Abstract
Defects in ubiquitin E3 ligases are implicated in the pathogenesis of several human diseases, including cancer, because of their central role in the control of diverse signaling pathways. RING E3 ligases promote the ubiquitination of proteins that are essential to a variety of cellular events. Identification of which ubiquitin ligases specifically affect distinct cellular processes is essential to the development of targeted therapeutics for these diseases. Here we discuss two novel RING E3 ligases, BCA2 and RNF11, that are closely linked to human breast cancer. BCA2 E3 ligase is coregulated with estrogen receptor and plays a role in the regulation of epidermal growth factor receptor (EGF-R) trafficking. RNF11 is a small RING E3 ligase that affects transforming growth factorβ and EGF-R signaling and is overexpressed in invasive breast cancers. These two proteins demonstrate the complexity of RING E3 ligase interactions in breast cancer and are potential targets for therapeutic interventions.
Keywords: RING ligase, breast cancer, BCA2, RNF11, ubiquitin
Introduction
Protein homeostasis at the cellular level is delicately balanced by de novo synthesis, posttranslational modification, and degradation [1]. The majority (80%) of cellular proteins is destroyed by the ubiquitin-proteasome system after ubiquitin tagging [2]. The ubiquitin system is hierarchically structured and confers specificity for protein substrates through a multitude of E3 ubiquitin ligases. A few E1 ubiquitin-activating enzymes exist; however, at least 50 E2 ubiquitin-conjugating enzymes and about 500 E3 ubiquitin ligases are present in the human genome [3–7]. E2 ubiquitin-conjugating enzymes are characterized by a conserved core sequence and by the ability to bind to E3 ligases. E2 ligases are therefore believed to have functional redundancy [7]. E3 ubiquitin ligases are currently grouped into two major categories: HECT-type and adaptor-type. The latter can contain a RING finger, a U-box, or a PHD domain [5,8–12]. RING E3 ligases mediate the direct transfer of E2-bound ubiquitin to substrates without thioester bond formation [11]. This is in contrast to HECT E3 ligases, which actively take up E2-bound ubiquitin by forming thioester linkages and then transferring ubiquitin to substrates.
Many of the known E3 ligases play a role in various diseases, including breast cancer development and progression. Among the first RING proteins to be associated with ubiquitination and cancer were the multisubunit complex E3 ligases ROC1 and APC11 [9,13]. The small RING finger protein ROC1 is an essential component of the multisubunit complex Skp1/cullin-1/F-box protein (SCF) E3 ligase [9]. This family of RING finger proteins interacts with cullins, creating a large number of multisubunit E3 Ub ligases of varying specificity [13]. SCF-type complexes ubiquitinate a broad range of substrates involved in cell signaling, cell cycle progression, signal transduction, and transcription [14]. In primary breast cancers, p27 levels are reduced by the SCF complex, leading to poor prognosis [5,14–16]. APC11 is a core catalytic subunit of the anaphase-promoting complex, which allows exit of cells from mitosis by destroying B-type cyclins complexed with Cdc2, resulting in the loss of Cdc2 kinase activity [17].
Closely associated with breast cancer are various monomeric RING finger E3 ligases, including Mdm2, Efp, c-Cbl, BRCA1/BARD1, COP1, and BCA2 [5,9,18,19]. Monomeric RING finger E3 ligases, unlike those participating in the SCF complex, have a substrate-binding domain and are capable of substrate ubiquitination and autoubiquitination. COP1, Mdm2, and BRCA1/BARD1 have a common substrate, namely, p53 [20,21].
COP1 has been found to be significantly overexpressed in 81% of breast carcinomas, and p53 is decreased in tumors expressing COP1 [19]. Germline mutations in the RING finger of BRCA1 predispose women to early-onset breast tumors [5], accounting for approximately 5% of breast cancers. The BRCA1-associated RING domain protein BARD1 acts with BRCA1 in double-strand break repair and ubiquitination. BRCA1 ubiquitin ligase activity is enhanced when it is dimerized with BRCA1-associated RING domain (BARD1), and it has been implicated as a mediator of apoptosis by binding to and stabilizing p53 [20,22]. Although COP1 seems to be a negative—and BRCA1/BARD1 seems to be a positive—regulator of p53 in breast cancer, the role of Mdm2 is less clear. Mdm2 was found to be an independent negative prognostic marker by studying protein expressions in a large number of microarrayed breast cancer cases [23]. However, Mdm2 expression in this tumor collection was not correlated to clinicopathological parameters. In a study that subclassified breast cancers, Mdm2 overexpression was associated with favorable prognostic markers: absence of lymph node involvement, low-grade nuclear atypia, and increased levels of estrogen receptor a protein [23,24]. Efp is an estrogen-inducible RING finger E3 ligase that ubiquitinates the 14-3-3σ protein. 14-3-3σ is a p53-regulated inhibitor of G2/M progression and has been shown to have tumor-suppressor function in breast cancer [25,26]. A survey of 151 breast cancers confirmed that Efp immunoreactivity is positively associated with lymph node status or estrogen receptor a status, and is negatively correlated with histologic grade or 14-3-3σ immunoreactivity. Efp immunoreactivity was significantly correlated with poor prognosis [27]. c-Cbl is an E3 ligase with a RING finger and an SH2 domain. It recognizes phosphorylated tyrosine residues in receptor tyrosine kinases through its SH2 domain, and it negatively regulates signaling by facilitating receptor ubiquitination [28–32]. In particular, c-Cbl acts as an E3 ligase, bringing together tyrosine-phosphorylated receptors and E2 ubiquitinconjugating enzymes. It thus induces the ubiquitination of activated tyrosine kinase receptors, including epidermal growth factor receptor (EGF-R) and platelet-derived growth factor receptor (PDGF-R). c-Cbl-dependent ubiquitination was shown to be important for early endosome to late endosome/lysosome sorting step during EGF-R turnover and, thus, was established as a major endogenous ubiquitin ligase responsible for EGF-R degradation [28,32]. Cbl has also been described as a suppressor of the HER2/Neu oncogene, which belongs to the EGF-R superfamily [29]. Overexpression of EGF-R, HER2/Neu, and EGF-R family heterodimer signaling is a major contributor to uncontrolled proliferation in many malignant diseases, including breast cancer [30]. The HER2 antibody trastuzumab (Herceptin; Genentech, San Francisco, CA) has been shown to direct HER2 to c-Cbl-regulated degradation, suggesting that mechanisms underlying the efficacy of trastuzumab immunotherapy might, in part, be due to EGF-induced degradation of HER2 by c-Cbl-mediated ubiquitination [31].
The following sections will focus on the role of two novel breast cancer-associated RING finger E3 ligases: the small RING finger protein RNF11 and the monomeric RING-H2 ubiquitin E3 ligase BCA2. Their potential usefulness as prognostic markers or therapeutic targets is also discussed.
RNF11 in Transforming Growth Factor (TGF)β-and EGF-R-Mediated Signaling
RNF11 encodes a RING-H2 domain and a PY motif, both of which mediate protein-protein interactions. In particular, the PPPPY sequence of the RNF11 PY motif is identical to that of Smad7, which has been shown to bind to WW domains of Smurf2—an E3 ubiquitin ligase that mediates the ubiquitination and degradation of the TGF receptor complex (Figure 1) [14,15].
Figure 1.
Model of RNF11 interactions in the TGFβ receptor signaling pathway. RNF11 acts against the degradation of the TGFβ receptor, enhancing its tumor-suppressor effects on breast cancer.
TGFβ is associated with both normal mammary gland development and breast carcinogenesis [33–35]. A dichotomous role for TGFβ in breast cancer as both a tumor suppressor in early-stage disease and as a tumor promoter in advanced disease has been described [36]. Many breast cancer cell lines show an increased expression of TGFβ but are refractory to TGFβ-induced cell cycle arrest. In addition, in many cancers, tumor cells lose responsiveness to TGFβ, leading to the idea that the disruption of this pathway may be an important factor in the development of cancer [37,38]. However, deletions of TβRII or Smads only represent a small fraction of cancers that are insensitive to TGFβ. Thus, there must be other uncharacterized proteins, such as RNF11, that contribute to the deregulation of the TGFβ signaling pathway and the accompanying TGFβ insensitivity evident in cancer cells.
Along those lines, we have found that RNF11 is capable of binding Smurf proteins that are involved in the TGFβ pathway [39–41]. Through its PY motif, RNF11 binds to the HECT-type E3 ligase Smurf2, making it a potential regulator of TGFβ signaling. RNF11 also competes with Smad7 binding to Smurf2, thereby disrupting the Smurf2/Smad7 complex (Figure 1) [42,43]. This prevents the negative effects of the Smurf2/Smad7 complex on TGFβ signaling and acts to restore TGFβ sensitivity to cells that have lost their responsiveness; overexpression of RNF11 increases the stability of the TGFβ receptor [41]. The interaction of RNF11 and Smurf2 could be crucial because it restores TGF signaling and because RNF11 could have multiple functions similar to those of Smad proteins. Perhaps RNF11 blocks some of the inhibitory effects of Smad7 on TGF signaling on cell proliferation and apoptosis, but leaves intact the positive effects of this pathway on malignant progression.
EGF-R is considered to be one of the main proteins that are elevated in breast cancers [44]. A strong correlation has been found between the presence of high levels of EGF-R in breast tumors and the aggressive potential of the tumor. EGF-R contains multiple phosphorylated residues, each of which directly interacts with specific domains in downstream signaling proteins. Under normal conditions, there are multiple mechanisms in the cell that act to tightly regulate these activities. These include positive regulation by growth factors, in addition to negative regulation of the receptor or of downstream signaling pathways through the action of phosphatases or other regulators of signaling pathways. Recently, ubiquitin ligases have been identified as such negative regulators. They primarily act following ligand stimulation, and internalized receptors are subject to two distinct fates: recycling back to the plasma membrane or degradation by lysosomal and proteosomal pathways.
Growing evidence indicates that ubiquitination of EGF-R and other receptor tyrosine kinase (RTKs) is critical for their lysosomal degradation through ubiquitin-dependent protein sorting, which retains RTKs in late endosomes and subsequently targets these receptors to intralumenal vesicles and lysosomal degradation [32,45–48]. RTKs can also be multi-monoubiquitinated, and monoubiquitination of EGF-R specifically is sufficient for its internalization and degradation [49]. Ubiquitinated receptors may be selectively recognized by proteins of the endocytic pathway that contain a ubiquitin-interacting motif, such as Epsin, EPS-15, Hrs, and STAM, and many other proteins [49,50]. We found that RNF11 associates with EPS-15 and AMSH, which is the molecule associated with the SH3 domain of STAM (AMSH). AMSH is an endosome-associated ubiquitin isopeptidase, and we have reported that its degradation is enhanced by RNF11 protein.
EPS-15 plays a major role in the clathrin-mediated internalization of EGF-R [51]. On activation, EGF-R phosphorylates EPS-15 at tyrosine 305. In an elaborate series of events, a multiprotein complex that includes hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and STAM binds to EGF-R, causing its internalization and degradation by lysosomal proteolysis [52,53]. This process requires an activated EGF-R and keeps EGF-R signaling in check. In contrast, EGF-R internalization is inhibited by AMSH protein. AMSH is a deubiquitinating enzyme that deubiquitinates ubiquitinated EGF-R and prevents the formation of the complex necessary for EGF-R internalization, which in turn prolongs EGF-R activation and promotes cellular proliferation. We have shown that RNF11 binds to AMSH and, in the presence of the E3 ligase Smurf2, promotes the ubiquitination and degradation of the AMSH protein [40]. In addition, it has been shown that RNF11 can bind directly to EPS-15. In order for EPS-15 to form a complex with Hrs and STAM, it must be monoubiquitinated following phosphorylation by EGF-R. Thus, it may be that, by its interaction with E2 and E3 ligases, RNF11 can mediate the mono-ubiquitination of EPS-15. This suggests that RNF11 could promote the internalization and degradation of EGF-R by two distinct protein-protein interactions by sequestering AMSH away from STAM and/or by mediating the mono-ubiquitination of EPS-15.
BCA2 E3 Ligase and Its Partners in Breast Cancer
BCA2 (synonymous to T3A12/ZNF364/Rabring7) has been identified as a novel breast cancer-associated gene by subtractive hybridization cloning of cDNA derived from matched normal and malignant breast cell RNA [54]. BCA2 has a close relative (RNF126) that shares 46% overall amino acid identity and 75% identity in RING domains. Both genes contain nine exons, encoding consensus AKT/14-3-3-binding sites, as well as zinc finger and RING finger domains (Figure 2A). A comparison of the RNF126 amino acid sequence with translated nucleotide sequences revealed probable homologues of BCA2 in vertebrate species, including humans, mice, birds, amphibians, and fish. This suggests that RNF126 might have a similar function as BCA2.
Figure 2.
Modular domains and binding sites in RNF126 and BCA2. (A) Schematic diagram of RNF126 and BCA2 protein sequences with originating exons marked in color, and with consensus zinc finger, AKT phosphorylation, and RING domains and sites indicated. Exons 4 and 9 bear unique codons that extend the length of RNF126 to 326 amino acids, 22 amino acids more than BCA2. Exon 4 of both RNF126 and BCA2 encodes a consensus AKT/14-3-3-binding site. (B) Detailed alignment of RNF126 and BCA2 domains and sites, as indicated with amino acid sequence locations that are numbered, with structurally important residues in purple or green and with potential phosphorylation sites marked in red. The RNF126 sequence has threonine residues, rather than serine residues, seen in BCA2 as likely sites of phosphorylation. (C) Amino acid regions unique to RNF126, including the serine tract and consensus SH3-binding sequence (Prosite).
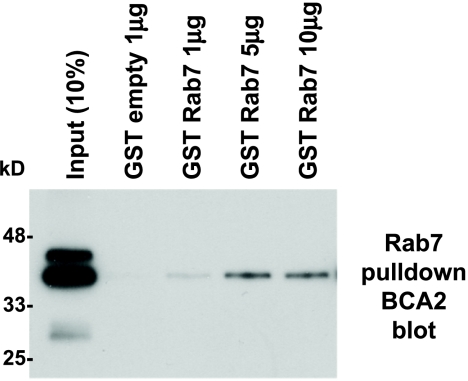
RING domain proteins have multiple motifs and domains that allow them to interact with multiple partner proteins. Others have shown that BCA2 is a target protein of Rab7 (a member of the Rab family of small G proteins) and that BCA2 also interacts with ALG2 [18,55,56]. Moreover, it was found that BCA2 significantly inhibited EGF degradation [56]. We have confirmed this interaction between the human Rabring7/BCA2 protein and the human Rab7 by using recombinant BCA2 (Xpress-tagged) and GST-Rab7 (Figure 3). Recombinant mutant AKT BCA2 protein does not bind to Rab7, raising the possibility that BCA2 phosphorylation by AKT [57] is essential to BCA2/Rab7 interaction. That interaction could affect receptor internalization and vesicle trafficking in the cytoplasm, separate from the E3 ligase function of BCA2 in the nucleus [18].
Figure 3.
BCA2 binds to Rab7. The GST-Rab7 fusion protein was incubated with Xpress-tagged recombinant BCA2 bacterial product at increasing concentrations of GST-Rab7.
Two partner proteins for RNF126, Traf6, and BAT3/Scythe have been discovered in high-throughput screening [58,59]. Although few other partners for either RNF126 or BCA2 have been identified, both proteins interact with one or more apoptosis-related proteins. Both BCA2 and RNF126 may interact with Scythe, and BCA2 interacts with ALG2. It may be that, in addition to or as a consequence of preserving gene structure, functional domains, and motifs, the general functions of these proteins have also been conserved throughout evolution.
Other potential binding motifs/partners can be deduced from the functional domains of BCA2 and RNF126 (Figure 2A), which include an AKT phosphorylation site within a potential 14-3-3-binding domain. 14-3-3 Molecules are a family of proteins that bind phosphorylated serine/threonine residues within a consensus-binding sequence. Several members of this gene family are downregulated in breast cancer, such as 14-3-3σ, which is a substrate of the E3 ligase Efp (see Refs. [18,25–27,57]). Both BCA2 and RNF126 have E3 ligase activity that could be affected by phosphorylation of their AKT sites (Figure 4) [18]. Those sites are located within 14-3-3-binding domains in exon 4 of BCA2 and RNF126 (Figure 2A). Mutation of the AKTsite of BCA2, unlike RING mutation, does not lead to complete loss of E3 ligase activity [18], suggesting that phosphorylation could contribute to BCA2 and perhaps RNF126 E3 ligase function in breast cancer.
Figure 4.
BCA2 and RNF126 E3 ligase activity. Autoubiquitination of recombinant BCA2 and RNF126. In vitro ubiquitination assays were performed with (+) and without (-) the E2-conjugating enzyme UbcH5b in the presence of ubiquitin. Shown is an anti-ubiquitin Western blot analysis of the reaction products visualized by chemiluminescence. High-molecular-weight products indicative of polyubiquitination are seen only when E2 is present in the reaction.
By employing multitumor tissue microarray technology, we found that normal breast, colon, and head and neck tumors lack detectable BCA2 expression (Figure 5A) [60]. Twenty of 20 breast cancers and 15 of 15 prostate cancers strongly expressed BCA2 (Figure 5B) [60]. Sixty percent of 17 renal cancers had strong BCA2 staining, and none of the clear cell renal tumors had detectable BCA2. Lower levels of BCA2 staining were seen in 57% of 16 colon cancers, in 65% of 22 pancreatic tumors, and in 73% of 17 bladder cancers. Weak expression of BCA2 was seen in 4 of 7 lung tumors and in 6 of 11 head and neck tumors.
Figure 5.
Expression of BCA2 protein in normal and tumor tissues. One hundred twenty-five tumor tissues and adjacent normal tissues from the same patient were assembled onto a tissue microarray slide, fixed, and probed for BCA2 expression by immunoperoxidase staining (positive reaction, brown). (A) The top panel shows representative cores (0.6 mm) of normal breast, colon and head and neck tissues stained for BCA2 protein expression (original magnification, x4). (B) The bottom panel shows strong nuclear and cytoplasmic expression of BCA2 in breast, prostate, and papillary renal cell cancers, but very weak staining in clear cell renal, colon, and head and neck carcinomas (original magnification, x40).
A second larger tumor tissue microarray study examined BCA2 expression in 945 invasive breast cancers [18]. Comparison with patient data revealed that BCA2 expression correlates with clinical variables such as lymph node status, regional recurrence, and estrogen receptor expression pattern [18,60]. Although BCA2 expression in the entire cohort did not correlate with overall survival, a subset of patients with regional recurrence history revealed that a higher BCA2 is linked with a significant 5-year survival benefit and that a low BCA2 is linked with lymph node metastasis. The correlation of BCA2 expression with estrogen receptor positivity indicates that BCA2 and estrogen receptor might be coregulated and cross-talked at the transcriptional level, perhaps in the nucleus.
Conclusions
When RING finger proteins were discovered, their role was recognized mainly in protein-protein interactions and protein dimerization [9]. As evidence evolved, their crucial function in mediating the transfer of ubiquitin to heterologous substrates as well as to themselves (autoubiquitination) and their crucial function as substrates with key roles in cell signaling and survival pathways emerged. RING E3 ligases have now become the subject of intense studies for their roles in disease and as potential therapeutic targets [5,6,9]. Several cancer-specific oncogenes code for ubiquitin ligases and/or their subunits. Interestingly, breast cancer, in particular, seems to be associated with the aberrant expression of several RING E3 ligases and their substrates.
All RING E3 ligases, including BCA2, contain a consensus protein sequence that would complex two zinc ions in the expressed protein (Figure 2A). This domain is likely essential to the ligase activity of all RING E3 ligases [5,18]. This dependence on the RING domain is demonstrated by the loss of E3 ligase activity in mutant RING proteins [18].
The crucial role of the RING-H2 finger and its zinc-complexing structure for BCA2, RNF126, and other RING E3 ligases is underscored by the fact that the zinc-ejecting compound disulfiram is able to inhibit BCA2 autoubiquitination [18,60,61]. Abrogation of E3 ligase activity by the zinc-ejecting compound has important implications for the therapeutic intervention of RING finger E3 ligase-mediated diseases. Disulfiram (Antabuse; Odyssey Pharmaceuticals, Inc., East Hanover, NJ) is in clinical use for the treatment of alcoholism owing to its ability to inhibit alcohol dehydrogenase (ADH) and has thus proven tolerability in humans [5,18]. If analogs with retained or even enhanced zinc-ejecting capabilities but without ADH inhibition could be developed, they might prove valuable for the treatment for cancers with imbalances of RING finger ligase signatures, such as breast cancer [5].
The biology of BCA2 demonstrates how E3 ligases can have multiple cellular functions. In the cytoplasm, BCA2 binds to Rab7 and plays a role in the lysosomal degradation of RTKs. In the nucleus, BCA2 augments ubiquitin-mediated degradation of target proteins destined for proteasomal degradation (Figure 6). The flexibility of RING E3 ligases is enhanced by the regulation by RTK signaling (e.g., BCA2 is regulated by AKT-mediated phosphorylation and auto-ubiquitination). These complex mechanisms need to be considered when designing therapeutic strategies. Rather than inhibiting ligase activity alone, a combined approach of E3 ligase inhibition and AKT inhibition might be most effective.
Figure 6.
Model of BCA2 and Rab7 interactions of intracellular vesicle trafficking in breast cancer. Receptor-mediated endocytosis internalizes activated EGF-R complexes and transports them to the early endosome for sorting. Rab GTPases regulate vesicle fusion events during this process. BCA2 plays crucial roles as a Rab7 target protein in vesicle traffic up to late endosome/lysosome and lysosome biogenesis.
Modulating E3 ligase activity can lead to tumor growth inhibition, as exemplified by geldanamycin analogs that induce CHIP activity or by zinc-ejecting agents that can inhibit RING finger ubiquitin ligases such as BCA2. Thus, the discovery and development of truly specific E3 ligase inhibitors are likely to be highly rewarding and could provide targeted therapies for many tumor types.
Footnotes
This work was supported by grants from the Canadian Institutes of Health Research and the Canadian Breast Cancer Research Alliance.
References
- 1.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, Stein R, Adams J. New insights into proteasome function: from archaebacteria to drug development. Chem Biol. 1995;2(8):503–508. doi: 10.1016/1074-5521(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 3.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 4.Handley-Gearhart PM, Stephen AG, Trausch-Azar JS, Ciechanover A, Schwartz AL. Human ubiquitin-activating enzyme, E1. Indication of potential nuclear and cytoplasmic subpopulations using epitope-tagged cDNA constructs. J Biol Chem. 1994;269(52):33171–33178. [PubMed] [Google Scholar]
- 5.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer. 2004;40(15):2217–2229. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Nalepa G, Wade Harper J. Therapeutic anti-cancer targets upstream of the proteasome. Cancer Treat Rev. 2003;29(1):49–57. doi: 10.1016/s0305-7372(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 7.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 8.Pray TR, Parlati F, Huang J, Wong BR, Payan DG, Bennett MK, Issakani SD, Molineaux S, Demo SD. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5(6):249–258. doi: 10.1016/s1368-7646(02)00121-8. [DOI] [PubMed] [Google Scholar]
- 9.Joazeiro CAP, Wa M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302(4):635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 11.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23(21):4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 12.Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13(1):7–12. doi: 10.1016/s0962-8924(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 13.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3(4):535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 14.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 15.Ohta T, Fukuda M. Ubiquitin and breast cancer. Oncogene. 2004;23(11):2079–2088. doi: 10.1038/sj.onc.1207371. [DOI] [PubMed] [Google Scholar]
- 16.Catzavelos C, Bhattacharya N, Ung YC, Wilson JA, Roncari L, Sandhu C, Shaw P, Yeger H, Morava-Protzner I, Kapusta L, et al. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer [see comments] Nat Med. 1997;3(2):227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 17.Morgan DO. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1(2):E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- 18.Burger AM, Gao Y, Amemiya Y, Kahn HJ, Kitching R, Yang Y, Sun P, Narod SA, Hanna WM, Seth AK. A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65(22):10401–10412. doi: 10.1158/0008-5472.CAN-05-2103. [DOI] [PubMed] [Google Scholar]
- 19.Dornan D, Bheddah S, Newton K, Ince W, Frantz GD, Dowd P, Koeppen H, Dixit VM, French DM. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64(20):7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 20.Feki A, Jefford CE, Berardi P, Wu JY, Cartier L, Krause KH, Irminger-Finger I. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24(23):3726–3736. doi: 10.1038/sj.onc.1208491. [DOI] [PubMed] [Google Scholar]
- 21.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21(3):307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2(1):1–8. [PubMed] [Google Scholar]
- 23.Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, Nielsen TO, Huntsman DG, Gilks CB. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol. 2006;19(1):69–74. doi: 10.1038/modpathol.3800484. [DOI] [PubMed] [Google Scholar]
- 24.Hori M, Shimazaki J, Inagawa S, Itabashi M. Overexpression of MDM2 oncoprotein correlates with possession of estrogen receptor alpha and lack of MDM2 mRNA splice variants in human breast cancer. Breast Cancer Res Treat. 2002;71(1):77–83. doi: 10.1023/a:1013350419426. [DOI] [PubMed] [Google Scholar]
- 25.Horie K, Urano T, Ikeda K, Inoue S. Estrogen-responsive RING finger protein controls breast cancer growth. J Steroid Biochem Mol Biol. 2003;85(2–5):101–104. doi: 10.1016/s0960-0760(03)00209-7. [DOI] [PubMed] [Google Scholar]
- 26.Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417(6891):871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Urano T, Tsukui T, Horie-Inoue K, Moriya T, Ishida T, Muramatsu M, Ouchi Y, Sasano H, Inoue S. Estrogen-responsive finger protein as a new potential biomarker for breast cancer. Clin Cancer Res. 2005;11(17):6148–6154. doi: 10.1158/1078-0432.CCR-05-0040. [DOI] [PubMed] [Google Scholar]
- 28.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286(5438):309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 29.Levkowitz G, Oved S, Klapper LN, Harari D, Lavi S, Sela M, Yarden Y. c-Cbl is a suppressor of the neu oncogene. J Biol Chem. 2000;275(45):35532–35539. doi: 10.1074/jbc.M002661200. [DOI] [PubMed] [Google Scholar]
- 30.Sridhar SS, Seymour L, Shepherd FA. Inhibitors of epidermal-growth-factor receptors: a review of clinical research with a focus on non-small-cell lung cancer. Lancet Oncol. 2003;4(7):397–406. doi: 10.1016/s1470-2045(03)01137-9. [DOI] [PubMed] [Google Scholar]
- 31.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60(13):3384–3388. [PubMed] [Google Scholar]
- 32.Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, et al. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem. 2003;278(31):28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 33.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 34.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: a tissue microarray study. Cancer Res. 2002;62(2):497–505. [PubMed] [Google Scholar]
- 35.Kalkhoven E, Roelen BA, de Winter JP, Mummery CL, van den Eijndenvan Raaij AJ, van der Saag PT, van der Burg B. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6(9):1151–1161. [PubMed] [Google Scholar]
- 36.Azhar M. Non-redundant tumour suppressor functions of transforming growth factor beta in breast cancer. J Biosci. 2001;26(1):9–12. doi: 10.1007/BF02708975. [DOI] [PubMed] [Google Scholar]
- 37.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 38.Miyazono K, Maeda S, Imamura T. Coordinate regulation of cell growth and differentiation by TGF-beta superfamily and Runx proteins. Oncogene. 2004;23(24):4232–4237. doi: 10.1038/sj.onc.1207131. [DOI] [PubMed] [Google Scholar]
- 39.Kitching R, Gish G, Burger A, Landberg G, Seth A. The RING-H2 protein RNF11 is differentially expressed in breast tumors and interacts with HECT-type E3 ligases. Biochim Biophys Acta. 2003;1639:104–112. doi: 10.1016/j.bbadis.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Li H-X, Seth A. An RNF11:Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene. 2004;23(10):1801–1808. doi: 10.1038/sj.onc.1207319. [DOI] [PubMed] [Google Scholar]
- 41.Subramaniam V, Lubovitz J, Li H-X, Burger A, Kitching R, Seth A. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br J Cancer. 2003;189:1538–1544. doi: 10.1038/sj.bjc.6601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone M, Jr, Wrana JL, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 43.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6(6):1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 44.Herbst RS, Fukuoka M, Baselga J. Gefitinib—a novel targeted approach to treating cancer. Nat Rev Cancer. 2004;4(12):956–965. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- 45.Jiang X, Huang F, Marusyk A, Sorkin A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol Biol Cell. 2003;14(3):858–870. doi: 10.1091/mbc.E02-08-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki S, Nishida K, Yoshida Y, Itoh M, Hibi M, Hirano T. Gab1 is required for EGF receptor signaling and the transformation by activated ErbB2. Oncogene. 2003;22(10):1546–1556. doi: 10.1038/sj.onc.1206284. [DOI] [PubMed] [Google Scholar]
- 47.Urbe S, Mills IG, Stenmark H, Kitamura N, Clague MJ. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol Cell Biol. 2000;20(20):7685–7692. doi: 10.1128/mcb.20.20.7685-7692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated micro-domains of early endosomes. Nat Cell Biol. 2002;4(5):394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 49.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 50.van Delft S, Govers R, Strous GJ, Verkleij AJ, van Bergen en Henegouwen PM. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272(22):14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 51.Confalonieri S, Salcini AE, Puri C, Tacchetti C, Di Fiore PP. Tyrosine phosphorylation of Eps15 is required for ligand-regulated, but not constitutive, endocytosis. J Cell Biol. 2000;150(4):905–912. doi: 10.1083/jcb.150.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem. 2003;278(14):12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- 53.Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, Kasai H, Takeshita T, Endo Y, Fujita T, Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its su ppressive effect on cytokine-induced cell growth. J Biol Chem. 1997;272(52):32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- 54.Burger A, Li H, Zhang XK, Pienkowska M, Venanzoni M, Vournakis J, Papas T, Seth A. Breast cancer genome anatomy: correlation of morphological changes in breast carcinomas with expression of the novel gene product Di12. Oncogene. 1998;16(3):327–333. doi: 10.1038/sj.onc.1201517. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki H, Fukunishi Y, Kagawa I, Saito R, Oda H, Endo T, Kondo S, Bono H, Okazaki Y, Hayashizaki Y. Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 2001;11(10):1758–1765. doi: 10.1101/gr.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell. 2003;14(9):3741–3752. doi: 10.1091/mbc.E02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connor MK, Subramaniam V, Li H-X, Seth A. Molecular characterization of Ring-finger protein 11. Mol Cancer Res. 2005;3(8):453–461. doi: 10.1158/1541-7786.MCR-04-0166. [DOI] [PubMed] [Google Scholar]
- 58.Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83(1):153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 59.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 60.Amemiya A, Burger A, Seth A. Molecular characterization of the BCA2 protein: a novel RING-type E3 ligase that is overexpressed in breast and prostate cancers. Proc Am Assoc Cancer Res. 2005;46:2329. [Google Scholar]
- 61.Kosarev P, Mayer KF, Hardtke CS. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 2002;3(4) doi: 10.1186/gb-2002-3-4-research0016. RESEARCH0016. [DOI] [PMC free article] [PubMed] [Google Scholar]