Abstract
The physiologic response to changes in cellular oxygen tension is ultimately governed by a heterodimeric transcription factor called hypoxia-inducible factor (HIF), which, in adaptation to compromised oxygen availability, transactivates a myriad of genes, including those responsible for de novo vascularization, production of oxygen-carrying red blood cells, and anaerobic metabolism. Accumulation of HIF is observed in most types of solid tumors and is frequently associated with poor prognosis and disease progression, underscoring the importance and relevance of HIF in cancer. The protein stability and, thereby, the activity of HIF are principally regulated by the von Hippel-Lindau (VHL) tumor-suppressor-containing E3 ubiquitin ligase complex (ECV) that targets the catalytic subunit HIFα for oxygen-dependent ubiquitin-mediated destruction. Individuals who inherit germline VHL mutation develop VHL disease, which is characterized by the development of hypervascular tumors in multiple yet specific organs. This review will examine recent progress in our understanding of the molecular mechanisms governing the function of ECV and the significance of consequential regulation of HIF in oncogenesis.
Keywords: VHL, HIF, ECV, ubiquitin, NEDD8
History of von Hippel-Lindau (VHL) Disease
VHL disease (OMIM 193300) is a familial cancer syndrome that was first described in the medical literature by a German ophthalmologist Eugen von Hippel, a Swedish neuropathologist Arvid Lindau, and a British surgeon E. Treacher Collins at the turn of the 20th century [1]. VHL disease is estimated to affect 1 in 36,000 individuals, displaying no ethnic, racial, cultural, or sexual bias, and is characterized by the presence of hypervascular tumors in multiple organs, including the central nervous system (cerebellum, brainstem, and spinal cord), retina, pancreas, adrenal gland, endolymphatic sac of the inner ear, epididymis (male), broad ligament (female), and kidneys [2]. Although most of the tumors associated with VHL disease are benign, kidney cancer is malignant and is of the clear cell type, which accounts for 75% of kidney cancers. Kidney cancer remains as the principal cause of morbidity and mortality for VHL patients [2,3].
VHL disease is caused by the inheritance of a defective copy of the VHL gene, which was identified by Latif et al. [4] (from the National Institutes of Health and Oxford University). Tumors arise in a VHL kindred when the remaining wild-type allele is mutated or lost in a susceptible cell. Thus, on a cellular level, VHL disease has an autosomal recessive pattern of inheritance requiring inactivation of both alleles. However, clinically, it is perceived as an autosomal-dominant disease because the occurrence of the second inactivating mutation on the wild-type allele is virtually guaranteed [2]. VHL inactivation has been established as an early and requisite step in renal clear-cell carcinoma (RCC) pathogenesis, as the loss of heterozygosity in the remaining wild-type VHL allele in the proximal renal tubular epithelial cells has been documented in early premalignant renal cysts in VHL patients [5]. In keeping with the two-hit model of Knudson [6], biallelic inactivation of the VHL gene is also observed in the majority of sporadic RCC, establishing VHL as the critical “gatekeeper” of the renal epithelium.
The VHL gene consists of three exons, producing two transcripts that are translated into three proteins. The first VHL mRNA of approximately 4.5 kb contains exons 1 to 3 and is translated into two proteins due to an internal translational initiation start site at codon 54 [7–9]. The larger product is a 213-amino-acid protein of approximately 24 to 30 kDa (VHL30), and the shorter product is an 18- to 19-kDa isoform (VHL19) of 160 amino acids. The second VHL mRNA contains exons 1 and 3 due to alternative splicing. Tumors that exclusively produce this exon 2-less transcript have been identified, suggesting that the protein product encoded by this alternatively spliced transcript is defective in tumor-suppressor activity [10]. Furthermore, VHL mRNA expression is ubiquitous and, thus, is not restricted to specific tissue types that have been associated with VHL disease [11,12]. In addition, the VHL expression pattern in fetal kidneys suggests a role in normal renal tubular development and differentiation [11,12].
VHL disease is classified into categories, depending on a patient's likelihood of developing pheochromocytoma [2]. Type 1 patients have a low risk of developing pheochromocytoma, but present with RCC. Type 2 patients have a high risk of developing pheochromocytoma, with type 2A patients having a low risk of developing RCC but with type 2B patients having a high risk of developing RCC. Type 1, type 2A, and type 2B patients also develop the two cardinal features of VHL disease: cerebellar and retinal hemangioblastomas. Type 2C patients develop pheochromocytoma exclusively. The mutations associated with type 1 disease are deletions, microinsertions, and nonsense mutations, whereas type 2 patients typically present with missense mutations.
VHL19 Is Not VHL30, But Is It a Tumor Suppressor?
The human VHL gene is translated into two wild-type VHL proteins: VHL30 and the internally translated VHL19 [7–9]. Reconstitution of RCC cells with either VHL30 or VHL19 suppressed tumor development in a nude mouse xenograft assay [8,9,13,14]. This observation led to the belief that VHL30 and VHL19 have overlapping functions as tumor suppressors. However, recent findings have challenged this notion. Firstly, VHL mutations associated with tumor development have been identified throughout an open reading frame, including several mutations within the first 53 amino acids that are predicted to produce functional VHL19 [15–17]. Secondly, VHL30 and VHL19 were shown to have different subcellular localization profiles. Although VHL30 is found in the nuclear, cytosolic, and membranous [associated with endoplasmic reticulum (ER)] fractions, VHL19 is excluded from the membrane fraction [7,18–20]. Although the functional significance of VHL30 association with ER is unclear, it may be related to the ability of VHL30, but not VHL19, to bind fibronectin and its requirement to promote the assembly of fibronectin extracellular matrix (ECM) [20,21]. Thirdly, Stickle et al. [22] have shown that mutant VHL-expressing RCC cells with intact hypoxia-inducible factor (HIF) regulation but defective fibronectin ECM assembly formed tumors in an SCID mouse xenograft assay, underscoring the significance of proper fibronectin ECM in the development of RCC. In addition, phosphorylation of the N-terminal acidic domain, which is lacking in VHL19, through casein kinase 2 was shown to attenuate the binding of VHL30 to fibronectin [23]. This result suggests that, although the first 53 amino acids of VHL30 are required for binding fibronectin, phosphorylation of this region either prevents the recruitment of fibronectin or releases bound fibronectin in the ER. In light of these recent findings, the question of whether VHL19 can support a tumor-suppressor role needs to be revisited.
ECV Complex and the Ubiquitin-Mediated Destruction of HIF
VHL (VHL30 or VHL19) is a component of an E3 ubiquitin ligase complex called ECV, consisting of elongin B, elongin C, Rbx1 (also known as ROC1/Hrt1), and Cullin 2 (Cul2) [3]. Structurally and functionally, ECV is analogous to the Skp1/Cdc53/F-box protein (SCF) complex. VHL consists of two functional domains: α and β [24]. The α domain is required for binding elongin C, which binds to Cul2 to nucleate the ECV complex. The β domain acts as a substrate-recognition/docking site. Disease-associated mutations in the VHL kindred frequently map to surface residues on either domain, suggesting that these domains are important for the tumor-suppressor function of VHL [24]. Rbx1, which is recruited by Cul2, is thought to recognize a cognate E2 ubiquitin-conjugating enzyme required for the E3 ligase function of ECV [25–27].
Several putative substrates of ECV, including atypical protein kinase C [28], VHL-interacting deubiquitinating enzyme [29], and the seventh (Rpb7) [30] and the large (Rbp1) [31] subunits of RNA polymerase II, have been identified. However, not every protein bound by VHL is subjected to polyubiquitylation. These include SP1 transcription factor [32], VHL-associated KRAB-A domain-containing protein transcription repressor [33], microtubules [34], and fibronectin [20,35]. These findings have led to the notion that VHL has multiple functions from transcription, to cytoskeletal organization, to ECM assembly through ubiquitin-dependent and ubiquitin-independent mechanisms. Although these are intriguing possibilities, especially with growing evidence supporting the role of VHL in the assembly of fibronectin ECM, whether other aforementioned functions are physiologically relevant and/or necessary for the tumor-suppressor activity of VHL remains to be (further) proven.
What is widely accepted as a bona fide ECV substrate is the α subunit of HIFα [36,37]. HIF is the major transcription factor that transactivates a number (more than 60 and growing) of hypoxia-inducible genes, including vascular endothelial growth factor (VEGF; also known as vascular permeability factor), erythropoietin (EPO), and glucose transporter-1 (GLUT1), to promote angiogenesis, production of oxygen-carrying erythrocytes, and anaerobic metabolism, respectively, in adaptation to reduced oxygen tension [38,39]. There are three members of the HIF family (HIF-1, HIF-2, and HIF-3) in humans [40,41]. HIF is a heterodimeric complex consisting of α and β subunits. The β subunit [also known as aryl hydrocarbon receptor nuclear translocator (ARNT)] is abundantly expressed independent of oxygen tension, whereas the α subunit is oxygen labile. Specifically, the α subunit is ubiquitylated on a stretch of residues within the oxygen-dependent degradation (ODD) domain and, consequently, is targeted for degradation through the 26S proteasome [42]. Under hypoxia, HIFα is stabilized and binds to the common ARNT to form an active HIF complex, which binds to hypoxia-responsive elements (HREs) within the promoter/enhancer of hypoxia-inducible genes. Thus, HIF regulation occurs at the level of the α subunit.
VHL, through its substrate-binding β domain, recruits the HIFα subunit for oxygen-dependent ubiquitylation [36,37,43,44]. In the presence of oxygen, HIFα is hydroxylated on conserved prolines (P) at positions 402 and 564 (the number according to HIF-1α) within the ODD domain by prolyl hydroxylase domain (PHD)-containing enzymes [45,46]. P564 hydroxylation is both necessary and sufficient for the binding of HIFα ODD to VHL [45,46]. Thus, ubiquitin-mediated destruction of HIFα only occurs in the presence of oxygen. Accordingly, under hypoxia, HIFα is no longer prolyl-hydroxylated and thus escapes recognition by VHL. The now stabile HIFα dimerizes with ARNT to bind HREs to the trigger transcriptional activation of numerous hypoxia-inducible genes.
In addition, a conserved C-terminal asparagine at position 803 on HIF-1α is hydroxylated by the factor-inhibiting HIF-1 enzyme in the presence of oxygen [47–49]. Unlike prolyl hydroxylation, which induces VHL binding to HIFα, asparagyl hydroxylation prevents the recruitment of p300/CBP transcriptional coactivators to HIFα Thus, asparagyl hydroxylation of HIF-1α attenuates the transcription of HIF target genes [50,51]. This would suggest that there are, at a minimum, two mechanisms that negatively regulate the expression of hypoxia-inducible genes under normoxia [1]: oxygen-dependent ubiquitylation of HIFα through ECV and [2] the inhibition of p300/CBP recruitment in any remaining HIFα that has evaded destruction by ECV.
Several lines of evidence support the significance of the VHL regulation of HIF in cancer. VHL-associated tumors are highly vascularized, displaying overproduction of angiogenic peptides, such as VEGF, which is one of many HIF-mediated genes. In addition, VHL-defective cells express inordinately high levels of numerous hypoxia-inducible transcripts even under normoxic conditions [14,36,52–54]. Reconstitution of cells devoid of VHL with wild-type VHL restored the cells' ability to regulate or, more precisely, downregulate the expression of hypoxia-inducible genes in the presence of oxygen [14,53,55–57]. The relative contribution of HIF-1 vs HIF-2 (and, more recently, HIF-3) to RCC is an emerging area of research. The introduction of an HIF-1α mutant that escapes VHL recognition into RCC cells reconstituted with wild-type VHL does not produce a tumorigenic phenotype in SCID mice [58]. However, the treatment of these VHL-restored RCC cells with an HIF-1α ODD peptide that can block VHL binding to HIFα substrates restored the tumorigenic phenotype [58]. This finding suggests that, although HIF-1α is dispensable, other HIFα subunits (or other ECV substrates) are associated with the tumor-suppressor function of VHL. In support of this notion, Kondo et al. [59] demonstrated that, unlike HIF-1α, nondegradable HIF-2α was able to restore the tumor phenotype in RCC cells expressing wild-type VHL. This suggests that HIF-2α is the relevant oncogenic player in the development of RCC. Interestingly, VHL mutations affecting HIF regulation were predominantly associated with the development of hemangioblastoma and RCC, but not pheochromocytoma [35,60]. For example, VHL mutants associated with type 2C VHL disease (i.e., exclusive development of pheochromocytoma) were shown to have “normal” E3 ubiquitin ligase activity and to retain proper HIF function [35,60]. These mutants, however, were incapable of binding and regulating the assembly of fibronectin ECM [35,60].
Mouse Model of VHL Disease
Conventional knockout of VHL in mice results in embryonic lethality due to defective placental vasculature, precluding the study of VHL inactivation/disease in adults [61]. Therefore, to generate a mouse model to study VHL disease, Rankin et al. [62] used the phosphoenolpyruvate carboxy-kinase (PEPCK) promoter to generate transgenic mice in which Cre-recombinase is expressed in renal proximal tubules and hepatocytes. Conditional inactivation of VHL in PEPCK-Cre mice resulted in glomerular and tubular renal cysts, increased serum EPO levels, and polycythemia [62]. Notably, elevation of EPO level was limited to the liver, whereas HIF downstream genes carbonic anhydrase 9 and multidrug resistance gene 1 were increased in the renal cortex. The inactivation of ARNT, but not HIF-1α, prevented conditional VHL knockout mice from developing renal cysts [62,63], further supporting the notion that another partner of ANRT (such as HIF-2α, but not HIF-1α) is the relevant oncogenic player in the transformation of renal proximal tubules.
Development of renal cysts in mice on VHL inactivation is similar to the human condition wherein loss of VHL has been observed in preneoplastic cysts [5], and suggests that other genetic events are required for the progression of premalignant cysts to RCC.
Role of NEDD8 in ECV Function
The E3 function of SCF and SCF-like ECV is dependent on the recruitment of their respective E2 ubiquitin-conjugating enzyme (Cdc34 and UbcH5a, respectively). Cullins are scaffold components of SCF/ECV, which, until recently, have been identified as singular proteins covalently modified by the ubiquitin-like molecule, NEDD8 [22,64]. NEDD8 is attached to substrates in a manner analogous to a ubiquitin conjugation process, requiring NEDD8-activating APP-BP1/Uba3 enzyme (E1; NAE) and NEDD8-conjugating enzyme UbcH12 (E2; NCE). Unlike the ubiquitin pathway that has multiple E2s, the NEDD8 pathway, to date, has just one E2. Importantly, the overall E3 ubiquitin ligase activity of the yeast SCF is enhanced by covalent modification of the Cullin orthologue Cdc53 by the NEDD8 orthologue, related-to-ubiquitin 1 [65,66]. Similarly, the activity of the mammalian SCFβTrCP and SCFSkp2 complexes is increased by neddylation of Cul1, which facilitates the ubiquitylation of IκBα and p27, respectively [67,68]. Accordingly, NEDD8 modification of Cul2 enhances the activity of ECV in vivo [69].
In search for mechanisms governing SCF function, the core Cullin/Rbx1 complex was shown to be required for the recruitment of Cdc34 by the yeast SCF [27,70]. Subsequently, the neddylated Cul1/Rbx1 complex was demonstrated to be significantly better at supporting the Cdc34-mediated assembly of polyubiquitin chains than the unneddylated Cul1/Rbx1 counterpart [71]. In support, NEDD8 modification of Cul1 was shown to directly enhance the binding of ubiquitin-conjugated E2 Ubc4 to SCFβTrCP [72]. In addition, p120CAND1 was identified to interact selectively with unneddylated Cul1 to cause Skp1 dissociation from the SCF complex. Conversely, neddylation of Cul1 prevented p120CAND1 binding, allowing SCF complex formation and activity [73–75]. However, it is unlikely that p120CAND1 or p120CAND1-like protein is involved in the NEDD8-dependent assembly of ECV because unneddylated Cul2 is also found in the ECV complex without causing the dissociation of VHL from the complex (M. Ohh, unpublished data). Although these reports reveal important insights into the NEDD8-mediated regulation of SCF, it is not entirely clear how the timing of E2 recruitment is coordinated with the engagement of the substrate through the F-box protein.
In addition to UbcH12, the NEDD8 modification of Cul2 requires Rbx1, which suggests Rbx1 to be an E3 NEDD8 ligase (R. I. Sufan and M. Ohh, unpublished data). Neddylated Cul2 preferentially binds UbcH5a (R. I. Sufan and M. Ohh, unpublished data). Interestingly, HIFα-engaged ECV preferentially contains neddylated Cul2, whereas ECV, consisting of mutant VHL incapable of recruiting HIFα, exclusively associates with unmodified Cul2 (R. I. Sufan and M. Ohh, unpublished data). These results support the notion that the oxygen-dependent binding of HIFα through VHL triggers Rbx1-mediated neddylation of Cul2, which promotes the engagement of UbcH5a to the ECV complex, thereby establishing a central role for the neddylation of Cul2 in the temporally coordinated activation of ECV with the recruitment of its substrate, HIFα. However, it is not yet known how the binding of HIFα triggers the NEDD8-mediated activation of ECV.
Emerging Models of HIF-Mediated Death and Adhesion
VHL-Associated Death Function
Tumors with elevated hypoxic tissue profile pose a serious problem to the efficacy of conventional radiation therapy and chemotherapy. Global gene expression profiling has revealed that RCC cells display VHL-dependent sensitivity to tumor necrosis factor (TNF) α-mediated cytotoxicity [76]. Reconstitution of RCC (VHL-/-) cells with wild-type VHL restored their sensitivity to TNFα cytotoxicity, at least in part, by downregulating the level of nuclear factor (NF) κB in the nucleus, resulting in the attenuated expression of NF-κB target antiapoptotic genes c-FLIP, Survivin, c-IAP-1, and c-IAP-2, which block the activities of caspases 8 and 3 [77]. Recently, An and Rettig [78] showed that the activation of N F-κB on the loss of VHL was dependent on the HIF pathway, which induces the expression of TGFα, with consequent activation of the EGFR/PI3-OH kinase/AKT/IκB kinase α/NF-κB signaling cascade. In keeping with the model of a classic tumor suppressor, VHL has a proapoptotic function that is HIF-mediated.
In contrast, Devarajan et al. [79] showed that, in comparison to VHL- cells, VHL+ cells display an upregulated expression of Bcl-2, reduced activation of caspase 9, and release of cytochrome c into the cytosol following chemical hypoxia. Thus, in this setting, VHL seems to have an antiapoptotic function. Although counterintuitive, the authors speculate that the loss of VHL may increase the sensitivity of cells to physiologic stresses, which may foster selective pressure for cells to circumvent death under such conditions. The clonal outgrowth of VHL- cells may then acquire additional genetic mutations contributing to neoplastic transformation. It is unknown whether the antiapoptotic function of VHL is HIF-dependent.
VHL-Associated Adhesion Function
VHL negatively regulates the activity of HIF, and failure in this regulation leads to tumor development in experimental settings. However, how or why the deregulation or, more precisely, the overactivation of HIF leads to tumorigenesis is still unclear. Krishnamachary et al. [80] and Esteban et al. [81] independently showed that the loss of VHL in RCC cells results in the loss of E-cadherin in an HIF-dependent manner. E-cadherins, homophilic adhesion molecules, and their associated catenins are the major constituents of cell junctions in polarized epithelial cells [82]. Increased expression of E-cadherin is associated with the differentiation of mesenchymal cells into tubular epithelial cells of adult nephrons. Conversely, loss of cell-cell adhesion is frequently associated with tumor progression, metastasis, and poor prognosis [82]. In support of this dogma, the loss of E-cadherin is associated with the progression of numerous carcinoma types [82]. Forced expression of E-cadherin suppresses tumor development and invasion in various in vitro and in vivo tumor model systems, establishing E-cadherin as a critical tumor suppressor of the epithelium [82]. Thus, HIF-dependent repression of E-cadherin in RCC, devoid of VHL, may provide the formerly “missing” biologic basis for the development, as well as the aggressive nature, of RCC.
This is not without some controversy. Krishnamachary et al. [80] argued that the regulation of E-cadherin expression is exclusively HIF-1-dependent, as it failed to see the recovery of E-cadherin level when 786-O cells, which were HIF-1-, were reconstituted with wild-type VHL. However, Esteban et al. [81], using RCC4 (HIF-1α+/+; HIF-2α+/+) and 786-O (HIF-1α-/-; HIF-2α+/+) cells, demonstrated dependency on both HIF-1 and HIF-2. In addition, activation of the HIF pathway on the loss of VHL transactivates the E-cadherin transcriptional repressors TCF3 (also known as E12/E47), ZFHX1A (δEF1 or ZEB1), and ZFHX1B (also known as SIP1 or ZEB2), which result in the downregulation of E-cadherin transcription [80]. Our group has observed a similar induction of E-cadherin transcriptional repressors on HIF-1 and/or HIF-2 activation, including ZFHX1B/SIP1 and Snail, but no significant changes on TCF3 or ZFHX1A were observed (A. J. Evans, O. R. Losada, R. C. Russell, and M. Ohh, unpublished data).
Recently, Calzada et al. [83] have shown that the introduction of wild-type VHL in RCC (VHL-/-) cells restores the assembly of intercellular junctions through an HIF-independent mechanism to promote the establishment of an epithelial-like cell shape in otherwise fibroblastic-like RCC cells. Kurban et al. [84] reported that the loss of ECM assembly correlates with increased tumor angiogenesis and matrix metalloproteinase-2 activity. Surprisingly, the loss of HIF regulation in RCC cells, while resulting in tumors with increased VEGF levels, displayed low microvessel density, tightly assembled ECM, and low invasive potential [84]. These results suggest that the loss of ECM integrity promotes tumor angiogenesis by providing a route for blood vessels to infiltrate the tumor.
VHL-p53 Connection
Recently, Roe et al. [85] reported an unexpected connection between p53 and VHL. VHL was shown to directly bind and stabilize p53 by suppressing Mdm2-mediated ubiquitination. VHL also induced the acetylation of p53 on genotoxic stress by promoting p53–p300 interaction, resulting in increased p53 transcriptional activity and p53-mediated cell cycle arrest and apoptosis [85]. This is truly a surprising finding given that VHL was shown previously to have negligible effect on the expression level of p53 in RCC cells [86]. Further independent validation will be critical to determine the role of VHL in p53 function.
Summary
As tumors grow, the diffusional capacity of oxygen from the nearest blood vessel is inevitably surpassed, creating pockets of hypoxia within the tumor. The hypoxic microenvironment triggers the stabilization, as well as the increased translation (through the mammalian target of rapamycin [mTOR]), of HIFα. HIFα binds to the constitutively expressed and stable ARNT to form an active HIF transcription factor that initiates the transcription of genes containing HREs within the promoter/enhancer regions. HIF-driven gene transcripts responsible for, but not limited to, the promotion of neovascularization, anaerobic metabolism, and cell survival are expressed in adaptation to the reduced and often compromised oxygen availability, underscoring the importance of HIF in the survival, growth, and metastasis of tumors. Not surprisingly, the degree of tumor hypoxia correlates with poor prognosis, as well as with resistance to conventional anticancer therapies.
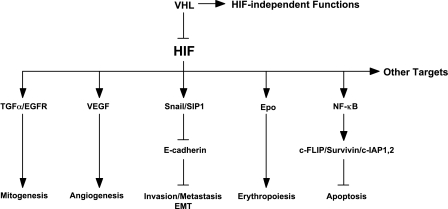
VHL or ECV (elongins/Cul2/VHL) is the major regulator of HIF by determining the stability of the catalytic HIFα subunit. ECV selectively targets HIFα that has undergone prolyl hydroxylation by PHDs in the presence of oxygen. Thus, ubiquitin-mediated destruction of HIFα occurs only under normoxic conditions. Interestingly, the engagement of HIFα to VHL is temporally coordinated with the neddylation of Cul2 through UbcH12 and Rbx1. The neddylated Cul2 then binds UbcH5a, which polyubiquitylates HIFα. Tumor hypoxia or inactivating mutation in VHL results in the stabilization of HIFα and in the consequential activation of HIF, triggering the expression of genes that ultimately drive neoplastic transformation—from loss of cell-cell contact inhibition, to dedifferentiation, to tipping of the balance toward survival over death (Figure 1).
Figure 1.
HIF-dependent VHL pathways in oncogenesis (see text for details).
Elucidation of HIF-dependent functions of VHL/ECV has revealed an unprecedented wealth of knowledge on the oxygen-sensing pathway and the pathophysiology of solid tumor development. Such information has afforded novel “smarter” avenues of anticancer strategies directed against important molecular targets revealed along the VHL-HIF pathway [87]. Imagine what we can learn from deciphering the other yet-defined HIF-dependent and HIF-independent tumor-suppressor functions of VHL.
Footnotes
This work was supported by the National Cancer Institute of Canada, with funds provided by the Canadian Cancer Society and the Kidney Foundation of Canada. M.O. is a Canada Research Chair in Molecular Oncology.
References
- 1.Maher E, Kaelin WG. von Hippel-Lindau disease. Medicine. 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Ohh M, Kaelin WG., Jr VHL and kidney cancer. Methods Mol Biol. 2003;222:167–183. doi: 10.1385/1-59259-328-3:167. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin WG., Jr Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 4.Latif F, Tory K, Gnarra J, Yao M, Duh F-M, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 5.Lubensky IA, Gnarra JR, Bertheau P, Walther MM, Linehan WM, Zhuang Z. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am J Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- 6.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iliopoulos O, Ohh M, Kaelin W. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc Natl Acad Sci USA. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci USA. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenship C, Naglich J, Whaley J, Seizinger B, Kley N. Alternate choice of initiation codon produces a biologically active product of the von Hippel Lindau gene with tumor suppressor activity. Oncogene. 1999;18:1529–1535. doi: 10.1038/sj.onc.1202473. [DOI] [PubMed] [Google Scholar]
- 10.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh F-M, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 11.Kessler P, Vasavada S, Rackley R, Stackhouse T, Duh F, Latif F, Lerman M, Zbar B, Williams B. Expression of the von Hippel-Lindau tumor-suppressor gene, VHL, in human fetal kidney and during mouse embryogenesis. Mol Med. 1995;1:457–466. [PMC free article] [PubMed] [Google Scholar]
- 12.Richards F, Schofield P, Fleming S, Maher E. Expression of the von Hippel-Lindau disease tumour suppressor gene during human embryogenesis. Hum Mol Genet. 1996;5:639–644. doi: 10.1093/hmg/5.5.639. [DOI] [PubMed] [Google Scholar]
- 13.Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumor suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 14.Gnarra JR, Zhou S, Merrill MJ, Wagner J, Krumm A, Papavassiliou E, Oldfield EH, Klausner RD, Linehan WM. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the VHL tumor suppressor gene product. Proc Natl Acad Sci. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crossey PA, Richards FM, Foster K, Green JS, Prowse A, Latif F, Lerman MI, Zbar B, Affara NA, Ferguson-Smith MA, et al. Identification of intragenic mutations in the von Hippel-Lindau disease tumor suppressor gene and correlation with disease phenotype. Hum Mol Genet. 1994;3:1303–1308. doi: 10.1093/hmg/3.8.1303. [DOI] [PubMed] [Google Scholar]
- 16.van der Harst E, de Krijger R, Dinjens W, Weeks L, Bonjer H, Bruining H, Lamberts S, Koper J. Germline mutations in the vhl gene in patients presenting with phaeochromocytomas. Int J Cancer. 1998;77:337–340. doi: 10.1002/(sici)1097-0215(19980729)77:3<337::aid-ijc5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Zbar B, Kishida T, Chen F, Schmidt L, Maher ER, Richards FM, Crossey PA, Webster AR, Affara NA, Ferguson-Smith MA, et al. Germline mutations in the von Hippel-Lindau (VHL) gene in families from North America, Europe, and Japan. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Corless CL, Kibel A, Iliopoulos O, Kaelin WGJ. Immunostaining of the von Hippel-Lindau gene product (pVHL) in normal and neoplastic human tissues. Hum Pathol. 1997;28:459–464. doi: 10.1016/s0046-8177(97)90035-6. [DOI] [PubMed] [Google Scholar]
- 19.Los M, Jansen GH, Kaelin WG, Lips CJM, Blijham GH, Voest EE. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab Invest. 1996;75:231–238. [PubMed] [Google Scholar]
- 20.Ohh M, Yauch RL, Lonergan KM, Whaley JM, Stemmer-Rachamimov AO, Louis DN, Gavin BJ, Kley N, Kaelin WG, Iliopoulos O, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 21.Esteban-Barragan MA, Avila P, Alvarez-Tejado M, Gutierrez MD, Garcia-Pardo A, Sanchez-Madrid F, Landazuri MO. Role of the von Hippel-Lindau tumor suppressor gene in the formation of beta1-integrin fibrillar adhesions. Cancer Res. 2002;62:2929–2936. [PubMed] [Google Scholar]
- 22.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24:3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lolkema MP, Gervais ML, Snijckers CM, Hill RP, Giles RH, Voest EE, Ohh M. Tumor suppression by the von Hippel-Lindau protein requires phosphorylation of the acidic domain. J Biol Chem. 2005;280:22205–22211. doi: 10.1074/jbc.M503220200. [DOI] [PubMed] [Google Scholar]
- 24.Stebbins CE, Kaelin WG, Pavletich NP. Structure of the VHL-elongin C-elongin B complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 25.Ohta T, Michel JJ, Schottelius AJ, Xiaong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 26.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 27.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 28.Okuda H, Hirai S, Takaki Y, Kamada M, Baba M, Sakai N, Kishida T, Kaneko S, Yao M, Ohno S, et al. Direct interaction of the betadomain of VHL tumor suppressor protein with the regulatory domain of atypical PKC isotypes. Biochem Biophys Res Commun. 1999;263:491–497. doi: 10.1006/bbrc.1999.1347. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Na X, Wang D, Schoen SR, Messing EM, Wu G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 30.Na X, Duan HO, Messing EM, Schoen SR, Ryan CK, di Sant'Agnese PA, Golemis EA, Wu G. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J. 2003;22:4249–4259. doi: 10.1093/emboj/cdg410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuznetsova AV, Meller J, Schnell PO, Nash JA, Ignacak ML, Sanchez Y, Conaway JW, Conaway RC, Czyzyk-Krzeska MF. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA. 2003;100:2706–2711. doi: 10.1073/pnas.0436037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen AJ, Li FP, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med. 1979;301:592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. The VHL protein recruits a novel KRAB-A domain protein to repress HIF1alpha transcriptional activity. EMBO J. 2003;22:1857–1867. doi: 10.1093/emboj/cdg173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, Kaelin WG., Jr von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- 36.Maxwell P, Weisner M, Chang G-W, Clifford S, Vaux E, Pugh C, Maher E, Ratcliffe P. The von Hippel-Lindau gene product is necessary for oxygen-dependent proteolysis of hypoxia-inducible factor a subunits. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 37.Ohh M, Park CW, Ivan M, Hoffman MA, Kim T-Y, Huang LE, Chau V, Kaelin WG. Ubiquitination of HIF requires direct binding to the von Hippel-Lindau protein beta domain. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 38.Semenza G. Perspectives on oxygen sensing. Cell. 1999;98:281–284. doi: 10.1016/s0092-8674(00)81957-1. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Bunn HF. Signal transduction. How do cells sense oxygen? Science. 2001;292:449–451. doi: 10.1126/science.1060849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maynard MA, Qi H, Chung J, Lee EH, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. Multiple splice variants of the human HIF-3alpha locus are targets of the VHL E3 ubiquitin ligase complex. J Biol Chem. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 41.Maynard MA, Ohh M. von Hippel-Lindau tumor suppressor protein and hypoxia-inducible factor in kidney cancer. Am J Nephrol. 2004;24:1–13. doi: 10.1159/000075346. [DOI] [PubMed] [Google Scholar]
- 42.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1 alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cockman M, Masson N, Mole D, Jaakkola P, Chang G, Clifford S, Maher E, Pugh C, Ratcliffe P, Maxwell P. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 44.Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 46.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 47.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 48.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1 alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wizigmann-Voos S, Breier G, Risau W, Plate K. Upregulation of vascular endothelial growth factor and its receptors in von Hippel-Lindau disease-associated and sporadic hemangio-blastomas. Cancer Res. 1995;55:1358–1364. [PubMed] [Google Scholar]
- 53.Iliopoulos O, Jiang C, Levy AP, Kaelin WG, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krieg M, Marti H, KH P. Coexpression of erythropoietin and vascular endothelial growth factor in nervous system tumors associated with von Hippel-Lindau tumor suppressor gene loss of function. Blood. 1998;92:3388–3393. [PubMed] [Google Scholar]
- 55.Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, Kaelin WG. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor protein. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 57.Stratmann R, Krieg M, Haas R, Plate K. Putative control of angiogenesis in hemangioblastomas by the von Hippel-Lindau tumor suppressor gene. J Neuropathol Exp Neurol. 1997;56:1242–1252. doi: 10.1097/00005072-199711000-00009. [DOI] [PubMed] [Google Scholar]
- 58.Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 59.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 60.Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum Mol Genet. 2001;10:1029–1038. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- 61.Gnarra J, Ward J, Porter F, Wagne J, Devor D, Grinberg A, Emmert-Buck M, Westphal H, Klausner R, Linehan W. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci USA. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rankin EB, Higgins DF, Walisser JA, Johnson RS, Bradfield CA, Haase VH. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol. 2005;25:3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 65.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1 p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osaka F, Saeki M, Katayama S, Aida N, Toh-E A, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Read M, Brownell J, Gladysheva T, Hottelet M, Parent L, Coggins M, Pierce J, Podust V, Luo R-S, Chau V, et al. Nedd8 modification of cul-1 activates SCFbTrCP dependent ubiquitination of IκBα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Podust V, Brownell J, Gladysheva T, Luo R, Wang C, Coggins M, Pierce J, Lightcap E, V C. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip 1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA, Kaelin WG. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3:177–182. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1 -CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 72.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1 -Cul1 -Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 75.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 76.Caldwell MC, Hough C, Furer S, Linehan WM, Morin PJ, Gorospe M. Serial analysis of gene expression in renal carcinoma cells reveals VHL-dependent sensitivity to TNFalpha cytotoxicity. Oncogene. 2002;21:929–936. doi: 10.1038/sj.onc.1205140. [DOI] [PubMed] [Google Scholar]
- 77.Qi H, Ohh M. The von Hippel-Lindau tumor suppressor protein sensitizes renal cell carcinoma cells to tumor necrosis factor-induced cytotoxicity by suppressing the nuclear factor-kappaB-dependent antiapoptotic pathway. Cancer Res. 2003;63:7076–7080. [PubMed] [Google Scholar]
- 78.An J, Rettig MB. Mechanism of von Hippel-Lindau protein-mediated suppression of nuclear factor kappa B activity. Mol Cell Biol. 2005;25:7546–7556. doi: 10.1128/MCB.25.17.7546-7556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Devarajan P, De Leon M, Talasazan F, Schoenfeld AR, Davidowitz EJ, Burk RD. The von Hippel-Lindau gene product inhibits renal cell apoptosis via Bcl-2-dependent pathways. J Biol Chem. 2001;276:40599–40605. doi: 10.1074/jbc.M103424200. [DOI] [PubMed] [Google Scholar]
- 80.Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, Semenza GL. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–2731. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 81.Esteban MA, Tran MGB, Harten SK, Hill P, Castellanos MC, Chandra A, Raval R, O'Brien TS, Maxwell PH. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–3575. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 82.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calzada MJ, Esteban MA, Feijoo-Cuaresma M, Castellanos MC, Naranjo-Suarez S, Temes E, Mendez F, Yanez-Mo M, Ohh M, Landazuri MO. von Hippel-Lindau tumor suppressor protein regulates the assembly of intercellular junctions in renal cancer cells through hypoxia-inducible factor-independent mechanisms. Cancer Res. 2006;66:1553–1560. doi: 10.1158/0008-5472.CAN-05-3236. [DOI] [PubMed] [Google Scholar]
- 84.Kurban G, Hudon V, Duplan E, Ohh M, Pause A. Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res. 2006;66:1313–1319. doi: 10.1158/0008-5472.CAN-05-2560. [DOI] [PubMed] [Google Scholar]
- 85.Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 86.Stickle NH, Cheng LS, Watson IR, Alon N, Malkin D, Irwin MS, Ohh M. Expression of p53 in renal carcinoma cells is independent of pVHL. Mutat Res. 2005;578:23–32. doi: 10.1016/j.mrfmmm.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 87.Maynard MA, Ohh M. Molecular targets from VHL studies into the oxygen-sensing pathway. Curr Cancer Drug Targets. 2005;5:345–356. doi: 10.2174/1568009054629672. [DOI] [PubMed] [Google Scholar]