more importantly, by the fact that a number of regulators of p53 function, such as Mdm2, MdmX, ARF, and PML, are also regulated directly by SUMO-1 conjugation and the PIAS family, or play a direct role in p53 sumoylation [122–126].
Like p53, both p63 and p73 are sumoylated. Minty et al. [127] demonstrated that the C-terminus of p73α associates with Ubc9, and that SUMO-1 covalently modifies both TA and ΔN isoforms of p73α at K627 (Figure 3B). The shorter C-terminal isoform p73β does not associate with Ubc9 and lacks the p73α lysine that is sumoylated. The authors reported that sumoylation of p73α does not affect its transcriptional activity, but instead alters its subcellular localization and promotes degradation [127]. Subsequently, PIAS1 was found to bind p73 in a region that includes the OD domain and, therefore, is able to interact with all p73 isoforms in the nucleus [128]. PIAS1 can only sumoylate the α isoforms of p73, and sumoylated p73 is located exclusively in the nuclear matrix. PIAS1 was also shown to stabilize p73α, but this stabilization was, in fact, independent of its sumoylation function. PIAS1 also inhibited TAp73α transcriptional activity, and this effect was dependent on the sumoylation function of PIAS1. The authors suggest that the C-terminal TAp73β isoform may have higher basal transcriptional activity due to the fact that it is not a substrate of SUMO-1, and that PIAS1 acts as a checkpoint regulator of G1 and G2 exit by negatively regulating TAp73α-mediated transcription of p21 through sumoylation. Further studies are necessary to elucidate whether sumoylation and/or PIAS binding plays a role in modulating the activity of TA or ΔNp73 isoforms in cancer.
Sumoylation of p63 is also thought to play an important role in regulating its biologic activity, and dysregulation of p63 sumoylation may represent an underlying mechanism of human developmental disorders associated with p63 mutations. Similar to p73, Ubc9 associates with the C-terminal domain of p63α and catalyzes SUMO-1 conjugation at K637, with K549 serving as a potential secondary sumoylation site [129,130] (Figure 3C). Another commonality between the sumoylation of p63 and the sumoylation of p73 is that it appears that sumoylation destabilizes p63α protein levels [100,129]. Studies have also reported that sumoylation modulates the transactivation activity of both TA and ΔN isoforms of p63. Sumoylation-defective TAp63α and ΔNp63α mutants have dramatically increased transcriptional activity [129,130]. However, TAp63α sumoylation-defective mutants mediate both the upregulation and the downregulation of different subsets of critical genes involved in cell differentiation and limb morphogenesis [130]. For example, sumoylation-defective mutants are unable to regulate target genes implicated in bone and tooth development, such as RUNX, and thus may contribute to the pathogenesis of SHFM and other p63-EEC-like syndromes. Furthermore, it has been reported that naturally occurring p63 mutations found in human developmental disorders, including SHFM, EEC, and LMS, have altered sumoyation status [100,129,130]. This appears to be attributed to the abrogation of Ubc9 binding and to the loss of all or part of the SUMO-1 modification site. Therefore, it has been proposed that sumoylation plays an important role in regulating p63 biologic activity and is an essential step in normal development. Whether sumoylation likewise affects the stability or activity of p63 isoforms in tumors is not known. There have been no reports of a p63-specific SUMO E3 ligase; however, the nucleoporin RanBP2 has been shown to associate with ΔNp63α [130]. Lastly, it is still not clear what effects SUMO-2 and SUMO-3 may have on the p53 family, and whether they have regulatory functions different from those of SUMO-1.
Neddylation of the p53 Family
NEDD8 (Rub1 in S. cerevisiae) is an 81-amino-acid polypeptide that shares 57% amino acid identity with ubiquitin. The NEDD8 conjugation pathway is composed of NEDD8 E1-activating enzyme (APP-BP1/Uba3), E2-conjugating enzyme (Ubc12), and E3 ligases (as summarized in Figure 2) (reviewed in Pan et al. [131]). An essential role for neddylation in cell cycle control and embryogenesis has been demonstrated by a variety of genetic model systems (i.e., fission yeast, Drosophila, and mammals) [131]. Until recently, the only known substrates of NEDD8 were the cullins—a family of structurally related proteins that function as molecular scaffolds responsible for the assembly of RING finger E3 ubiquitin ligase complexes. Neddylation of cullins has been shown to enhance the ubiquitination activity of these cullin-based RING E3s [132–134]. In 2004, two tumor-suppressor proteins, pVHL and p53, were identified as substrates for NEDD8 [135,136], providing further insight into the biological role of NEDD8. These findings also raise the possibility that other tumor-suppressor proteins are covalently modified by NEDD8.
The first demonstration that the NEDD8 pathway influences p53 function came from a study in 2001, which demonstrated that the mechanism of p53 degradation by the human adenovirus protein E4orf6 was mediated through a cullin-containing E3 ubiquitin ligase complex consisting of Cul5, elongins B and C, and Rbx1 [137]. Recently, NEDD8 was shown to play a more direct role in regulating p53 activity, as both Mdm2 and p53 were found to be covalently modified by NEDD8 (Figure 3A) [136]. Using non-neddylatable p53 mutants in conjunction with the well-characterized ts41 CHO cell line, which possesses a thermolabile NEDD8 E1 enzyme, Xirodimas et al. demonstrated that Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Furthermore, NEDD8 conjugation of Mdm2, which appears to be catalyzed by an autoneddylation process, impairs the ability of Mdm2 to inhibit p53. In light of this finding and of the observations that the interaction of TAp73 and Mdm2 is not consistent with a role for Mdm2-mediated ubiquitination, we investigated whether Mdm2 promotes the neddylation of p73. We have found that TAp73α and TAp73β, but not ΔNp73β, which lacks a Mdm2-binding site, are covalently modified by NEDD8 in an Mdm2-dependent manner (Watson and Irwin, unpublished data). Furthermore, neddylation of TAp73β through Mdm2 inhibits TAp73β transcriptional activity, and this may be due to changes in subcellular localization (Watson and Irwin, unpublished data). Other studies have also implicated the NEDD8 pathway in the regulation of p73 activity, and Bernassola et al. [85] have suggested that a cullin-containing E3 ligase regulates p73 stability [138]. Lastly, it is not clear whether p63 is also regulated by NEDD8 modification and how these modifications may affect p63/p73 roles in tumorigenesis and development.
Conclusion and Outstanding Questions
UBL modification plays important roles in regulating the p53 family, and perturbations in these pathways have implications for both tumorigenesis and developmental abnormalities. First, p53-specific E3 ligases such as Mdm2, Pirh2, and COP1 are amplified in human cancers. Second, mutations in p63 found in a number of developmental abnormalities appear to affect SUMO-1-mediated regulation of p63 activity and, potentially, p63 ubiquitination. Third, some chemotherapeutic agents specifically mediate ubiquitination and degradation of the antiapoptotic ΔN isoforms of p63 and p73. In light of the accumulating evidence suggesting that the relative expression and stability of the different N-terminal isoforms of p63/p73 may contribute to a role in tumorigenesis, elucidating pathways that differentially regulate the activity and stability of TA and ΔN isoforms, such as through TA-specific or ΔN-specific E3 ligases, may have important therapeutic implications.
Many outstanding questions regarding UBL modification and the p53 family remain. First, p63 and p73 appear to be regulated by an autoregulatory feedback loop analogous to the p53-Mdm2, Pirh2, and COP1 pathways; however, specific p63-inducible or p73-inducible E3 ligases have yet to be identified. Second, pathways regulating ΔNp63 and ΔNp73 destabilization following DNA-damaging agents have yet to be clearly elucidated. Third, in light of the recognition that p53 exists as multiple isoforms potentially having different functions, the question arises as to whether various p53 isoforms are differentially regulated by the p53-specific E3 ubiquitin ligases. In addition, it is still unclear whether p53 family proteins undergo multiple ubiquitin and UBL modifications simultaneously, and whether the regulation of UBL modifications modulates other posttranslational modifications. Specifically, because acetylation occurs on lysines, it is possible that “competition” for each lysyl residue could lead to dramatically different functional outcomes. Finally, because proteasome inhibitors are being developed as therapeutic agents in cancer, understanding the regulatory pathway involving ubiquitination, sumoylation, and neddylation of the p53 family that is involved in tumorigenesis and chemosensitivity is critical to predicting the tumor types that may respond to such therapy.
Acknowledgement
We thank the members of the Irwin laboratory for helpful discussions and comments.
Footnotes
This work was supported by the Terry Fox Foundation of the National Cancer Institute of Canada. M.S.I. is a recipient of the Canada Research Chair.
References
- 1.Marin MC, Jost C, Irwin MS, DeCaprio JA, Caput D, Kaelin WG. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias J-M, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 4.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 5.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53 [see comments] Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. ([published erratum appears in Nat Med 1998;4(9):982]) [DOI] [PubMed] [Google Scholar]
- 6.Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, Chen X. Aquaporin 3, a glycerol and water transporter, is regulated by p73 of the p53 family. FEBS Lett. 2001;489:4–7. doi: 10.1016/s0014-5793(00)02437-6. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. The p53 family member genes are involved in the Notch signal pathway. J Biol Chem. 2002;277:719–724. doi: 10.1074/jbc.M108080200. [DOI] [PubMed] [Google Scholar]
- 10.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 11.Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, Oren M, Hainaut P. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–6728. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh A, Stewart D, Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24:7987–7997. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 14.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D, Iggo R, Frebourg T. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene. 1996;12:813–818. [PubMed] [Google Scholar]
- 16.Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 17.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Laurenzi VD, Catani MV, Terrinoni A, Corazzari M, Melino G, Costanzo A, Levrero M, Knight RA. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta [letter] Cell Death Differ. 1999;6:389–390. doi: 10.1038/sj.cdd.4400521. [DOI] [PubMed] [Google Scholar]
- 19.Ishimoto O, Kawahara C, Enjo K, Obinata M, Nukiwa T, Ikawa S. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- 20.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 21.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding. Implications for tumorigenesis. J Biol Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 22.Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa T, Takahashi M, Ozaki T, Watanabe Ki K, Todo S, Mizuguchi H, Hayakawa T, Nakagawara A. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol Cell Biol. 2002;22:2575–2585. doi: 10.1128/MCB.22.8.2575-2585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 25.Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002;62:3598–3602. [PubMed] [Google Scholar]
- 26.Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol. 2004;24:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32:138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- 28.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 29.Irwin MS, Miller FD. p73: regulator in cancer and neural development. Cell Death Differ. 2004;11(1):S17–S22. doi: 10.1038/sj.cdd.4401452. [DOI] [PubMed] [Google Scholar]
- 30.van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, Mills AA, Miller FD, Kaplan DR. p63 is an essential proapoptotic protein during neural development. Neuron. 2005;48:743–756. doi: 10.1016/j.neuron.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 32.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 33.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 36.Gong J, Costanzo A, Yang H, Melino G, Kaelin WG, Levero M, Wang J. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–808. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Z-M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 38.Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 39.Irwin MS, Kondo KK, Marin MC, Cheng LS, Hahn WC, Kaelin WG. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 40.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Keyes WM, Vogel H, Koster MI, Guo X, Qi Y, Petherbridge KM, Roop DR, Bradley A, Mills AA. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc Natl Acad Sci USA. 2006;103:8435–8440. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez G, Garcia JM, Pena C, Silva J, Garcia V, Martinez L, Maximiano C, Gomez ME, Rivera JA, Garcia-Andrade C, et al. ΔTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2005;24:805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- 46.Douc-Rasy S, Barrois M, Echeynne M, Kaghad M, Blanc E, Raguenez G, Goldschneider D, Terrier-Lacombe MJ, Hartmann O, Moll U, et al. DeltaN-p73alpha accumulates in human neuroblastic tumors. Am J Pathol. 2002;160:631–639. doi: 10.1016/s0002-9440(10)64883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan M, Chen Y. Aberrant expression of DeltaNp73 in benign and malignant tumours of the prostate: correlation with Gleason score. J Clin Pathol. 2005;58:1175–1179. doi: 10.1136/jcp.2005.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putzer BM, Tuve S, Tannapfel A, Stiewe T. Increased DeltaN-p73 expression in tumors by upregulation of the E2F1-regulated, TA-promoter-derived DeltaN′-p73 transcript. Cell Death Differ. 2003;10:612–614. doi: 10.1038/sj.cdd.4401205. [DOI] [PubMed] [Google Scholar]
- 49.Casciano I, Ponzoni M, Lo Cunsolo C, Tonini GP, Romani M. Different p73 splicing variants are expressed in distinct tumour areas of a multifocal neuroblastoma [letter] Cell Death Differ. 1999;6:391–393. doi: 10.1038/sj.cdd.4400522. [DOI] [PubMed] [Google Scholar]
- 50.Concin N, Hofstetter G, Berger A, Gehmacher A, Reimer D, Watrowski R, Tong D, Schuster E, Hefler L, Heim K, et al. Clinical relevance of dominant-negative p73 isoforms for responsiveness to chemotherapy and survival in ovarian cancer: evidence for a crucial p53–p73 cross-talk in vivo. Clin Cancer Res. 2005;11:8372–8383. doi: 10.1158/1078-0432.CCR-05-0899. [DOI] [PubMed] [Google Scholar]
- 51.Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL, Wei F, Chen BS, Huang XP, Han YS, et al. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102:580–583. doi: 10.1002/ijc.10739. [DOI] [PubMed] [Google Scholar]
- 52.Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63:7113–7121. [PubMed] [Google Scholar]
- 53.Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114:2063–2072. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- 54.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 55.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 56.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dulloo I, Sabapathy K. Transactivation-dependent and-independent regulation of p73 stability. J Biol Chem. 2005;280:28203–28214. doi: 10.1074/jbc.M501702200. [DOI] [PubMed] [Google Scholar]
- 58.Wu L, Zhu H, Nie L, Maki CG. A link between p73 transcriptional activity and p73 degradation. Oncogene. 2004;23:4032–4036. doi: 10.1038/sj.onc.1207538. [DOI] [PubMed] [Google Scholar]
- 59.Ying H, Chang DL, Zheng H, McKeon F, Xiao ZX. DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol Cell Biol. 2005;25:6154–6164. doi: 10.1128/MCB.25.14.6154-6164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 61.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 62.Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 63.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 67.Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302:635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 68.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 70.Bottger A, Bottger V, Sparks A, Liu WL, Howard SF, Lane DP. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 71.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 72.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 73.Kubbutat M, Jones S, Vousden K. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 74.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 75.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 77.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 78.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 79.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 80.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 81.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dornan D, Bheddah S, Newton K, Ince W, Frantz GD, Dowd P, Koeppen H, Dixit VM, French DM. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 83.Duan W, Gao L, Druhan LJ, Zhu WG, Morrison C, Otterson GA, Villalona-Calero MA. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J Natl Cancer Inst. 2004;96:1718–1721. doi: 10.1093/jnci/djh292. [DOI] [PubMed] [Google Scholar]
- 84.Balint E, Bates S, Vousden KH. Mdm-2 binds p73 alpha without targeting degradation. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 85.Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- 87.Westfall MD, Joyner AS, Barbieri CE, Livingstone M, Pietenpol JA. Ultraviolet radiation induces phosphorylation and ubiquitin-mediated degradation of DeltaNp63alpha. Cell Cycle. 2005;4:710–716. doi: 10.4161/cc.4.5.1685. [DOI] [PubMed] [Google Scholar]
- 88.Bottger A, Bottger V, Garcia-Echeverria C, Chene P, Hochkeppel HK, Sampson W, Ang K, Howard SF, Picksley SM, Lane DP. Molecular characterization of the hdm2-p53 interaction. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 89.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 90.Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, Cox LS, Poon RY. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9:829–832. doi: 10.1016/s0960-9822(99)80367-4. [DOI] [PubMed] [Google Scholar]
- 91.Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, Kaelin WG, Jr, Oren M, Chen J, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gu J, Chen D, Rosenblum J, Rubin RM, Yuan ZM. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol Cell Biol. 2000;20:1243–1253. doi: 10.1128/mcb.20.4.1243-1253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53-homologue p63. J Biol Chem. 2001;19:19. doi: 10.1074/jbc.M107173200. [DOI] [PubMed] [Google Scholar]
- 94.Little NA, Jochemsen AG. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene. 2001;20:4576–4580. doi: 10.1038/sj.onc.1204615. [DOI] [PubMed] [Google Scholar]
- 95.Wang X, Arooz T, Siu WY, Chiu CH, Lau A, Yamashita K, Poon RY. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001;490:202–208. doi: 10.1016/s0014-5793(01)02124-x. [DOI] [PubMed] [Google Scholar]
- 96.Miyazaki K, Ozaki T, Kato C, Hanamoto T, Fujita T, Irino S, Watanabe K, Nakagawa T, Nakagawara A. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun. 2003;308:106–113. doi: 10.1016/s0006-291x(03)01347-0. [DOI] [PubMed] [Google Scholar]
- 97.Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fomenkov A, Zangen R, Huang YP, Osada M, Guo Z, Fomenkov T, Trink B, Sidransky D, Ratovitski EA. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle. 2004;3:1285–1295. doi: 10.4161/cc.3.10.1155. [DOI] [PubMed] [Google Scholar]
- 99.Ozaki T, Watanabe K, Nakagawa T, Miyazaki K, Takahashi M, Nakagawara A. Function of p73, not of p53, is inhibited by the physical interaction with RACK1 and its inhibitory effect is counteracted by pRB. Oncogene. 2003;22:3231–3242. doi: 10.1038/sj.onc.1206382. [DOI] [PubMed] [Google Scholar]
- 100.Bakkers J, Camacho-Carvajal M, Nowak M, Kramer C, Danger B, Hammerschmidt M. Destabilization of DeltaNp63alpha by Nedd4-mediated ubiquitination and Ubc9-mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle. 2005;4:790–800. doi: 10.4161/cc.4.6.1694. [DOI] [PubMed] [Google Scholar]
- 101.Rossi MDSM, Pollice A, Santoro R, La Mantia G, Guerrini L, Calabro V. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle. 2006;5 doi: 10.4161/cc.5.16.2861. (NA) [DOI] [PubMed] [Google Scholar]
- 102.Ohtsuka T, Ryu H, Minamishima YA, Ryo A, Lee SW. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene. 2003;22:1678–1687. doi: 10.1038/sj.onc.1206306. [DOI] [PubMed] [Google Scholar]
- 103.Asher G, Shaul Y. p53 proteasomal degradation: polyubiquitination is not the whole story. Cell Cycle. 2005;4:1015–1018. doi: 10.4161/cc.4.8.1900. [DOI] [PubMed] [Google Scholar]
- 104.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci USA. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsvetkov P, Asher G, Reiss V, Shaul Y, Sachs L, Lotem J. Inhibition of NAD(P)H:quinone oxidoreductase 1 activity and induction of p53 degradation by the natural phenolic compound curcumin. Proc Natl Acad Sci USA. 2005;102:5535–5540. doi: 10.1073/pnas.0501828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hosoda M, Ozaki T, Miyazaki K, Hayashi S, Furuya K, Watanabe K, Nakagawa T, Hanamoto T, Todo S, Nakagawara A. UFD2a mediates the proteasomal turnover of p73 without promoting p73 ubiquitination. Oncogene. 2005;24:7156–7169. doi: 10.1038/sj.onc.1208872. [DOI] [PubMed] [Google Scholar]
- 108.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 109.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 110.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 111.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 112.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2000;21:21. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 113.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 114.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 117.Melchior F, Hengst L. SUMO-1 and p53. Cell Cycle. 2002;1:245–249. [PubMed] [Google Scholar]
- 118.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 119.Megidish T, Xu JH, Xu CW. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1) J Biol Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- 120.Nelson V, Davis GE, Maxwell SA. A putative protein inhibitor of activated STAT (PIASy) interacts with p53 and inhibits p53-mediated transactivation but not apoptosis. Apoptosis. 2001;6:221–234. doi: 10.1023/a:1011392811628. [DOI] [PubMed] [Google Scholar]
- 121.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 123.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miyauchi Y, Yogosawa S, Honda R, Nishida T, Yasuda H. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J Biol Chem. 2002;277:50131–50136. doi: 10.1074/jbc.M208319200. [DOI] [PubMed] [Google Scholar]
- 125.Pan Y, Chen J. Modification of MDMX by sumoylation. Biochem Biophys Res Commun. 2005;332:702–709. doi: 10.1016/j.bbrc.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 126.Xirodimas DP, Chisholm J, Desterro JM, Lane DP, Hay RT. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 2002;528:207–211. doi: 10.1016/s0014-5793(02)03310-0. [DOI] [PubMed] [Google Scholar]
- 127.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 128.Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight RA, et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol Cell Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghioni P, D'Alessandra Y, Mansueto G, Jaffray E, Hay RT, La Mantia G, Guerrini L. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle. 2005;4:183–190. doi: 10.4161/cc.4.1.1359. [DOI] [PubMed] [Google Scholar]
- 130.Huang YP, Wu G, Guo Z, Osada M, Fomenkov T, Park HL, Trink B, Sidransky D, Fomenkov A, Ratovitski EA. Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle. 2004;3:1587–1596. doi: 10.4161/cc.3.12.1290. [DOI] [PubMed] [Google Scholar]
- 131.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 132.Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 133.Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA, Kaelin WG., Jr An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3:177–182. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24:3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 137.Querido E, Morisson MR, Chu-Pham-Dang H, Thirlwell SW, Boivin D, Branton PE. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J Virol. 2001;75:699–709. doi: 10.1128/JVI.75.2.699-709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oberst A, Rossi M, Salomoni P, Pandolfi PP, Oren M, Melino G, Bernassola F. Regulation of the p73 protein stability and degradation. Biochem Biophys Res Commun. 2005;331:707–712. doi: 10.1016/j.bbrc.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 139.Chan WM, Mak MC, Fung TK, Lau A, Siu WY, Poon RY. Ubiquitination of p53 at multiple sites in the DNA-binding domain. Mol Cancer Res. 2006;4:15–25. doi: 10.1158/1541-7786.MCR-05-0097. [DOI] [PubMed] [Google Scholar]